Abstract
Pathogens are increasingly emerging in human-altered environments as a serious threat to biodiversity. In this context of rapid environmental changes, improving our knowledge on the interaction between ecology and evolution is critical. The objective of this study was to evaluate the influence of an immunocompetence gene, the major histocompatibility complex (MHC) class IIβ, on the pathogen infection levels in wild Atlantic salmon populations, Salmo salar, and identify selective agents involved in contemporary coevolution. MHC variability and bacterial infection rate were determined throughout the summer in juvenile salmon from six rivers belonging to different genetic and ecological regions in Québec, Canada. A total of 13 different pathogens were identified in kidney by DNA sequence analysis, including a predominant myxozoa, most probably recently introduced in North America. Infection rates were the highest in southern rivers at the beginning of the summer (average 47.6±6.3% infected fish). One MHC allele conferred a 2.9 times greater chance of being resistant to myxozoa, while another allele increased susceptibility by 3.4 times. The decrease in frequency of the susceptibility allele but not other MHC or microsatellite alleles during summer was suggestive of a mortality event from myxozoa infection. These results supported the hypothesis of pathogen-driven selection in the wild by means of frequency-dependent selection or change in selection through time and space rather than heterozygous advantage, and underline the importance of MHC standing genetic variation for facing pathogens in a changing environment.
Keywords: major histocompatibility complex, infection, local adaptation, myxozoa, bacteria, salmon
1. Introduction
Recent evidence suggests that parasites and infectious diseases are increasingly emerging in wild populations (Harvell et al. 1999; Daszak et al. 2000; Dobson & Foufopoulos 2001). Emerging diseases are due, in part, to increased human activity and most particularly to pollution, environmental degradation, climate change, habitat fragmentation, animal transfer and hatcheries (Harvell et al. 1999; Daszak et al. 2000; Dobson & Foufopoulos 2001). In the aquatic environment and in fish populations, in particular, several diseases such as whirling disease, furunculosis and infectious salmon anaemia have been associated with aquaculture-related activities (Dobson & Foufopoulos 2001). For instance, louse infestations associated with salmon farms are blamed for depressed wild pink salmon populations, potentially causing over 80 per cent mortality in some cases (Krkošek et al. 2007). These emerging pathogens have the potential to significantly reduce the abundance of threatened species (Dobson & Foufopoulos 2001; Lafferty et al. 2004) and as such, represent a serious threat to which wild populations have to adapt rapidly. In such a context of rapid environmental changes, linking ecological and evolutionary processes then becomes critical to evaluate the influence of standing genetic variation on contemporary adaptation (see also Porlier et al. 2009; Zheng et al. 2009).
Adaptation can occur over a very short time scale (Thompson 1998), suggesting that wild populations could potentially adapt to the increasing number of novel pathogens emerging in the environment. In vertebrates, genes of the major histocompatibility complex (MHC) are involved in pathogen resistance by encoding cell-surface proteins that bind peptide fragments derived from pathogens and present them to T cells that activate a specific immune response (Potts & Wakeland 1990). These genes are among the most polymorphic in vertebrates and most of the variability is concentrated at the peptide-binding region (PBR) involved in pathogen binding (Hughes & Yeager 1998). Theory predicts that, at the population level, high MHC diversity should confer resistance to a diverse array of pathogens (reviewed in Bernatchez & Landry 2003; Sommer 2005; Piertney & Oliver 2006). Recent studies conducted at a large spatial scale supported a positive association between MHC diversity and pathogen diversity in the wild, independent of historical colonization processes, in humans (Prugnolle et al. 2005) and Atlantic salmon (Dionne et al. 2007). Yet, the exact underlying mechanism involved in pathogen resistance at the individual level and how this translates into the maintenance of MHC diversity at the population level is not well understood (Sommer 2005; Piertney & Oliver 2006).
At the population level, MHC diversity is thought to be maintained by balancing selection through (i) heterozygote advantage, (ii) frequency-dependent selection, or (iii) variable selection in time and space (Nei & Hughes 1991; Hedrick 2002). The heterozygote advantage (or overdominance) hypothesis suggests that the fitness of heterozygote individuals is greater than that of both homozygotes because of their ability to present a broader spectrum of antigens to T cells (Hughes & Nei 1988). MHC heterozygote advantage was observed in humans for resistance to hepatitis B and HIV infections (Thursz et al. 1997; Carrington et al. 1999) and in mice for resistance to Salmonella infections (Penn et al. 2002; McClelland et al. 2003). On the other hand, frequency-dependent selection occurs when the advantage of possessing an MHC allele depends on its frequency in the population and is usually referred to as negative frequency-dependent selection or rare allele advantage (Takahata & Nei 1990). The variable selection in time and space hypothesis reasons that MHC polymorphism is maintained by shifting pathogen compositions that occur through time and space (Hedrick 2002). Support for the latter two hypotheses is provided by the association of certain MHC alleles with resistance to specific diseases, such as those found for malaria and HIV in humans (Hill et al. 1991; Carrington et al. 1999), Marek's disease in chicken (Briles et al. 1977) and furunculosis, infectious salmon anaemia and infectious haemopoietic necrosis virus in salmonids (Langefors et al. 2001; Grimholt et al. 2003; Miller et al. 2004). Further support for the latter hypothesis was also found in a temporal study conducted on the great reed warbler, Acrocephalus arundinaceus, where change in frequency of two MHC class I alleles contrasted with the stochastic variability of microsatellite markers (Westerdahl et al. 2004). When contrasting the heterozygote advantage and the frequency-dependent selection hypotheses in pathogen resistance, the latter has received more support in the literature (Sommer 2005). However, the exact pathogen resistance mechanism occurring at the individual level in the wild is yet to be clarified. Moreover, although the importance of pre-existing genetic variation, referred to as ‘standing genetic variation’, has been emphasized for adaptation and rapid evolution (Barrett & Schulter 2008), its importance for pathogen resistance in the wild is yet to be evaluated. Finally, although associations between pathogenic infection and population abundance have been observed in the wild (Krkošek et al. 2007; Miller & Vincent 2008), investigating demographic consequences of immune maladaptation is still in its infancy.
Anadromous populations of Atlantic salmon, Salmo salar, reproduce and spend the first years of their life in rivers along the North American and European Atlantic coasts, where a major decline in abundance has been observed during the last decades (Caron et al. 2005). After a feeding period of 1–3 years at sea, the majority of Atlantic salmon returns to spawn in natal rivers (Stabell 1984) and evidence suggests that they adapt to local conditions (reviewed in Taylor 1991; García de Leániz et al. 2007). Recently, adaptation to local temperature regime and bacterial community prevailing in the natal river has been supported through large-scale variations in MHC class IIβ diversity specifically at the PBR, suggesting the influence of pathogen-driven balancing selection on MHC diversity in Atlantic salmon (Dionne et al. 2007). Also, indirect evidence was found for lower fitness of immigrants coming from genetically and thermally distinct regions, suggesting a potential influence of local adaptation in limiting gene flow (Dionne et al. 2008). Evidence of adaptation through disassortative mating at the PBR in MHC class II was also found, possibly providing better pathogen resistance for the progeny (Landry et al. 2001). Evidence of selection at MHC class II was also found at the juvenile stage. Indeed, observed genotype frequencies of hatchery juveniles introduced in a river were significantly different after six months than the expected frequencies from parental crosses (de Eyto et al. 2007). However, uncertainty still persists whether early juvenile stages are able to mount specific immune responses (Bakke & Harris 1998). More importantly, information about selective agents, namely pathogens infecting Atlantic salmon in the wild along with the associated infection rates, is lacking. This lack of knowledge is even more pronounced for juvenile stages, as most pathogen resistance studies have been conducted on adults in laboratory conditions (Langefors et al. 2001; Grimholt et al. 2003; Miller et al. 2004).
The objectives of this study were, first, to document infection rates and identify pathogens infecting juvenile Atlantic salmon inhabiting rivers from distinct genetic groups and ecological regions; second to evaluate the influence of MHC class IIβ alleles in conferring resistance or susceptibility to pathogen infection in the wild; and third, to test which of the heterozygote advantage, frequency-dependent selection or variable selection in time and space hypotheses are prevailing at the individual level. This was achieved by testing whether (i) heterozygotes at MHC class II are less infected than homozygotes and whether (ii) there is an association between specific MHC alleles and the resistance or susceptibility of juveniles to the most prevalent infections. A final objective consisted of an indirect test for pathogen-induced mortality by following MHC allele frequency changes during the summer for alleles found to be associated with the most prevalent infections in the wild. In order to dissociate the influence of selection from other evolutionary forces, MHC variability was compared with that observed at putative neutral microsatellite loci.
2. Material and methods
(a) Sampling
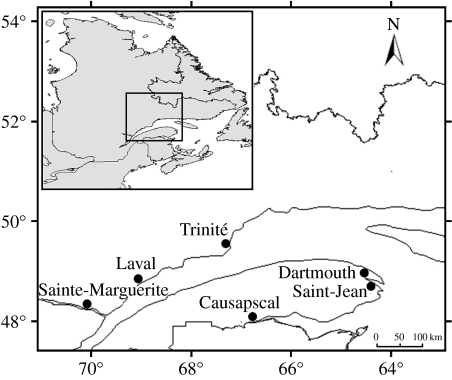
Sampling was conducted in six rivers from Québec, Canada: the Trinité (TR), Laval (LA), and Sainte-Marguerite (SM) rivers on the north shore, and the Causapscal (CA), Dartmouth (DA), and Saint-Jean (SJ) rivers on the south shore of the St Lawrence Estuary (figure 1). These rivers belong to three different Atlantic salmon genetic regions according to a Bayesian clustering analysis based on neutral microsatellite markers: TR and LA in region 4; SM in region 5; and all southern rivers in region 6 (Dionne et al. 2008). These genetic regions are also ecologically differentiated, mainly in terms of their historical summer temperature index (average temperature regime for multiple rivers in regions 4, 5 and 6, respectively: 1066±132; 1542±160; and 1343±77 degree days; Dionne et al. 2008). Rivers draining the north shore of the St Lawrence Estuary are characterized by lower bacterial diversity in the water than those draining the south shore (2.5±2.4 and 38.9±12.8 ×103 intergrated optical density (IOD) mm−2, respectively, see details in Dionne et al. 2007). Each river was sampled on three occasions (June, July and August) during summer 2005. On each occasion, we sampled on average 50 parr per river (age 1+ and 2+ in an approximate ratio of 1 : 1 per sample), for a total of 898 juvenile salmon collected. Fish were handled using gloves to avoid external contamination. Each fish was individually inserted in a sterile sampling bag (Whirl-Pak, Nasco, Fort Atkinson, WI, USA), measured for fork length and quick-frozen on dry ice in order to preserve tissues for DNA and pathogen analyses. In the laboratory and under sterile conditions, the adipose fin was clipped and stored in 95 per cent ethanol for subsequent DNA analysis while the whole fish was kept at −80°C.
Figure 1.
Location of the six Atlantic salmon rivers where sampling took place in Québec, Canada. The Sainte-Marguerite, Laval and Trinité rivers are located on the north shore while the Causapscal, Saint-Jean and Dartmouth rivers are located on the south shore of the St Lawrence Estuary.
(b) Microsatellite and MHC class IIβ genotyping
DNA was extracted from fin clips using the Qiagen DNeasy Tissue Kit (Mississauga, Ontario, Canada) following the guidelines of the manufacturer. Microsatellite polymorphism was quantified at 13 loci as detailed in the appendix of Dionne et al. (2007): Ssa85; Ssa202; Ssa197 (O'Reilly et al. 1996); Ssosl417 (Slettan et al. 1995); SsaD85 (T. King, unpublished data); SsaD71; SsaD144 (King et al. 2005); MST-3 (Presa & Guyomard 1996); Sssp1605; Sssp2210; Sssp2215; Sssp2216; and SsspG7 (Paterson et al. 2004). MHC class IIβ exon 2 was amplified using fluorescently labelled primers (Dionne et al. 2007) and alleles were separated via denaturing gradient gel electrophoresis (DGGE), as detailed in the appendix of Dionne et al. (2007).
(c) Bacterial infection analyses
Kidney tissue was selected to evaluate bacterial pathogenic infection in juvenile salmon as this organ is affected by multiple well-known fish diseases such as furunculosis, bacterial kidney disease, proliferative kidney disease and vibriosis (Bakke & Harris 1998). Also, a healthy kidney is normally free of bacteria, so the presence of bacteria implies pathogenic infection (Uhland et al. 2000). A total of 421 age 2+ parr were selected for infection analyses. The kidney of each fish was carefully dissected under a fume hood and under sterile conditions, with approximately 25 mg of tissue used for DNA extraction. Total DNA was extracted from the kidney using the QIAamp DNA Mini Kit following the guidelines of the manufacturer for the bacteria DNA protocol (Qiagen). Bacterial DNA was then specifically amplified using 16S rDNA universal primers (Crump et al. 2003) in a nested PCR protocol, as detailed in the analysis of bacterial diversity in water samples (Appendix 2 in Dionne et al. 2007). The 16S rDNA is a conserved region that is widely used to amplify and identify all bacteria present in a given environment (Woo et al. 2008). Distinct amplified bacterial sequences were separated using DGGE (BioRad, Hercules, CA, USA) under the conditions outlined in Appendix 2 of Dionne et al. (2007). Eleven different 16S rDNA fragments, amplified from 11 different bacteria species/strains, were included as standards on each DGGE gel to allow band comparison based on the positions of the standard sequences. Negative and positive controls (infected salmon with a known bacterium) were included in each PCR and were run on each gel to detect and avoid potential contamination and ensure proper amplification, which proved to be adequate. Each bacterial band was cut from the gel and sequenced using the 16S-357F and 16S-519R primers and the Big Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). Sequencing reactions were purified using the DyeEx 2.0 Spin Kit (Qiagen) and electrophoresis was conducted on a 3730 Capillary DNA Sequencer (Applied Biosystems). Nucleotide sequences were edited in BioEdit 7.0.5.3 (Hall 1999) and identified using the Basic Local Alignment Search Tool in the National Center for Biotechnology Information.
(d) Statistical analyses
(i) Microsatellite basic analyses
The potential occurrence of null alleles and scoring errors due to stuttering or large allele dropout in the dataset was assessed using the software Micro-Checker (van Oosterhout et al. 2004). Deviation from Hardy–Weinberg expectations was tested for each locus and river sampled, and linkage disequilibrium was tested for each pair of loci using Genepop 3.4 (Raymond & Rousset 1995). The LnRH neutrality test was performed on each microsatellite locus to verify the neutrality of each marker (Schlötterer 2002; see Dionne et al. (2007) for calculation details). Temporal stability of microsatellite allele frequency distributions per river across the three sampling periods was assessed using the Fisher's exact probability test in Genepop 3.4 (Raymond & Rousset 1995).
(ii) Infection analyses
In order to evaluate the overall pattern of variation in infection rate of wild juvenile salmon between regions and sampling periods, a two-factor logistic regression was conducted on infection status of the fish (infected or uninfected) using SAS 9.1 (SAS Institute). A second general analysis, a two-factor Poisson regression analysis, was conducted using SAS to evaluate the regional and temporal variations in pathogen diversity (number of different pathogen strains found in each fish). It was not possible to include individual rivers as a fixed factor in these analyses because of a lack of power due to the limited number of infected fish in certain rivers.
(iii) Infection and MHC polymorphism
Levels of heterozygosity were calculated for infected and uninfected fish using Genepop 3.4 (Raymond & Rousset 1995) and compared between MHC and microsatellite markers. Under the heterozygote advantage hypothesis, heterozygosity is expected to be lower for infected compared with uninfected fish at MHC but not at microsatellite markers. To test this hypothesis further, MHC p-distance for each fish was calculated using Mega 3.1 (Kumar et al. 2000) for both the overall polymorphic sites and for the PBR only. This p-distance represents a genetic measure of allelic divergence within individuals, which incorporates the proportion of different amino acids between two MHC alleles of any given fish (Nei & Kumar 2000). The PBR was identified based on codons involved in pathogen binding in humans (Brown et al. 1988, 1993) and corresponded to those presented in Landry & Bernatchez (2001) and Dionne et al. (2007). For comparison, a comparable individual distance measure for microsatellites, representing the difference in mutational steps, was estimated by calculating mean difference in the number of repeats between two microsatellite alleles per locus for each individual (hereafter referred to as ‘repeat distance’). A two-factor multivariate analysis of variance (MANOVA) was then conducted using SAS to evaluate the potential implication of the MHC p-distance (overall and PBR), the microsatellite repeat distance and the fork length in differentiating between infected and uninfected salmon among the different sampling periods. Fork length was used here as a phenotypic index representing general body condition, which might influence indirectly the probability for a fish to be infected in the wild.
To evaluate the potential association between specific MHC alleles and either resistance or susceptibility to the most prevalent infection, a Fisher's exact probability test was first conducted using SAS and odds ratios were calculated for each allele. A high odds ratio would indicate a high probability for a fish to be infected when having a given allele. Secondly, a multivariate logistic regression was conducted on fish infection status (infected or uninfected with the most prevalent infection), with region, sampling period and all MHC alleles as factors integrated in a stepwise process using SAS. The Fisher's exact probability test and the multivariate logistic regression were also conducted at the amino acid level to identify specific codon positions potentially associated with resistance or susceptibility to the most prevalent infection. Finally, frequency of the resistance and susceptibility alleles identified in the previous analyses was compared among sampling periods, along with the other MHC and microsatellite alleles, in order to test for differential mortality caused by the predominant infection during summer. Mantel–Haenszel Χ2-tests were conducted on all juveniles sampled to evaluate trends in allele frequency changes using SAS and sequential Bonferroni correction was applied to evaluate significance (Rice 1989). All analyses in SAS were conducted in SAS software 9.1 (SAS Institute).
3. Results
(a) Microsatellite polymorphism
No evidence for null alleles or scoring errors due to stuttering or large allele dropout was found in the overall dataset. Deviations from Hardy–Weinberg equilibrium were identified in 13 out of 234 comparisons (all heterozygote deficits), a number close to the 12 significant tests expected by chance at α=0.05. There was no evidence of significant deviations associated with either particular loci or populations, which translated into small global FIS values ranging from −0.010 to 0.018 depending on the population. No significant linkage disequilibrium was detected within populations or between locus pairs across populations. The LnRH neutrality test identified microsatellite Sssp1605 as a locus potentially under selection, which was thus removed from further analyses (Schlötterer 2002, see Dionne et al. (2007) for calculation details).
(b) Infection types and prevalence
Overall, 12.1 per cent of the juvenile salmon analysed were infected with one or more pathogens (51/421 salmon). The observed infection rates in rivers of both north and south shores were highest in June and declined rapidly thereafter according to the logistic regression and the post hoc comparisons (p<0.0001 between June and the two other summer periods; table 1). Indeed, a proportion of 20.6±6.9 and 47.6±6.3 per cent of juvenile salmon were found to be infected at the beginning of the summer in north and south shore rivers, respectively, while infection rates dropped, respectively, to 3.2±1.9 and 2.3±1.6 per cent by the end of the summer (table 2). There was also a nearly significant trend for infection rates to be higher in St Lawrence south shore than in north shore rivers (p=0.058). Pathogen diversity, in terms of number of different pathogen species per individual, also followed the same pattern with higher pathogen diversity in June (p<0.0001) and a tendency for higher pathogen diversity in south shore when compared with the north shore rivers, although the latter was not significant (p=0.23). Note that values of pathogen diversity are close to zero due to the high number of uninfected fish. Sex ratio was slightly male biased in infected salmon (male/female=1.27) compared with uninfected individuals (male/female=1.09), although both were not departing significantly from equal sex ratios (Χ2-tests: p=0.40 and 0.39 for infected and uninfected groups).
Table 1.
Logistic regression analysis on infection status (infected or uninfected) of juvenile Atlantic salmon and Poisson regression analysis on pathogen diversity (number of different pathogens) infecting each individual according to the region (north or south shore) and the sampling period during summer (June, July or August).
| factor | F | p-value | |
|---|---|---|---|
| infection status | region | 3.61 | 0.058 |
| period | 20.17 | <0.0001 | |
| region×period | 1.47 | 0.231 | |
| pathogen diversity | region | 1.46 | 0.227 |
| period | 16.52 | <0.0001 | |
| region×period | 0.82 | 0.442 |
Table 2.
Mean infection rate and pathogen diversity infecting juvenile Atlantic salmon from north and south shore rivers of the St Lawrence Estuary. (s.d., standard deviation.)
| infection rate (±s.d.) | pathogen diversity (±s.d.) | |||
|---|---|---|---|---|
| sampling period | north shore | south shore | north shore | south shore |
| June | 0.206±0.069 | 0.476±0.063 | 0.205±0.077 | 0.524±0.091 |
| July | 0.024±0.017 | 0.111±0.039 | 0.048±0.024 | 0.112±0.042 |
| August | 0.032±0.019 | 0.023±0.016 | 0.033±0.019 | 0.023±0.016 |
A total of 13 different pathogens were identified based on their DNA sequences of 151 bp on average, as indicated by the best match with known sequences from GenBank (table 3). The nearest taxonomic neighbour represented the genus (or order for myxozoa) corresponding to all best matches in GenBank with the unknown sequence (homology above 80%) and the proportion of sequence similarity represented highest homology with species in that genus group. A total of 92.2 per cent of the infected juvenile salmon had a single pathogen in the kidney (47/51 juvenile salmon). Bacterial kidney infections were present in 29.4 per cent of the infected fish (15/51 juvenile salmon) and 3.6 per cent of the salmon sampled (15/421 juvenile salmon). Each identified bacteria had low prevalence in the populations studied, infecting on an average 0.36±0.81 per cent juvenile salmon per river over all time periods (range: 0.0–4.4%, table 3). However, one predominant pathogen was present in 76.5 per cent of the infected fish (39/51 juvenile salmon) and was found to infect 9.3 per cent of the juvenile salmon sampled (39/421 juvenile salmon). This represented an average prevalence of 8.9±6.4 per cent infected juvenile salmon per river during summer (range: 0.0–16.4%), with higher infection rates in the St Lawrence south shore than in north shore rivers (south shore prevalence=14.2±2.3%, north shore prevalence=3.7±3.9%; t-test, p=0.011). This predominant pathogen was identified as a myxozoa, class Myxosporea, order Bivalvulidae based on GenBank known sequences (85% similarity with available species in the Bivalvulidae order). This pathogen was also compared with unpublished myxozoa sequences available from C. Whipps (2008; SUNY College of Environmental Science & Forestry, Syracuse, NY) and fell within the Myxobolus clade as defined by Fiala (2006), yielding matches (78% similarity) to Myxobolus species infecting salmonids, i.e. M. arcticus and M. kisutchi (C. Whipps 2008, personal communication). Histological observations of infected kidneys yielded no myxospore to allow further identification (C. Whipps 2008, personal communication).
Table 3.
Pathogen identity found in the kidney of juvenile Atlantic salmon from six rivers in Québec: Sainte-Marguerite (SM), Laval (LA), Trinité (TR), Causapscal (CA), Dartmouth (DA) and Saint-Jean (SJ). (GenBank sequence accession number for the myxozoa and each bacterium is indicated. Nearest taxonomic neighbour (genus or order for myxozoa) and proportion of sequence similarity with GenBank available species for each associated genus is indicated. Prevalence indicates the proportion of salmon infected per river over all time periods during summer.)
| no. | pathogen type | sequence accession number | nearest taxonomic neighbour | sequence similarity (%) | prevalence (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| north | south | |||||||||
| SM | LA | TR | CA | DA | SJ | |||||
| 1 | myxozoa | FJ560442 | Bivalvulidae | 85 | 3.3 | 7.7 | 0.0 | 14.3 | 16.4 | 11.8 |
| 2 | bacteria | FJ560443 | Mycoplasma sp. 1 | 85 | 0.0 | 0.0 | 1.7 | 0.0 | 1.5 | 0.0 |
| 3 | bacteria | FJ560444 | Mycoplasma sp. 2 | 96 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 |
| 4 | bacteria | FJ560445 | Sphingomonas sp. | 98 | 0.0 | 0.0 | 1.7 | 0.0 | 0.0 | 0.0 |
| 5 | bacteria | FJ560446 | Methylobacterium sp. 1 | 99 | 0.0 | 0.0 | 1.7 | 0.0 | 0.0 | 0.0 |
| 6 | bacteria | FJ560447 | Methylobacterium sp. 2 | 96 | 0.0 | 0.0 | 1.7 | 0.0 | 0.0 | 0.0 |
| 7 | bacteria | FJ560448 | Rickettsiella sp. | 96 | 0.0 | 0.0 | 1.7 | 0.0 | 0.0 | 0.0 |
| 8 | bacteria | FJ560449 | Pseudomonas sp. | 95 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 0.0 |
| 9 | bacteria | FJ560450 | Shewanella sp. | 96 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 0.0 |
| 10 | bacteria | FJ560451 | Bosea sp. | 93 | 0.0 | 0.0 | 0.0 | 1.3 | 0.0 | 1.5 |
| 11 | bacteria | FJ560452 | Piscirickettsia sp. | 83 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.4 |
| 12 | bacteria | FJ560453 | Lactobacillus sp. | 100 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 0.0 |
| 13 | bacteria | FJ560454 | Burkholderia sp. | 87 | 0.0 | 0.0 | 0.0 | 2.6 | 0.0 | 0.0 |
(c) Infection and MHC polymorphism
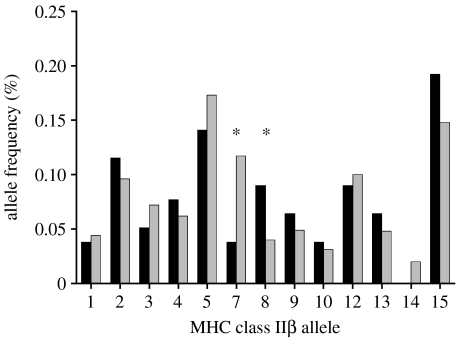
A total of 13 different MHC class IIβ alleles were found among all sampled juvenile salmon (figure 2). The number of different amino acids between MHC alleles averaged 9.3±2.7 (range: 4–17), while the proportion of different amino acids averaged 0.12±0.04 (range: 0.03–0.22). The observed heterozygosity at microsatellite loci was 0.83 and 0.86 for infected and uninfected fish, respectively, and both groups exhibited a slight yet significant deficit in heterozygotes compared with the expected values (HE: 0.86 and 0.88, respectively; p<0.0001 for both groups). A deficit in heterozygotes was expected as each group represented a pool of mixed individuals from different populations (Wahlund effect; all rivers pooled because of low number of infected fish). A deficit in heterozygotes was also observed at MHC class IIβ for uninfected fish (HO: 0.84, HE: 0.90, p<0.0001). However, in infected fish, an excess in heterozygotes was observed at MHC class IIβ (HO: 0.96, HE: 0.90, p=0.048), contrasting with neutral expectations. Overall, the observed microsatellite heterozygosity levels were similar for infected and uninfected juvenile salmon (0.83±0.08 and 0.86±0.08, respectively; t-test, p=0.45). However, MHC heterozygosity was higher for infected than for uninfected salmon (0.96 and 0.84, respectively). The MANOVA revealed an overall significant difference in MHC p-distance, microsatellite repeat distance or fork length between infected and uninfected juvenile salmon (p=0.027) and across the different time periods (p=0.029), but there was no significant interaction between infection status and time period (p=0.20). In the univariate tests, MHC p-distance, calculated both using all polymorphic sites or PBR only, was higher for infected than for uninfected juvenile salmon, although this trend was only significant within the PBR (p=0.068 and 0.019, respectively; table 4). On the other hand, no significant difference in microsatellite repeat distance was observed between infected and uninfected juvenile salmon (p=0.829). Univariate tests also revealed that fork length was the factor varying across time, with juvenile salmon becoming longer with time during summer as expected following summer growth (p=0.008). There was also a non-significant tendency for infected juvenile salmon to be smaller than the uninfected salmon (p=0.07).
Figure 2.
Allele frequency of each MHC class IIβ allele found in infected (black bars) and uninfected (grey bars) juvenile Atlantic salmon sampled in six rivers in the province of Québec, Canada. (Asterisks indicate alleles significantly associated with infection probability according to the Fisher's exact probability test.)
Table 4.
Two-factor MANOVA comparing individual genetic distances (MHC p-distance and microsatellite repeat distance) and fork lengths between the different infection status of juvenile Atlantic salmon (infected or uninfected) and among the different sampling periods during summer (June, July and August). (MHC p-distance was calculated either on all polymorphic sites or on PBR only. The p-values indicate significance in the univariate tests for each dependent variable compared between infected and uninfected fish (infection status) and among sampling periods (period). s.d., standard deviation. Values in bold represent significant tests.)
| dependent variable | infected fish (±s.d.) | uninfected fish (±s.d.) | p-value (infection status) | p-value (period) |
|---|---|---|---|---|
| MHC p-distance polymorphic sites | 0.11±0.009 | 0.09±0.003 | 0.068 | 0.964 |
| MHC p-distance PBR sites | 0.30±0.03 | 0.24±0.007 | 0.019 | 0.699 |
| microsatellite repeat distance | 4.42±0.25 | 4.48±0.07 | 0.829 | 0.889 |
| fork length | 8.67±0.24 | 9.13±0.07 | 0.070 | 0.008 |
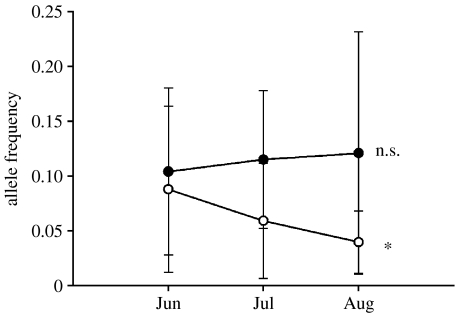
An association was uncovered between the occurrence of specific MHC alleles and the probability of juvenile salmon to be infected by myxozoa, the most prevalent pathogen, according to the Fisher's exact probability test (table 5). Allele 7 had a low odds ratio and was therefore identified as a potential resistance allele decreasing the risk of myxozoa infection (p=0.02, odds ratio=0.28). Conversely, allele 8 had a high odds ratio and represented a potential susceptibility allele increasing the chance of myxozoa infection (p=0.03, odds ratio=2.57). In order to take into account multiple testing in this analysis, 200 odds ratios per allele were calculated based on 200 simulated datasets generated by randomly permuting MHC individual genotypes with infection status (infected or uninfected by myxozoa) using Poptool 3.0.3 (Hood 2008). Simulations indicated that the observed odds ratios for alleles 7 and 8 were outside the theoretical expectation (p=0.020 and 0.025, respectively), with four and five simulated odds ratios out of 200 respectively lower and higher than the observed values for alleles 7 and 8. The Fisher's exact probability test at the amino acid level identified a valine in position 65 and a leucine in position 68 as amino acid positions associated with resistance of allele 7 and susceptibility of allele 8 to myxozoa infection, respectively (p<0.05). Phenylalanine or tyrosine and phenylalanine or histidine were otherwise found at positions 65 and 68, respectively. The multivariate logistic regression gave concordant results and revealed that the infection status of salmon depended on the region of origin, the sampling period and the presence of allele 8 (p=0.018, p<0.001 and p=0.019, respectively). Indeed, in concordance with the previous global logistic regression on infection rate, myxozoa infection was predominant in St Lawrence south shore relative to north shore rivers and was highest in June and decreased thereafter. Also, salmon with allele 8 had 3.4 times more chance to be infected with myxozoa than those not having this allele, when controlling for spatial and temporal variations in infection rate. Allele 7 was only retained by the model when the factor region was excluded from the analysis (p=0.031), indicating that the frequency of allele 7 was correlated with the regional variation, being slightly more frequent and less variable among north shore than south shore rivers (0.12±0.05 and 0.11±0.11, respectively). Still, according to the logistic regression and when controlling for spatial and temporal variations in infection rate, salmon carrying allele 7 had 2.9 times greater chance of being uninfected by myxozoa than salmon not carrying this allele. A comparison of allele frequencies for each microsatellite locus across time indicated no significant temporal change during summer (p>0.05). The frequency of MHC allele 7 increased slightly through time but this trend was not significant (p=0.30), while the frequency of MHC allele 8 decreased significantly through summer (p=0.003; figure 3). All other MHC alleles did not show any trend in frequency change during the summer (p>0.05).
Table 5.
Fisher's exact probability test on myxozoa infection status of juvenile Atlantic salmon as a function of MHC class IIβ alleles. (Values in bold represent alleles significantly associated with the probability of being infected by myxozoa in the wild according to the Fisher's exact probability test and the permutation test of significance.)
| MHC allele | p-value | odds ratio |
|---|---|---|
| 1 | 0.74 | 1.19 |
| 2 | 0.39 | 1.39 |
| 3 | 1.00 | 0.81 |
| 4 | 0.15 | 1.99 |
| 5 | 1.00 | 0.95 |
| 7 | 0.02 | 0.28 |
| 8 | 0.03 | 2.57 |
| 9 | 0.60 | 1.22 |
| 10 | 0.74 | 1.19 |
| 12 | 0.83 | 1.05 |
| 13 | 0.58 | 1.29 |
| 14 | 0.38 | 1.90 |
| 15 | 0.47 | 1.32 |
Figure 3.
Average allele frequency of the identified MHC resistance allele 7 (filled circles) and MHC susceptibility allele 8 (open circles) to myxozoa across the summer in juvenile Atlantic salmon from six rivers in Québec. (Error bars represent standard deviation associated with the different populations. Asterisk indicates significance while n.s. indicates a non-significant change in allele frequency according to the Mantel–Haenszel Χ2-tests.)
4. Discussion
The goal of this study was to clarify the mechanism of pathogen resistance at the individual level and evaluate the spatio-temporal intensity of immune challenges in the wild by screening bacterial infections in juvenile Atlantic salmon and relating these to MHC and microsatellite variability. An overall kidney infection rate of 12.1 per cent was found in wild juvenile Atlantic salmon within the six rivers studied. Infection rate and pathogen diversity were greater at the beginning of the summer when an average proportion as high as one in every two juveniles was found to be infected in St Lawrence south shore rivers and one in every five in north shore rivers. The sex ratio of the infected individuals was slightly male biased, albeit not significantly, which suggested that both sexes had similar infection rates in the wild. A myxozoa parasite of the genus Myxobolus, most probably recently introduced in North America, was discovered and showed an average prevalence of 14.2 per cent in south shore rivers and 3.7 per cent in north shore rivers during summer. One MHC class IIβ allele was associated with resistance and another with susceptibility to the myxozoa, suggesting a role of MHC standing genetic variation in influencing infection levels towards an emerging parasite. As no evidence for heterozygote advantage was found, these results better support the frequency-dependent selection or the variable selection in time and space hypotheses. In addition, a significant decrease in frequency of the susceptibility allele but not other MHC or microsatellite alleles during summer suggested the occurrence of a mortality event due to myxozoa infection. Overall, these findings suggest that the myxozoa parasite can exert a selective pressure on its host, but that standing genetic variation at MHC in host populations could contribute to promoting adaptation over time.
Spatial variation in infectivity suggested a trend for higher infection rates and pathogen diversity in juvenile salmon from south shore compared with north shore rivers in the St Lawrence system. This result was concordant with the evidence of a higher pathogen selection pressure in southern warm than northern cold habitats found in a large-scale study conducted on wild Atlantic salmon populations (Dionne et al. 2007). Indeed, the bacterial community in each of the six rivers was analysed during the same year and bacterial diversity was higher in south shore than in north shore rivers (38.9±12.8 and 2.5±2.4×103 IOD mm−2, respectively; see Dionne et al. (2007) for bacterial community analyses). Temporal variation in infectivity also suggested higher infection rates in June when water temperature rise rapidly from approximately 6°C to over 18°C (Ministère des Ressources Naturelles et de la Faune du Québec). Pathogenic bacteria and other parasites may increase in abundance and virulence once a temperature threshold is reached, as is the case for myxozoa infections that mostly occur at more than 15°C (Uhland et al. 2000), and as previously observed in the laboratory for other fish pathogens (e.g. Larsen et al. 2004). The observed decrease in infection rate thereafter suggested that either juveniles lost their infection with time, following the pathogen natural life cycle or through immune resistance, or mortality occurred following infection. Although these hypotheses are not mutually exclusive, our data are more consistent with the latter hypothesis, as a significant decrease in frequency of a MHC susceptibility allele towards myxozoa infection, as well as a trend towards the increase in a MHC resistance allele, were observed during the summer while no significant change was detected for other MHC and microsatellite alleles. Overall, we found evidence for spatio-temporal variation in infection rate concordant with variation in pathogen selection pressure in the environment, and also with the immune competence associated with specific MHC alleles (see below).
Twelve species of potentially pathogenic bacteria were found in kidneys of wild juvenile salmon, all of which had a low prevalence in the populations. Some of them were known pathogens, namely Mycoplasma infecting farmed cod and other vertebrates (Messick et al. 2002; Nylund et al. 2006), the pathogenic bacteria Rickettsiella mainly infecting arthropods (Cordeaux et al. 2007), the opportunistic fish pathogen Pseudomonas causing septicaemia in stressful conditions (Thune et al. 1993), Shewanella responsible for ulcer disease in some molluscs and fish (Cai et al. 2006) and Piscirickettsia causing epizootic diseases in salmonids (Fryer & Hedrick 2003). Other bacteria, namely Sphingomonas, Methylobacterium, Bosea, Lactobacillus and Burkholderia, have been previously found in soil and freshwater environments and some such as Burkholderia can become opportunistic pathogens (Berriatua et al. 2001). Although prevalence was low for these bacteria, some of them such as Piscirickettsia are qualified as epizootic, affecting a small number of individuals but, under favourable environmental conditions, possessing the capacity to spread rapidly in the population (Fryer & Hedrick 2003). As such, these could constitute latent pathogens representing a potential health risk for salmon populations in the future. The most prevalent pathogen infecting juvenile salmon was a myxozoa of the Myxobolus clade, a group of fish parasites first discovered in farmed salmonids in Europe (Hoffman 1990). Myxobolus infections, as well as other documented myxozoa infections, have been detected in the USA since the 1950s and epidemiological and molecular evidence suggests that these parasites were initially introduced through fish transported from Europe (Hedrick et al. 1993; Andree et al. 1999; Kent et al. 2001; Miller & Vincent 2008). Myxobolus infections have been recently reported in eastern Canada in some fish species such as the log-perch, Percina caprodes, and the banded killifish, Fundulus diaphanus (Cone & Marcogliese 2004; Cone et al. 2006). However, to our knowledge, this is the first published report of a myxozoa infection in wild Atlantic salmon in eastern Canada. As such, this most probably represents an emerging salmon parasite in this system. Myxozoa parasites are responsible for multiple diseases in salmonids, including the well-documented whirling disease caused by Myxobolus cerebralis, which can severely compromise survival in some species (Kent et al. 2001). Even though MHC allele 7 covaried with regional variability, MHC alleles 7 and 8 were associated with resistance and susceptibility to myxozoa infection, respectively, after controlling for spatio-temporal variability in infection rate. Previous studies conducted in the laboratory also found an association between an MHC allele and incidence of diseases, namely furunculosis, infectious salmon anaemia and infectious haemopoietic necrosis virus in Atlantic salmon (Langefors et al. 2001; Grimholt et al. 2003; Miller et al. 2004). However, very few studies have attempted to test for an association between MHC alleles and parasite resistance in wild populations (but see Harf & Sommer (2005) and references therein on small mammals and Madsen & Ujvari (2006) on pythons). In addition to identifying an association between MHC alleles and susceptibility, we have also attempted to disentangle the influence of spatio-temporal variability in pathogen selection pressure from immune resistance processes in determining infection rates in the wild, which has always been a challenge in the past (Wegner et al. 2003). The significant shift in the frequency of the susceptibility allele 8 over the summer supports the hypothesis of ‘real-time’ myxozoa selection pressure on juvenile Atlantic salmon causing mortality over the summer. An increasing number of studies have demonstrated the effect of natural selection on MHC genes over an evolutionary time scale in the wild (Miller et al. 2001; Prugnolle et al. 2005; van Oosterhout et al. 2006; Dionne et al. 2007), but few have identified differential mortality associated with MHC allelic composition during a single generation (Piertney & Oliver 2006; but see de Eyto et al. 2007). Assuming that this susceptibility allele frequency change is strictly associated with mortality, we can speculate that the deleterious consequences of maladaptation in these populations resulted roughly in a 5 per cent reduction in population abundance from June to August (allele frequency changes from 0.088 to 0.040, figure 3). In another system, the introduction of M. cerebralis was associated with a drastic decline in rainbow trout (Oncorhynchus mykiss) recruitment over 18 years (Miller & Vincent 2008). Altogether, these results then suggest that Myxobolus parasites could have demographic consequences on infected populations, and underline the importance of immune adaptation for population persistence. Admittedly, more in-depth studies will be necessary to rigorously assess the deleterious consequences of immune maladaptation in salmon populations. Such studies will become increasingly important in the face of climate change, which may cause rapid variations in pathogen communities and selection pressures.
The heterozygote advantage hypothesis was not supported by our data, which might not be surprising in the context of a predominant pathogen, as suggested by Wegner et al. (2004). Admittedly, not all possible pathogens were identified in this study, which might have influenced the relationship observed between heterozygosity and infection rate. However, what was interesting was the detection of the reverse pattern, with significantly more MHC heterozygotes and higher MHC amino acid genetic distance in infected than in uninfected fish. A similar trend was also observed in Atlantic salmon with furunculosis in the laboratory (Langefors et al. 2001). One possible explanation could be the low frequency of the MHC susceptibility allele 8 (overall frequency less than 0.05), which would result in this allele being mostly restricted to heterozygous individuals, with the result that heterozygotes appear more infected than expected. In such a case, MHC amino acid genetic distance should follow this pattern only if the susceptibility allele is more differentiated in terms of amino acids compared with other alleles, which was indeed found at PBR but not at overall polymorphic sites (t-test comparing p-distances associated with allele 8 with p-distances associated with all other allele comparisons: p=0.029 and 0.39, respectively), in concordance with the results of the MANOVA. Additionally, higher MHC heterozygosity in infected fish could also be partially explained by spatial variability, as a tendency for higher MHC heterozygosity was observed in south shore rivers where infections were more frequent (HO south shore, 0.90±0.05; north shore, 0.81±0.05; t-test: p=0.085). This suggested that the heterozygote advantage hypothesis should be tested with care in the wild, as frequency and identity of MHC alleles conferring susceptibility as well as spatial variability in MHC heterozygosity could influence the relationship between MHC heterozygosity (or other MHC individual genetic distance) and infection rate.
Our results, showing an association between MHC alleles and infection level, are more compatible with the negative frequency-dependent selection and the variable selection in time and space hypotheses (Takahata & Nei 1990; Hedrick 2002), but possibly better support the latter. Although rare alleles were not found to be more advantageous than the others, this study suggested that at the individual level, when facing an emerging pathogen, possessing the right alleles to initiate an appropriate immune response may be important for contemporary adaptation. As the pathogen community is changing, different alleles could be selected for through time, maintaining MHC polymorphism at the population level over an evolutionary time frame, as suggested by the variable selection in time and space hypothesis (Hedrick 2002) and as evidenced in the temporal study of Westerdahl et al. (2004). Indeed, at the population level, balancing selection maintaining MHC diversity could represent the combined influence of multiple directional selection pressures associated with each pathogen over time. However, allele frequency and infection rates would need to be followed across generations to find a stronger support for these hypotheses and evaluate whether a resistance process will develop over an evolutionary time frame in these myxozoa-exposed populations.
Overall, this study represents one of the very few attempts to test for an association between MHC alleles and infection level in wild populations by differentiating the influence of spatio-temporal pathogen selection pressure from individual immune competence. In doing so, we documented the prevalence and type of pathogen representing a contemporary or a potential future selection pressure for Atlantic salmon in the wild. Moreover, this study identified a potential MHC-selective mortality event, and an emerging parasite was identified as the most probable selective agent, possibly representing the first steps of a coevolution process between a host and its pathogen in the wild. Finally, these results suggest the implication of MHC standing genetic variation in facing pathogen challenges in the wild and underline the importance of maintaining diversity at MHC genes in natural populations to increase chances of host contemporary adaptation in a changing environment.
Acknowledgments
Special thanks to F. Caron, D. Fournier, M. Dorais, J.-P. Lebel, R. Isabel, M. Talbot and M. Valentine from ‘Ministère des Ressources Naturelles et de la Faune’ (MRNF) and A. Boivin and J.-F. Bourque from the ‘Centre Inter-universitaire de Recherche sur le Saumon Atlantique’ (CIRSA) for their assistance in the field. R. Firth, J. Roy, R. Lamy and C. Bernier are also gratefully acknowledged for their help during sampling. We also thank K. Giguère, L. Papillon, V. Albert, M. Gauthier, L. Landry, R. Martel and T. Ming for their laboratory assistance. A special thanks to B. Wilson for his great help with the pathogen detection protocol and to C. Whipps for his generous help in identifying the myxozoa parasite. Funding for this project was provided by Natural Sciences and Engineering Research Council of Canada (NSERC, collaborative project) to L.B., J.J.D. and K.M.M. and from MRNF. M.D. was financially supported by an NSERC postgraduate scholarship. Diversitas is also acknowledged for its financial support to M.D. This is a contribution to the research programmes of Québec-Océan and CIRSA.
Footnotes
One contribution of 14 to a Theme Issue ‘Eco-evolutionary dynamics’.
References
- Andree K.B., El-Matbouli M., Hoffman R.W., Hedrick R.P. Comparison of 18S and ITS-1 rDNA sequences of selected geographic isolates of Myxobolus cerebralis. Int. J. Parasitol. 1999;29:771–775. doi: 10.1016/s0020-7519(99)00035-1. doi:10.1016/S0020-7519(99)00035-1 [DOI] [PubMed] [Google Scholar]
- Bakke T.A., Harris P.D. Diseases and parasites in wild Atlantic salmon (Salmo salar) populations. Can. J. Fish. Aquat. Sci. 1998;55:247–266. doi:10.1139/cjfas-55-S1-247 [Google Scholar]
- Barrett R.D.H., Schulter D. Adaptation from standing genetic variation. Trends Ecol. Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. doi:10.1016/j.tree.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Bernatchez L., Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years. J. Evol. Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. doi:10.1046/j.1420-9101.2003.00531.x [DOI] [PubMed] [Google Scholar]
- Berriatua E., Ziluaga I., Miguel-Virto C., Uribarren P., Juste R., Laevens S., Vandamme P., Govan J.R.W. Outbreak of subclinical mastitis in a flock of dairy sheep associated with Burkholderia cepacia complex infection. J. Clin. Microbiol. 2001;39:990–994. doi: 10.1128/JCM.39.3.990-994.2001. doi:10.1128/JCM.39.3.990-994.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briles W.E., Stone H.A., Cole R.K. Marek's disease: effects of B histocompatibility alloalleles in resistant and susceptible chicken lines. Science. 1977;195:193–195. doi: 10.1126/science.831269. doi:10.1126/science.831269 [DOI] [PubMed] [Google Scholar]
- Brown J.H., Jardetzky T., Saper M.A., Samraoui B., Bjorkman P.J., Wiley D.C. A hypothetical model of the foreign antigen binding site of class II histocompatibility molecules. Nature. 1988;332:845–850. doi: 10.1038/332845a0. doi:10.1038/332845a0 [DOI] [PubMed] [Google Scholar]
- Brown J.H., Jardetzky T.S., Gorga J.C., Stern L.J., Urban R.G., Strominger J.L., Wiley D.C. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. doi:10.1038/364033a0 [DOI] [PubMed] [Google Scholar]
- Cai J., Chen H., Thompson K.D., Li C. Isolation and identification of Shewanella alga and its pathogenic effects on post-larvae of abalone Haliotis diversicolor supertexta. J. Fish Dis. 2006;29:505–508. doi: 10.1111/j.1365-2761.2006.00732.x. doi:10.1111/j.1365-2761.2006.00732.x [DOI] [PubMed] [Google Scholar]
- Caron F., Fontaine P.-M., Cauchon V. Ministère des Ressources naturelles et de la Faune; Québec, Canada: 2005. États des stocks de saumon au Québec en 2005. [Google Scholar]
- Carrington M., et al. HLA and HIV-I: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. doi:10.1126/science.283.5408.1748 [DOI] [PubMed] [Google Scholar]
- Cone D.K., Marcogliese D.J. Remodelling of mixed bone during infections of Myxobolus scleroperca (Myxozoa) in Percina caprodes (Rafinesque) (Percidae) from the St Lawrence River, Quebec. J. Fish Dis. 2004;27:551–554. doi: 10.1111/j.1365-2761.2004.00566.x. doi:10.1111/j.1365-2761.2004.00566.x [DOI] [PubMed] [Google Scholar]
- Cone D.K., Marcogliese D.J., Barse A.M., Burt D.B. The myxozoan fauna of Fundulus diaphanus (Cyprinodontidae) from freshwater localities in eastern North America: prevalence, community structure, and geographic distribution. J. Parasitol. 2006;92:52–57. doi: 10.1645/GE-590R.1. doi:10.1645/GE-590R.1 [DOI] [PubMed] [Google Scholar]
- Cordeaux R., Paces-Fessy M., Raimond M., Michel-Salzat A., Zimmer M., Bouchon D. Molecular characterization and evolution of arthropod-pathogenic Rickettsiella bacteria. Appl. Environ. Microbiol. 2007;73:5045–5047. doi: 10.1128/AEM.00378-07. doi:10.1128/AEM.00378-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump B.C., Kling G.W., Bahr M., Hobbie J.E. Bacterioplankton community shifts in an Arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 2003;69:2253–2268. doi: 10.1128/AEM.69.4.2253-2268.2003. doi:10.1128/AEM.69.4.2253-2268.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P., Cunningham A.A., Hyatt A.D. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. doi:10.1126/science.287.5452.443 [DOI] [PubMed] [Google Scholar]
- de Eyto E., et al. Natural selection acts on Atlantic salmon major histocompatibility (MH) variability in the wild. Proc. R. Soc. B. 2007;274:861–869. doi: 10.1098/rspb.2006.0053. doi:10.1098/rspb.2006.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne M., Miller K.M., Dodson J.J., Caron F., Bernatchez L. Clinal variation in MHC diversity with temperature: evidence for the role of host–pathogen interaction on local adaptation in Atlantic salmon. Evolution. 2007;61:2154–2164. doi: 10.1111/j.1558-5646.2007.00178.x. doi:10.1111/j.1558-5646.2007.00178.x [DOI] [PubMed] [Google Scholar]
- Dionne M., Caron F., Dodson J.J., Bernatchez L. Landscape genetics and hierarchical genetic structure in Atlantic salmon: the interaction of gene flow and local adaptation. Mol. Ecol. 2008;17:2382–2396. doi: 10.1111/j.1365-294X.2008.03771.x. doi:10.1111/j.1365-294X.2008.03771.x [DOI] [PubMed] [Google Scholar]
- Dobson A., Foufopoulos J. Emerging infectious pathogens of wildlife. Phil. Trans. R. Soc. B. 2001;356:1001–1012. doi: 10.1098/rstb.2001.0900. doi:10.1098/rstb.2000.0758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala I. The phylogeny of Myxosporea (Myxozoa) based on small subunit ribosomal RNA gene analysis. Int. J. Parasitol. 2006;36:1521–1534. doi: 10.1016/j.ijpara.2006.06.016. doi:10.1016/j.ijpara.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Fryer J.L., Hedrick R.P. Piscirickettsia salmonis: a Gram-negative intracellular bacterial pathogen of fish. J. Fish Dis. 2003;26:251–262. doi: 10.1046/j.1365-2761.2003.00460.x. doi:10.1046/j.1365-2761.2003.00460.x [DOI] [PubMed] [Google Scholar]
- García de Leániz C., et al. A critical review of adaptive genetic variation in Atlantic salmon: implication for conservation. Biol. Rev. 2007;82:173–211. doi: 10.1111/j.1469-185X.2006.00004.x. doi:10.1111/j.1469-185X.2006.00004.x [DOI] [PubMed] [Google Scholar]
- Grimholt U., Larsen S., Nordmo R., Midtlyng P., Kjøglum S., Storset A., Sæbø S., Stet R.J.M. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. doi:10.1007/s00251-003-0567-8 [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Harf R., Sommer S. Association between major histocompatibility complex class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the southern Kalahari. Mol. Ecol. 2005;14:85–91. doi: 10.1111/j.1365-294X.2004.02402.x. doi:10.1111/j.1365-294X.2004.02402.x [DOI] [PubMed] [Google Scholar]
- Harvell C.D., et al. Emerging marine diseases—climate links and anthropogenic factors. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. doi:10.1126/science.285.5433.1505 [DOI] [PubMed] [Google Scholar]
- Hedrick P.W. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56:1902–1908. doi: 10.1111/j.0014-3820.2002.tb00116.x. doi:10.1111/j.0014-3820.2002.tb00116.x [DOI] [PubMed] [Google Scholar]
- Hedrick R.P., MacConnell E., de Kinkelin P. Proliferative kidney disease of salmonid fish. Annu. Rev. Fish Dis. 1993;3:277–290. doi:10.1016/0959-8030(93)90039-E [Google Scholar]
- Hill A.V.S., et al. Common West African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. doi:10.1038/352595a0 [DOI] [PubMed] [Google Scholar]
- Hoffman G.L. Myxobolus cerebralis, a worldwide cause of salmonid whirling disease. J. Aquat. Anim. Health. 1990;2:30–37. doi:10.1577/1548-8667(1990)002<0030:MCAWCO>2.3.CO;2 [Google Scholar]
- Hood, G. M. 2008 Poptools version 3.0.3. See http://www.cse.csiro.au/poptools
- Hughes A.L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. doi:10.1038/335167a0 [DOI] [PubMed] [Google Scholar]
- Hughes A.L., Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu. Rev. Genet. 1998;32:415–425. doi: 10.1146/annurev.genet.32.1.415. doi:10.1146/annurev.genet.32.1.415 [DOI] [PubMed] [Google Scholar]
- Kent M.L., et al. Recent advances in our knowledge of the myxozoa. J. Eukaryot. Microbiol. 2001;48:395–413. doi: 10.1111/j.1550-7408.2001.tb00173.x. doi:10.1111/j.1550-7408.2001.tb00173.x [DOI] [PubMed] [Google Scholar]
- King T.L., Eackles M.S., Letcher B.H. Microsatellite DNA markers for the study of Atlantic salmon (Salmo salar) kinship, population structure, and mixed-fishery analyses. Mol. Ecol. Notes. 2005;5:130–132. doi:10.1111/j.1471-8286.2005.00860.x [Google Scholar]
- Krkošek M., Ford J.S., Morton A., Lele S., Myers R.A., Lewis M.A. Declining wild salmon populations in relation to parasites from farm salmon. Science. 2007;318:1772–1775. doi: 10.1126/science.1148744. doi:10.1126/science.1148744 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I., Nei M. Pennsylvania State University; University Park, PA: 2000. Mega: molecular evolutionary genetics analysis, v. 2.0. [Google Scholar]
- Lafferty K.D., Porter J., Ford S.E. Are diseases increasing in the ocean? Annu. Rev. Ecol. Evol. Syst. 2004;35:31–54. doi:10.1146/annurev.ecolsys.35.021103.105704 [Google Scholar]
- Landry C., Bernatchez L. Comparative analysis of population structure across environments and geographical scales at major histocompatibility complex and microsatellite loci in Atlantic salmon (Salmo salar) Mol. Ecol. 2001;10:2525–2539. doi: 10.1046/j.1365-294x.2001.01383.x. doi:10.1046/j.1365-294X.2001.01383.x [DOI] [PubMed] [Google Scholar]
- Landry C., Garant D., Dushesne P., Bernatchez L. ‘Good genes as heterozygosity’: the major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar) Proc. R. Soc. B. 2001;268:1279–1285. doi: 10.1098/rspb.2001.1659. doi:10.1098/rspb.2001.1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langefors A.H., Lohm J., Grahn M., Andersen O., Schantz T. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. R. Soc. B. 2001;268:479–485. doi: 10.1098/rspb.2000.1378. doi:10.1098/rspb.2000.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.H., Blackburn N., Larsen J.L., Olsen J.E. Influences of temperature, salinity and starvation on the motility and chemotactic response of Vibrio anguillarum. Microbiology. 2004;150:1283–1290. doi: 10.1099/mic.0.26379-0. doi:10.1099/mic.0.26379-0 [DOI] [PubMed] [Google Scholar]
- Madsen T., Ujvari B. MHC class I variation associates with parasite resistance and longevity in tropical pythons. J. Evol. Biol. 2006;19:1973–1978. doi: 10.1111/j.1420-9101.2006.01158.x. doi:10.1111/j.1420-9101.2006.01158.x [DOI] [PubMed] [Google Scholar]
- McClelland E.E., Penn D.J., Potts W.K. Major histocompatibility complex heterozygote superiority during coinfection. Infect. Immun. 2003;71:2079–2086. doi: 10.1128/IAI.71.4.2079-2086.2003. doi:10.1128/IAI.71.4.2079-2086.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messick J.B., Walker P.G., Raphael W., Berent L., Shi X. ‘Candidatus Mycoplasma haemodidelphidis’ sp. nov., ‘Candidatus Mycoplasma haemolamae’ sp. nov. and Mycoplasma haemocanis comb. nov., haemotrophic parasites from a naturally infected opossum (Didelphis virginiana), alpaca (Lama pacos) and dog (Canis familiaris): phylogenetic and secondary structural relatedness of their 16S rRNA genes to other mycoplasmas. Int. J. Syst. Evol. Microbiol. 2002;52:693–698. doi: 10.1099/00207713-52-3-693. doi:10.1099/ijs.0.01861-0 [DOI] [PubMed] [Google Scholar]
- Miller K.M., Kaukinen K.H., Beacham T.D., Withler R.E. Geographic heterogeneity in natural selection on an MHC locus in sockeye salmon. Genetica. 2001;111:237–257. doi: 10.1023/a:1013716020351. doi:10.1023/A:1013716020351 [DOI] [PubMed] [Google Scholar]
- Miller K.M., Winton J.R., Schulze A.D., Purcell M.K., Ming T.J. Major histocompatibility complex loci are associated with susceptibility of Atlantic salmon to infectious hematopoietic necrosis virus. Environ. Biol. Fishes. 2004;69:307–316. doi:10.1023/B:EBFI.0000022874.48341.0f [Google Scholar]
- Miller K.P., Vincent E.R. Rapid natural selection for resistance to an introduced parasite of rainbow trout. Evol. Appl. 2008;1:336–341. doi: 10.1111/j.1752-4571.2008.00018.x. doi:10.1111/j.1752-4571.2008.00018.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Hughes A.L. Polymorphism and evolution of the major histocompatibility complex loci in mammals. In: Selander R., Clark A., Whittams T., editors. Evolution at the molecular level. Sinauer Associates; Sunderland, MA: 1991. pp. 222–247. [Google Scholar]
- Nei M., Kumar S. Oxford University Press; New York, NY: 2000. Molecular evolution and phylogenetics. [Google Scholar]
- Nylund A., Ottem K.F., Watanabe K., Karlsbakk E., Krossøy B. Francisella sp. (family Francisellaceae) causing mortality in Norwegian cod (Gadus morhua) farming. Arch. Microbiol. 2006;185:383–392. doi: 10.1007/s00203-006-0109-5. doi:10.1007/s00203-006-0109-5 [DOI] [PubMed] [Google Scholar]
- O'Reilly P.T., Hamilton L., McConnell S.K.J., Wright J.M. Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can. J. Fish. Aquat. Sci. 1996;53:2292–2298. doi:10.1139/cjfas-53-10-2292 [Google Scholar]
- Paterson S., Piertney S.B., Knox D., Gilbey J., Verspoor E. Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Mol. Ecol. Notes. 2004;4:160–162. doi:10.1111/j.1471-8286.2004.00598.x [Google Scholar]
- Penn D.J., Damjanovich K., Potts W.K. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA. 2002;99:11260–11264. doi: 10.1073/pnas.162006499. doi:10.1073/pnas.162006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piertney S.B., Oliver M.K. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. doi:10.1038/sj.hdy.6800724 [DOI] [PubMed] [Google Scholar]
- Porlier M., Bélisle M., Garant D. Non-random distribution of individual genetic diversity along an environmental gradient. Phil. Trans. R. Soc. B. 2009;364:1543–1554. doi: 10.1098/rstb.2009.0010. doi:10.1098/rstb.2009.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts W.K., Wakeland E.K. Evolution of diversity at the major histocompatibility complex. Trends Ecol. Evol. 1990;5:181–187. doi: 10.1016/0169-5347(90)90207-T. doi:10.1016/0169-5347(90)90207-T [DOI] [PubMed] [Google Scholar]
- Presa P., Guyomard R. Conservation of microsatellites in three species of salmonids. J. Fish Biol. 1996;49:1326–1329. doi:10.1111/j.1095-8649.1996.tb01800.x [Google Scholar]
- Prugnolle F., Manica A., Charpentier M., Guégan J.-F., Guernier V., Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 2005;15:1022–1027. doi: 10.1016/j.cub.2005.04.050. doi:10.1016/j.cub.2005.04.050 [DOI] [PubMed] [Google Scholar]
- Raymond M., Rousset F. Genepop (version 3.2): population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Rice W.R. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. doi:10.2307/2409177 [DOI] [PubMed] [Google Scholar]
- Schlötterer C. A microsatellite-based multilocus screen for the identification of local selective sweeps. Genetics. 2002;160:753–763. doi: 10.1093/genetics/160.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slettan A., Olsaker I., Lie O. Atlantic salmon, Salmo salar, microsatellites at the SSOSL25, SSOSL85, SSOSL311, SSOSL417 loci. Anim. Genet. 1995;26:281–282. doi: 10.1111/j.1365-2052.1995.tb03262.x. [DOI] [PubMed] [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2005;2:1–18. doi: 10.1186/1742-9994-2-16. doi:10.1186/1742-9994-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell O.B. Homing and olfaction in salmonids: a critical review with special references to the Atlantic salmon. Biol. Rev. 1984;59:333–388. doi:10.1111/j.1469-185X.1984.tb00709.x [Google Scholar]
- Takahata N., Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.B. A review of local adaptation in salmonidae, with particular reference to Pacific and Atlantic salmon. Aquaculture. 1991;98:185–207. doi:10.1016/0044-8486(91)90383-I [Google Scholar]
- Thompson J.N. Rapid evolution as an ecological process. Trends Ecol. Evol. 1998;13:329–332. doi: 10.1016/s0169-5347(98)01378-0. doi:10.1016/S0169-5347(98)01378-0 [DOI] [PubMed] [Google Scholar]
- Thune R.L., Stanley L.A., Cooper R.K. Pathogenesis of Gram-negative bacterial infections in warmwater fish. Annu. Rev. Fish Dis. 1993;3:37–68. doi:10.1016/0959-8030(93)90028-A [Google Scholar]
- Thursz M.R., Thomas H.C., Greenwood B.M., Hill A.V. Heterozygote advantage for HLA class-II type in hepatitis B virus infection. Nat. Genet. 1997;17:11–12. doi: 10.1038/ng0997-11. doi:10.1038/ng0997-11 [DOI] [PubMed] [Google Scholar]
- Uhland C., Mikaelian I., Martineau D. Les Presses de l'Université de Montréal; Montréal, Canada: 2000. Maladies des poissons d'eau douce du Québec: guide de diagnostic. [Google Scholar]
- van Oosterhout C., Hutchinson W.F., Wills D.P.M., Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. doi:10.1111/j.1471-8286.2004.00684.x [Google Scholar]
- van Oosterhout C., Joyce D.A., Cummings S.M., Blais J., Barson N.J., Ramnarine I.W., Mohamed R.S., Persad N., Cable J. Balancing selection, random genetic drift, and genetic variation at the major histocompatibility complex in two wild populations of guppies (Poecilia reticulata) Evolution. 2006;60:2562–2574. doi:10.1554/06-286.1 [PubMed] [Google Scholar]
- Wegner K.M., Reusch T.B.H., Kalbe M. Multiple parasites are driving major histocompatibility complex polymorphism in the wild. J. Evol. Biol. 2003;16:224–232. doi: 10.1046/j.1420-9101.2003.00519.x. doi:10.1046/j.1420-9101.2003.00519.x [DOI] [PubMed] [Google Scholar]
- Wegner K.M., Kalbe M., Schaschl H., Reusch T.B.H. Parasites and individual major histocompatibility complex diversity—an optimal choice? Microbes Infect. 2004;6:1110–1116. doi: 10.1016/j.micinf.2004.05.025. doi:10.1016/j.micinf.2004.05.025 [DOI] [PubMed] [Google Scholar]
- Westerdahl H., Hansson B., Bensch S., Hasselquist D. Between-year variation of MHC allele frequencies in great reed warblers: selection or drift? J. Evol. Biol. 2004;17:485–492. doi: 10.1111/j.1420-9101.2004.00711.x. doi:10.1111/j.1420-9101.2004.00711.x [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Teng J.L., Tse H., Yuen K.Y. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008;14:908–934. doi: 10.1111/j.1469-0691.2008.02070.x. doi:10.1111/j.1469-0691.2008.02070.x [DOI] [PubMed] [Google Scholar]
- Zheng C., Ovaskainen O., Hanski I. Modelling single nucleotide effects in phosphoglucose isomerase on dispersal in the Glanville fritillary butterfly: coupling of ecological and evolutionary dynamics. Phil. Trans. R. Soc. B. 2009;364:1519–1532. doi: 10.1098/rstb.2009.0005. doi:10.1098/rstb.2009.0005 [DOI] [PMC free article] [PubMed] [Google Scholar]