Abstract
The identification of receptors that detect environmental stimuli lays a foundation for exploring the mechanisms and neural circuits underlying sensation. The mouse vomeronasal organ (VNO), which detects pheromones and other semiochemicals, has 2 known families of chemoreceptors, V1Rs and V2Rs. Here, we report a third family of mouse VNO receptors comprising 5 of 7 members of the formyl peptide receptor (FPR) family. Unlike other FPRs, which function in the immune system, these FPRs are selectively expressed in VNO neurons in patterns strikingly similar to those of V1Rs and V2Rs. Each FPR is expressed in a different small subset of neurons that are highly dispersed in the neuroepithelium, consistently coexpress either Gαi2 or Gαo, and lack other chemoreceptors examined. Given the presence of formylated peptides in bacteria and mitochondria, possible roles for VNO FPRs include the assessment of conspecifics or other species based on variations in normal bacterial flora or mitochondrial proteins.
Keywords: fpr, GPCR olfactory, pheromone
Semiochemicals that transmit messages within and between species are detected by the olfactory systems of both vertebrate and invertebrate organisms. The stereotyped responses elicited by these chemicals and their importance to the perpetuation of the species suggest the involvement of dedicated chemosensory receptors and hard-wired neural circuits that assure appropriate responses to specific chemosensory stimuli.
Mice can detect a variety of semiochemicals. These include pheromones that induce changes in reproductive hormone levels or stimulate sexual or aggressive behaviors (1–4). They also include genetically determined individuality cues present in urine that can influence the choice of a mating partner or cause a failure of embryo implantation (“pregnancy block”) (5, 6). In addition, predator odors can elicit innate fear responses involving both hormonal and behavioral changes (7). Many of these semiochemicals are not yet defined at the molecular level. Those with known structures include several volatile urinary compounds reported to affect reproductive physiology and behavior (8), major urinary proteins (MUPs) that stimulate male–male aggression (9) and may serve as individuality cues (10), several peptides that bind to major histocompatibility complex (MHC) proteins and can act as individuality cues that interfere with pregnancy (5), and a compound in fox feces that stimulates a fear response (7).
In mice, many semiochemicals are detected by sensory neurons in the VNO. The VNO is a tubular olfactory structure at the base of the nasal septum that connects to the nasal cavity via a small duct (2, 3). However, some semiochemicals are also, or instead, detected in the nasal olfactory epithelium (OE) (7, 11, 12), which is also responsible for the detection of odorants (4). Consistent with an important role in the detection of pheromones and other semiochemicals, sensory signals generated in the VNO travel to limbic areas such as the amygdala and hypothalamus, which control basic drives, hormone levels, and instinctive behaviors. In contrast, OE signals are sent to higher cortical areas important in odor perception and to limbic areas (2, 4).
The VNO contains 2 known families of chemosensory receptors, the V1R and V2R families (13–16), which have about 240 and 120 members, respectively (17, 18) (J. Young and B. Trask, personal communication). Like chemosensory receptors in the OE [≈1,000 different odorant receptors (ORs) and 14 trace amine-associated receptors (TAARs)] (19, 20), V1Rs and V2Rs belong to the G protein-coupled receptor (GPCR) superfamily and members of each family are diverse in protein sequence, suggesting that each family may detect a variety of chemicals. Within the VNO neuroepithelium, V1Rs are coexpressed with the G protein Gαi2 in neurons in an apical zone whereas V2Rs are coexpressed with Gαo in neurons in a basal zone (13–15). Because of difficulties in obtaining functional expression of V1Rs and V2Rs in cell lines, little is known about their ligands. However, one volatile pheromone is detected by a particular V1R (21), a male exocrine gland-secreting peptide (ESP) is detected by a specific V2R (22), and MUPs and MHC-binding peptides activate Gαo-expressing neurons, suggesting the possible involvement of V2Rs (9, 23).
Here, we report the existence of a third family of candidate chemosensory receptors in the VNO. By conducting a high throughput screen for GPCRs expressed in mouse VNO neurons, we found that 5 of 7 members of the formyl peptide receptor (FPR) family are expressed by VNO neurons. The other 2 FPRs are instead expressed in the immune system, where they are believed to stimulate chemotaxis to sites of infection or tissue damage upon recognition of their ligands, such as formylated pepides from bacteria or mitochondria (24). The expression patterns of the VNO FPRs are remarkably similar to those of V1Rs and V2Rs: they are selectively expressed in the VNO and each FPR is expressed in a different small subset of neurons that are highly dispersed, consistently express Gαi2 or Gαo, and appear to lack other chemoreceptors. These findings suggest that the VNO FPRs are likely to function as chemosensory receptors. Phylogenetic analyses indicate that genes encoding VNO FPRs evolved recently in the rodent lineage, raising the possibility that these receptors impart a novel chemosensory function to rodents.
Results
A High Throughput Screen for Receptors Expressed in VNO Neurons.
To explore whether there might be additional families of receptors in VNO neurons, we used a high throughput approach. Preliminary experiments revealed endogenous β-galactosidase activity in VNO neurons, but not other VNO cell types (supporting information (SI) Fig. S1). To obtain an enriched population of VNO neurons, we labeled dissociated VNO cells with a fluorescent β-galactosidase substrate, fluorescein di-(β-galactopyranoside) and then used fluorescence-activated cell sorting to isolate the labeled neurons. We next prepared cDNA from the RNA of the isolated neurons and used the cDNA in real-time quantitative PCR (qPCR) reactions in 384-well plates. In these experiments, we tested primer pairs specific for 365 GPCRs not previously implicated in the detection of odors, pheromones, or tastes (20).
Surprisingly, cDNAs encoding several members of the formyl peptide receptor (FPR) family were amplified from VNO neuron cDNA. This family has 7 members in mice named FPR1, FPR-rs1, FPR-rs2, FPR-rs3, FPR-rs4, FPR-rs6, and FPR-rs7 (Fpr-rs5 is a pseudogene) and 3 in humans termed FPR1, FPR2 (FPRL1), and FPR3 (FPRL2) (24). All 3 human FPR genes and 2 of the 7 mouse Fpr genes (Fpr1 and Fpr-rs2) are expressed by neutrophils or myeloid lineage cells and are believed to play an important role in innate immune responses by recognizing formylated peptides released from bacteria or mitochondria at sites of infection or tissue destruction (24).
Members of the Formyl Peptide Receptor Family Are Selectively Expressed in the VNO.
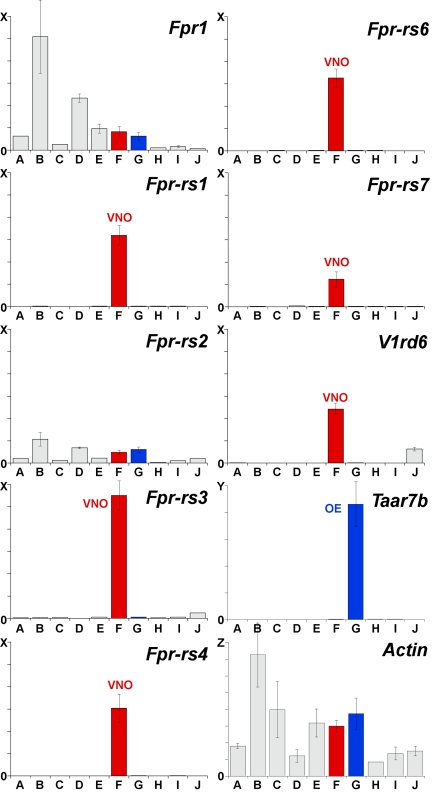
To verify the expression of Fpr genes in the VNO and compare it with their expression in other tissues, we conducted qPCR reactions with cDNAs prepared from 10 different mouse tissues, including both male and female VNO (Fig. 1). For the two Fpr genes expressed in immune cells (Fpr1 and Fpr-rs2), VNO cDNA showed only low levels of expression that were less than those seen for some other tissues. In sharp contrast, the other 5 Fpr genes showed relatively high expression in VNO cDNA, but not in cDNA from any other tissue, including the olfactory epithelium. Similar results were obtained using male and female VNO cDNA. Interestingly, the expression levels of the 5 Fpr genes in the VNO resembled that of a V1r gene, V1rd6. These results indicate that 5 of 7 mouse Fpr genes are selectively expressed in the VNO and, furthermore, that their levels of expression resemble that of a VNO chemosensory receptor.
Fig. 1.
Five of 7 mouse Fpr genes are expressed in the VNO. qPCR was conducted using primers specific for each mouse Fpr gene, a mouse V1r gene (V1rd6), a mouse Taar gene (Taar7b), or the mouse β-Actin gene and, as templates, cDNAs prepared from DNase-treated RNAs from different mouse tissues: A, heart; B, spleen; C, intestine; D, liver; E, brain; F, VNO (red); G; olfactory epithelium (blue); H, circumvallate taste papillae; I, olfactory bulb; and J, testis). Results of triplicate experiments are shown (±SD). No PCR products were seen in control experiments lacking reverse transcriptase. cDNAs for five of the seven mouse Fpr genes (Fpr-rs1, Fpr-rs3, Fpr-rs4, Fpr-rs6, and Fpr-rs7) were selectively amplified from VNO cDNA (red bars), similar to V1rd6 cDNAs. Scales on the y axis differ as follows: X = 60,000 copies, Y = 100,000 copies, and Z = 8,000,000 copies. Each column represents signal from cDNA prepared from 1 μg total RNA, based on reactions using 10 ng RNA.
To further investigate Fpr gene expression in different tissues, we searched the mouse EST (expressed sequence tag) database at NCBI (National Center for Biotechnology Information) using BLASTN with each mouse Fpr coding region sequence as query. (http://www.ncbi.nlm.nih.gov). We found sequences encoding both immune system FPRs (FPR1 and FPR-rs2) in ESTs from a variety of tissues, probably because of the presence of blood-containing neutrophils and monocytes in all tissues. Strikingly, however, we found no ESTs for any of the VNO FPRs. We also failed to identify ESTs for 2 VNO receptors, V1Rb2 or V1Re11, a result that likely reflects the lack of large-scale sequencing of VNO cDNAs. Together, these results indicate that 5 of 7 members of the Fpr gene family are predominantly or exclusively expressed in the VNO.
Fpr Genes Are Expressed in Small Subsets of Dispersed VNO Neurons.
We next used RNA in situ hybridization to ask whether Fpr genes are truly expressed in VNO neurons. Serial sections were collected from male or female VNOs and different sections were hybridized to digoxigenin-labeled cRNA probes for each of the 7 mouse Fpr genes or, as controls, V1r or V2r probes. Preliminary experiments indicated that sequences with <87% identity do not cross hybridize under the high stringency in situ hybridization conditions used. Therefore, it was possible to distinguish the expression of all Fpr genes except Fpr-rs1 versus Fpr-rs2 (87% identical) and Fpr-rs6 versus Fpr-rs7 (96% identical).
Probes for each of these genes labeled a small subset of neurons that were scattered in the VNO neuroepithelium (Figs. 2 and 3). We detected no differences in this labeling pattern along the anterior–posterior axis of the VNO nor did we detect differences in male versus female VNOs. The hybridization patterns seen with individual Fpr probes resembled those seen using V1r and V2r probes in these experiments (Table S1) and in previous studies (13–16). No VNO expression was evident for either of the Fpr genes expressed in the immune system: the Fpr1 probe did not label any cells and rare and weak labeling with the Fpr-rs2 probe appeared to be the result of cross-hybridization to Fpr-rs1 RNA. These experiments indicated that all 5 Fpr genes expressed in the VNO are expressed in VNO neurons.
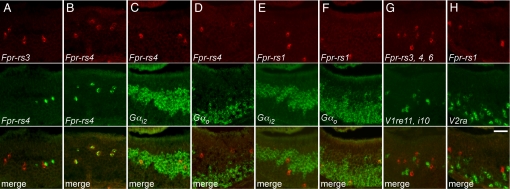
Fig. 2.
Fpr genes are expressed by subsets of dispersed VNO neurons. Digoxigenin-labeled cRNA probes were hybridized to coronal sections through the mouse VNO. Representative sections are shown for antisense probes for different Fprs and one V1r (V1ra2), and a sense probe for one Fpr. Similar to the V1r probe, each antisense Fpr probe labeled a subset of neurons dispersed in the VNO. (Scale bar = 200 μm.)
Fig. 3.
The expression of individual Fpr genes defines unique subsets of VNO neurons. The expression patterns of pairs of genes were compared using two-color RNA in situ hybridization (columns A–H). Probes for different Fprs labeled different neurons (A) whereas different probes for the same Fpr labeled the same neurons (B). Neurons labeled for Fpr-rs4 were colabeled for Gαi2 (C), but not Gαo (D), while cells labeled for Fpr-rs1 were colabeled for Gαo (F), but not Gαi2 (E). Neurons labeled by a mix of Fpr-rs3, Fpr-rs4, and Fpr-rs6 probes were not labeled by a mixed V1r probe (G) nor were those labeled for Fpr-rs1 colabeled by a V2r probe (H). (Scale bar = 50 μm.)
To obtain information on the size of neuronal subsets expressing individual Fpr genes, we counted the number of neurons labeled by different Fpr probes in multiple tissue sections and compared these data with counts obtained with V1r and V2r probes (Table S1). Taking into account the number of mouse genes with ≥87% identity to each probe, we calculate that individual Fpr genes were expressed, on average, in 3.7 neurons per 14 μm section. This was remarkably similar to results obtained with probes that matched V1rs and V2rs, which indicated that individual V1r genes and V2r genes were expressed, on average, in 3.3 and 6.8 neurons per 14 μm section, respectively. Thus, both the number and patterning of neurons expressing individual Fpr genes resemble what is seen for members of the V1r and V2r chemosensory receptor gene families.
Individual Fpr Genes Are Consistently Coexpressed with Gαi2 or GαO.
V1Rs are coexpressed with Gαi2 in an apical zone of the VNO while V2Rs are coexpressed with Gαo in a complementary basal zone. To determine whether FPRs are similarly coexpressed with Gαi2 or Gαo, we performed dual in situ hybridization using Gαi2 and Gαo probes together with individual Fpr probes or, as controls, V1r or V2r probes (Fig. 3, C–F, Table S2).
The great majority of neurons labeled for Fpr-rs3 (94.8%), Fpr-rs4 (98.5%), and Fpr-rs6/7 (97.8%) were also labeled for Gαi2 whereas only 4.3%, 2.0%, and 4.8%, respectively, were also labeled for Gαo. In contrast, most neurons labeled for Fpr-rs1 (93.6%) were colabeled for Gαo and only 3.5% for Gαi2. These results were comparable to those obtained using a V1ra2 probe, which showed 97.8% of hybridized neurons colabeled for Gαi2 and only 4.8% for Gαo. Thus, similar to the expression of V1Rs and V2Rs in the VNO, individual FPRs are consistently coexpressed with either Gαi2 or Gαo, but not both. However, in contrast to the selective coexpression of V1Rs with Gαi2 and V2Rs with Gαo, FPR-rs1 is coexpressed with Gαo whereas the other VNO FPRs are coexpressed with Gαi2.
Different Fpr Genes Are Expressed in Different VNO Neurons.
One common theme among chemosensory receptors in both the VNO and olfactory epithelium is that each neuron appears to express only 1 receptor gene (13–15, 25). The only known exceptions are members of 1 V2r gene subfamily (the V2r2 subfamily), which appear to be coexpressed at a low level with most or all other V2r genes (26). To determine whether different Fpr genes are expressed in different neurons, we hybridized VNO sections to pairs of differentially labeled Fpr probes.
Using all pairwise combinations of Fpr-rs1, Fpr-rs3, Fpr-rs4, and Fpr-rs6 probes, we found that more than 99% of cells labeled by each Fpr probe were unlabeled by another Fpr probe (2609/2612 cells total) (Fig. 3 A and B, Table S2). These results indicate that, similar to V1Rs and V2Rs, and to chemosensory receptors in the olfactory epithelium, different FPRs are expressed in different subsets of VNO neurons.
Neurons Expressing FPRs Appear to Lack Other Chemosensory Receptors.
The above experiments demonstrated that FPR family members have VNO expression patterns similar to those seen for V1R and V2R chemosensory receptors. This suggested that there may be subsets of VNO neurons that use different FPRs rather than V1Rs or V2Rs to detect chemosensory ligands. However, another possibility was that neurons with FPRs also express V1Rs or V2Rs. To examine this possibility, we used double fluorescence in situ hybridization to compare VNO neurons that hybridize to Fpr versus V1r or V2r probes. Because FPR-rs1 is coexpressed with Gαo, we compared its expression to that of V2Rs, but since the other FPRs are coexpressed with Gαi2, we compared their expression to that of V1Rs. (Fig. 3 G and H, Table S2).
Neurons labeled by a mix of Fpr-rs3, Fpr-rs4, and Fpr-rs6 probes were compared with those labeled by 10 different V1r probes, (alone or in mixes), representing 10 of the 12 V1R subfamilies (27). On the basis of our observation that sequences with ≥87% identity cross-hybridize under the hybridization stringency we used, and our determination of intact V1r genes in the genome with this level of identity to our V1r probes, we calculated that the 10 V1r probes used in these experiments would hybridize to RNAs encoded by 43 (18%) of the 239 intact V1r genes in the mouse genome. Of neurons labeled for Fprs shown to be coexpressed with Gαi2, only 0.3% were colabeled for the V1rs tested (6/2081 cells total). If FPR+ neurons instead coexpressed one V1R chosen at random, we would expect to see more colabeled cells (51/2081, on the basis of the particular probe combinations used, Table S2). Using a Poisson distribution, the probability that we would see 6/2081 colabeled cells rather than 51/2081 colabeled cells is extremely low (1.7 × 10−15). Of neurons that were labeled by the Fpr-rs1 probe, none (0/106) was colabeled by a V2ra probe predicted to hybridize to RNAs encoded by 17 V2r genes (14% of the 121 V2r intact genes in the genome).
Without comparing the expression of VNO Fpr genes with that of all V1r and V2r genes, the methods used cannot exclude the possibility that FPRs are coexpressed with specific, unexamined members of the V1R or V2R family, a caveat that also holds true for the expression of individual V1Rs and V2Rs in different neurons. However, these results are consistent with a model in which there are multiple subsets of VNO neurons that use different FPRs, rather than V1Rs or V2Rs, to detect chemosensory stimuli.
VNO FPRs Evolved Recently and Underwent Neofunctionalization.
The VNO is an evolutionarily old structure found not only in most mammals, but also in reptiles. V1Rs and V2Rs are present even in fish, suggesting a still older origin (28, 29). Do any mammals other than mice have larger families of FPRs than humans, suggesting the possible existence of VNO FPRs? To explore this question, we searched for genes encoding FPR-related proteins in NCBI genome sequence databases for a variety of mammals using TBLASTN and individual mouse FPRs as queries. On the basis of sequence relationships among FPRs versus other receptors, we only considered proteins with at least 45% identity to 1 or more mouse FPRs.
These analyses identified 7 genes encoding FPR-related proteins in rat, 2–3 in primates, 1–2 each in dog, cat, and horse, and none in cow, sheep, or pig. We also found 6 genes encoding proteins with ≈40% identity to FPRs in opossum, 3 in platypus, and 1 in zebrafish, but these proteins also shared ≈40% identity to several chemokine receptors, so it is unclear whether these are FPR family members. Incomplete genome sequence data could explain the small number of Fpr genes found in some species. However, genome sequence coverage is extensive for dog, cow, and horse, suggesting that these species are unlikely to have large families of FPRs and could instead have even fewer than humans, whose three FPRs are expressed in the immune system. These results suggest that VNO FPRs are likely to be present in only certain mammals, possibly only in rodents.
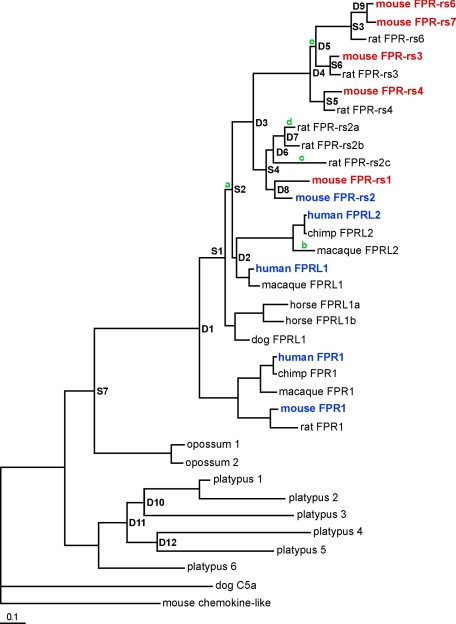
To gain insight into relationships among FPRs found in different species, we constructed a phylogenetic tree of the FPRs using Bayesian analysis (Fig. 4). We then analyzed evolutionary rates and selection of Fpr genes along branches of the tree by calculating ratios of non-synonymous to synonymous substitutions. These analyses suggest that, in rodents, an ancestral Fpr gene underwent a period of positive selection consistent with neofunctionalization, laying the foundation for expansion and diversification of this gene family. Numerous gene duplications increased the size of the Fpr gene family in mice and rats, giving rise to a series of FPRs that lack clear orthologs in other species. These FPRs are located within a rodent-specific branch of the FPR phylogenetic tree (Fig. 4). Importantly, this group of FPRs includes all of those that are selectively expressed in the mouse VNO. Although additional Fpr genes may be revealed in other mammals as additional genome sequence data become available, these phylogenetic analyses indicate that an expansion of the FPR family to generate VNO receptors occurred recently along the rodent lineage and represents the de novo evolution of a novel chemosensory receptor family in rodents, but not other mammals.
Fig. 4.
Phylogenetic analysis of FPR family members in mammals. Fpr coding sequences were compiled from BLAST searches of whole genome databases and their encoded proteins used to generate a phylogenetic tree using Bayesian analysis. Branch points leading to new genes through duplication (D) or speciation (S) were assigned by soft parsimony. Green letters indicate branches showing evidence of lineage-specific positive selection, as determined by analyzing ratios of non-synonymous to synonymous substitutions in Fpr genes. Mouse VNO FPRs are highlighted in red and mouse and human immune system FPRs in blue. VNO FPRs are in a rodent-specific branch of the phylogenetic tree, which expanded due to several recent gene duplications (D3–D9). All tree nodes have a posterior probability above 0.9, except D2 (0.64), S2 (0.77), S7 (0.5), D10 (0.62), D11 (0.78), and D12 (0.86). The branch length scale bar indicates substitutions per site.
Discussion
In these studies, we identified a third family of candidate chemosensory receptors in the mouse VNO. By conducting a high throughput search for receptors expressed in VNO neurons, we found evidence that these neurons express certain members of the FPR family, a receptor family previously implicated in the innate immune response. Subsequent experiments demonstrated that five of seven members of this family are selectively expressed in VNO neurons whereas the two FPRs found in the immune system are not. The expression patterns of the VNO FPRs resemble those of V1R and V2R chemosensory receptors, suggesting that there are multiple subsets of VNO neurons that use individual FPRs rather than V1Rs or V2Rs to detect chemosensory stimuli. Phylogenetic analyses indicate that VNO FPRs evolved recently in the rodent lineage and may thus impart novel chemosensory functions to rodents.
FPRs as Candidate Chemosensory Receptors in the VNO.
Three lines of evidence suggest that VNO FPRs are likely to serve as chemosensory receptors in the VNO. First, the expression patterns of the VNO FPRs are strikingly similar to those of V1Rs and V2Rs. Each FPR is expressed in only a small subset of VNO neurons. Neurons with the same FPR are dispersed in the neuroepithelium and found throughout its entire length. Different FPRs are expressed in different neurons. And neurons with the same FPR uniformly coexpress either the Gαi2 or Gαo G protein.
Second, no evidence was found for the coexpression of FPRs with V1Rs or V2Rs. Without comparing the expression of VNO FPRs with that of all ≈360 V1Rs and V2Rs, it cannot be excluded that the FPRs are coexpressed with a small number of V1R or V2R family members. However, comparisons of neurons expressing FPRs versus a large number of different V1Rs or V2Rs did not uncover any evidence for their coexpression.
Third, VNO FPRs appear to be expressed predominantly or exclusively in the VNO. Similar to what has been observed for V1Rs and V2Rs, no evidence was found for the expression of VNO FPRs in other tissues, consistent with a dedicated role for these receptors in chemosensation.
Potential Functions of VNO FPRs.
Given that the VNO expresses over 300 V1Rs and V2Rs, what might an additional five FPRs add to VNO chemosensory detection? Our phylogenetic analyses indicate that the VNO FPRs resulted from recent gene duplications and positive selection and thus are likely to provide a selective advantage to the animal. This advantage could derive from an ability of FPRs to recognize sensory ligands that are not detected by either V1Rs or V2Rs. Consistent with this idea, VNO FPRs do not share significant sequence similarity with either V1Rs or V2Rs nor do they resemble ORs or TAARs expressed in the OE. Alternatively, FPRs might recognize some of the same ligands as V1Rs or V2Rs, but generate signals that are conveyed to different brain regions, thus allowing for different responses to the same ligands.
What sensory ligands might VNO FPRs recognize? Since immune system FPRs recognize a number of formylated as well as nonformylated peptides (24), it is quite possible that VNO FPRs recognize similar types of ligands. Sequence relationships among FPR family members are consistent with this idea. The two mouse immune system FPRs, FPR1 and FPR-rs2, are 59% identical and both recognize the formylated peptide fMLF in addition to differentially recognizing other peptides. FPR-rs2 is even more related to the VNO FPR, FPR-rs1, (81% identity) and shows comparable relatedness to the other VNO FPRs (59–66% identity). BLASTP searches of the NCBI mouse protein database with several VNO FPRs indicated that, after immune system FPRs, these proteins are most related to other GPCRs with peptide ligands, consistent with the idea that VNO FPRs may detect peptides. Thus far, we have been unable to identify ligands for VNO FPRs using heterologous expression in HEK293 cells and test ligands varying from fMLF to natural substances, such as mouse urine. However, this failure could derive from difficulties in heterologous expression, as is seen for other chemosensory receptors.
The VNO plays a major role in the detection of semiochemicals that stimulate hormonal or behavioral responses, allow recognition of individual conspecifics on the basis of genetic polymorphisms, and, at least in some species (rat), the detection of predator odors that elicit innate fear responses. FPRs could conceivably be involved in the recognition of semiochemicals that elicit one or more of these responses or they might be involved in other innate responses that are not yet described. Little is known about the ligands of V1Rs and V2Rs. At least one V1R recognizes a volatile pheromone (21) whereas one sex-specific exocrine peptide is linked to a specific V2R (22) and MUPs and MHC binding peptides that elicit innate responses both activate Gαo+ VNO neurons, suggesting that they may also be recognized by V2Rs (9, 23). The fact that only one of the five VNO FPRs is coexpressed with Gαo suggests that they are unlikely to detect MUPs or MHC peptides, which vary among conspecifics, but instead may recognize other types of peptides that elicit innate behavioral or physiological responses.
One intriguing possibility is that VNO FPRs specifically recognize formylated peptides. N-terminal formyl groups can be found on peptides derived from bacteria, mitochondria, and plant chloroplasts (30). Immune system FPRs expressed by neutrophils and monocytes are thought to play a role in the innate immune response by recognizing formylated peptides released from bacteria or mitochondria at sites of infection or tissue damage, thereby stimulating chemotaxis (24). The recognition of formylated peptides by VNO neurons could potentially contribute an additional dimension to chemosensory recognition in the VNO and be advantageous in a number of possible ways. For example, VNO FPRs might signal the edibility of specific plants, decay in a potential food source, or bacterial infection in a conspecific. Another interesting possibility is that VNO FPRs detect formylated peptides that are derived from mitochondria or normal bacterial flora in the animal and are released, for example, in feces. In this scheme, differences in mitochondrial peptides or normal flora among animals could provide individuality cues that permit discrimination among members of the same species or signal the presence of other types of animals, such as predators.
The VNO has two zones, one expressing V1Rs and Gαi2 and the other expressing V2Rs and Gαo. These two zones project axons to different parts of the accessory olfactory bulb, which in turn have largely overlapping, but partially distinct, projections to the amygdala and bed nucleus of the stria terminalis (31). Curiously, one VNO FPR is coexpressed with Gαo, but the other four are coexpressed with Gαi2, raising the possibility that sensory inputs from different VNO FPRs might ultimately be targeted to different brain areas with distinct functions.
Lineage-Specific Evolution of FPRs in the VNO.
Based on our phylogenetic analyses, FPRs may play a role in the VNO of rodents but not of other mammals. Rats also have an expanded FPR family similar to that of mice, but no evidence for a comparable expansion was found by analyzing intact Fpr genes in genome sequence data from horse, cat, cow, sheep, or pig. Humans, which lack a functional VNO, have three FPR genes, all of which function in the immune system (24). In contrast, our studies indicate that the expanded Fpr gene family of mice exhibits functional dichotomy wherein different members are expressed in different cell types that mediate entirely different functions. Fpr genes are found in a single genomic cluster adjacent to ≈39 V1r and V2r genes (24). It is possible that a gene duplication event placed an Fpr gene in close proximity to a vomeronasal receptor promoter, altering its expression pattern and function, and laying the foundation for neofunctionalization of a novel chemosensory receptor family.
Given the proposed role of the VNO in species-selective social ecology, one might predict that VNO chemosensory receptors would be especially likely to undergo rapid evolution and diversification in individual species. Indeed, V1R and V2R families, which evolved chemosensory functions earlier in vertebrate evolution (28), exhibit dramatic species-dependent heterogeneity (29). Considering the tremendous diversity of ecological niches and the animals that inhabit them, as well as the inherent developmental flexibility observed in some sensory systems, it may well be that additional new families of chemosensory receptor families have evolved and expanded in animals whose genome sequences are still unknown, perhaps endowing those species with sensory capabilities lacking in others.
Methods
VNO Sensory Neuron Isolation.
VNO sensory neurons were obtained using cell staining and sorting techniques previously described for isolating olfactory sensory neurons (20). VNO tissue was pooled from 16 adult C57BL/6J animals, yielding ≈140 VNO sensory neurons per animal.
qPCR.
DNase-treated RNA obtained from various mouse tissues or untreated RNA from FACS-sorted VNO sensory neurons were used to prepare cDNA for qPCR as described previously (20). Fpr primer pairs are listed in SI Text, and their specificity was verified with control PCR reactions involving Fpr-containing plasmids.
Fluorescence in Situ Hybridization.
Single color and two color fluorescence in situ hybridization experiments were performed as described previously (20) except that hybridizations were done at 65 °C to increase stringency; under these conditions probes did not cross-hybridize to sequences with 85% identity (Fpr-rs3 and Fpr-rs4) or less, cross-hybridized weakly to those with 87% identity (Fpr-rs1 and Fpr-rs2), and cross-hybridized strongly to those with 96% identity (Fpr-rs6 and Fpr-rs7). cRNA riboprobes were used that matched the full coding regions of all Fprs, G proteins, and V1rs, and a portion of V2ra (base pairs 1529–2345). Cells were counted blind to probe identity. Each image used in a figure was photographed to capture its entire dynamic range. Brightness, contrast, and gamma adjustments were made to permit adequate visualization of the images after printing.
Phylogenetic Analyses.
A more detailed description of methodology applied can be found in the SI. Protein sequences encoded by Fpr genes and outgroup sequences were aligned using MUSCLE (32), and a phylogenetic tree was built using MrBayes (33) with the mixed model for amino acid substitutions. In the resulting tree, speciation and duplication events were identified by comparison with accepted species relationships using soft parsimony (34). Positive selection was detected by the ratio of nonsynonymous to synonymous substitution rates using various models in PAML (35) and using SWAPSC (36).
Supplementary Material
Acknowledgments.
We thank Janet Young and Barbara Trask for unpublished data on V1r genes, Janet Young for critique of the manuscript, and members of the Buck laboratory for helpful discussions and comments. This work was supported by the Howard Hughes Medical Institute (L.B.B.) and by grants from the National Institutes of Health (National Institute on Deafness and Other Communication Disorders) (L.B.B.) and the National Science Foundation (D.A.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904464106/DCSupplemental.
References
- 1.Brennan PA, Keverne EB. Something in the air? New insights into mammalian pheromones. Curr Biol. 2004;14(2):R81–R89. doi: 10.1016/j.cub.2003.12.052. [DOI] [PubMed] [Google Scholar]
- 2.Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci. 2003;4(7):551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- 3.Halpern M, Martinez-Marcos A. Structure and function of the vomeronasal system: an update. Progress in neurobiology. 2003;70(3):245–318. doi: 10.1016/s0301-0082(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 4.Kandel ER, Schwartz JH, Jessell TM. Principles of neural science. New York: McGraw-Hill; 2000. pp. 625–647. [Google Scholar]
- 5.Boehm T, Zufall F. MHC peptides and the sensory evaluation of genotype. Trends in neurosciences. 2006;29(2):100–107. doi: 10.1016/j.tins.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Restrepo D, Lin W, Salcedo E, Yamazaki K, Beauchamp G. Odortypes and MHC peptides: Complementary chemosignals of MHC haplotype? Trends in neurosciences. 2006;29(11):604–609. doi: 10.1016/j.tins.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Kobayakawa K, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450(7169):503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- 8.Novotny MV. Pheromones, binding proteins and receptor responses in rodents. Biochemical Society transactions. 2003;31(Pt 1):117–122. doi: 10.1042/bst0310117. [DOI] [PubMed] [Google Scholar]
- 9.Chamero P, et al. Identification of protein pheromones that promote aggressive behaviour. Nature. 2007;450(7171):899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- 10.Hurst JL, et al. Individual recognition in mice mediated by major urinary proteins. Nature. 2001;414(6864):631–634. doi: 10.1038/414631a. [DOI] [PubMed] [Google Scholar]
- 11.Mandiyan VS, Coats JK, Shah NM. Deficits in sexual and aggressive behaviors in Cnga2 mutant mice. Nature neuroscience. 2005;8(12):1660–1662. doi: 10.1038/nn1589. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, et al. Pheromone detection in male mice depends on signaling through the type 3 adenylyl cyclase in the main olfactory epithelium. J Neurosci. 2006;26(28):7375–7379. doi: 10.1523/JNEUROSCI.1967-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83(2):195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 14.Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90(4):763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- 15.Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90(4):775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- 16.Ryba NJ, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19(2):371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- 17.Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome research. 2007;17(2):166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young JM, Kambere M, Trask BJ, Lane RP. Divergent V1R repertoires in five species: Amplification in rodents, decimation in primates, and a surprisingly small repertoire in dogs. Genome research. 2005;15(2):231–240. doi: 10.1101/gr.3339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65(1):175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 20.Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442(7103):645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- 21.Boschat C, et al. Pheromone detection mediated by a V1r vomeronasal receptor. Nature neuroscience. 2002;5(12):1261–1262. doi: 10.1038/nn978. [DOI] [PubMed] [Google Scholar]
- 22.Kimoto H, Haga S, Sato K, Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437(7060):898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- 23.Leinders-Zufall T, et al. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306(5698):1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- 24.Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 2006;17(6):501–519. doi: 10.1016/j.cytogfr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 26.Martini S, Silvotti L, Shirazi A, Ryba NJ, Tirindelli R. Co-expression of putative pheromone receptors in the sensory neurons of the vomeronasal organ. J Neurosci. 2001;21(3):843–848. doi: 10.1523/JNEUROSCI.21-03-00843.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez I, Del Punta K, Rothman A, Ishii T, Mombaerts P. Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nature neuroscience. 2002;5(2):134–140. doi: 10.1038/nn795. [DOI] [PubMed] [Google Scholar]
- 28.Grus WE, Zhang J. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Molecular biology and evolution. 2009;26(2):407–419. doi: 10.1093/molbev/msn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nature reviews. 2008;9(12):951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 30.Giglione C, Pierre M, Meinnel T. Peptide deformylase as a target for new generation, broad spectrum antimicrobial agents. Mol Microbiol. 2000;36(6):1197–1205. doi: 10.1046/j.1365-2958.2000.01908.x. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Marcos A. On the organization of olfactory and vomeronasal cortices. Progress in neurobiology. 2009;87(1):21–30. doi: 10.1016/j.pneurobio.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 34.Berglund-Sonnhammer AC, Steffansson P, Betts MJ, Liberles DA. Optimal gene trees from sequences and species trees using a soft interpretation of parsimony. Journal of molecular evolution. 2006;63(2):240–250. doi: 10.1007/s00239-005-0096-1. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 36.Fares MA, Elena SF, Ortiz J, Moya A, Barrio E. A sliding window-based method to detect selective constraints in protein-coding genes and its application to RNA viruses. J Mol Evol. 2002;55(5):509–521. doi: 10.1007/s00239-002-2346-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.