Abstract
The Cryptococcus species complex contains two sibling taxa, Cryptococcus neoformans and Cryptococcus gattii. Both species are basidiomycetous yeasts and major pathogens of humans and other mammals. Genotyping methods have identified major haploid molecular types of C. neoformans (VNI, VNII, VNB and VNIV) and of C. gattii (VGI, VGII, VGIII and VGIV). To investigate the phylogenetic relationships among these haploid genotypes, we selected 73 strains from 2000 globally collected isolates investigated in our previous typing studies, representing each of these genotypes and carried out multigene sequence analyses using four genetically unlinked nuclear loci, ACT1, IDE, PLB1 and URA5. The separate or combined sequence analyses of all four loci revealed seven clades with significant support for each molecular type. However, three strains of each species revealed some incongruence between the original molecular type and the sequence-based type obtained here. The topology of the individual gene trees was identical for each clade of C. neoformans but incongruent for the clades of C. gattii indicating recent recombination events within C. gattii. There was strong evidence of recombination in the global VGII population. Both parsimony and likelihood analyses supported three major clades of C. neoformans (VNI/VNB, VNII and VNIV) and four major clades of C. gattii (VGI, VGII, VGIII and VGIV). The sequence variation between VGI, VGIII and VGIV was similar to that between VNI/VNB and VNII. MATa was for the first time identified for VGIV. The VNIV and VGII clades are basal to the C. neoformans or the C. gattii clade, respectively. Divergence times among the seven haploid monophyletic lineages in the Cryptococcus species complex were estimated by applying the hypothesis of the molecular clock. The genetic variation found among all of these haploid monophyletic lineages indicates that they warrant varietal status.
Introduction
The Cryptococcus species complex includes two basidiomycetous encapsulated yeast species, Cryptococcus neoformans, an opportunistic pathogen, and Cryptococcus gattii, a primary pathogen. Both species are the most common fungal agents of infection of the central nervous system [1]. Two varieties of C. neoformans are recognized, C. neoformans var. grubii (serotype A), which is found worldwide, and C. neoformans var. neoformans (serotype D), which occurs mainly in Europe and South America [1], [2], [3]. C. gattii [4], was previously known as C. neoformans var. gattii (serotype B and C), and thought to be restricted to tropical and subtropical zones [2], [5] until a recent outbreak of cryptococcosis occurred on Vancouver Island, Canada, which has expanded the range of this yeast to temperate regions [6].
In recent years, an array of molecular studies, including PCR-fingerprinting [7], Amplified Fragment Length Polymorphisms (AFLP) analysis [8], and analysis of the orotidine monophosphate pyrophosphorylase (URA5) [9] and phospholipase (PLB1) [10] genes by Restriction Fragment Length Polymorphism (RFLP), have grouped all globally obtained strains into eight distinct molecular types: VNI ( = AFLP1) and VNII ( = AFLP1A and AFLP1B) (C. neoformans var. grubii, serotype A); VNIV ( = AFLP2) (C. neoformans var. neoformans, serotype D); VNIII ( = AFLP3) (hybrid, serotype AD); and VGI ( = AFLP4), VGII ( = AFLP6), VGIII ( = AFLP5) and VGIV ( = AFLP7), all corresponding to serotypes B and C (C. gattii), indicating that they have evolved independently in parallel. Similar sub-groups, representing the same level of genetic heterogeneity have been found in subsequent sequencing studies [11], [12], [13], [14], [15]. The degree of variation among these molecular types indicated that the varieties and species are genetically distinct and undergoing evolutionary divergence [11]. AFLP and MLST analyses recently identified strains closely related to the VNI group as being unique to Botswana, and suggested for those a new molecular type, VNB [15]. In addition, hybrid strains of C. gattii have been reported but no specific molecular type(s) were designated [8, Trilles et al., unpublished data]. More recently, naturally occurring hybrids between C. neoformans and C. gattii were reported as DaBα AFLP8 [16].
Here, we investigated the phylogenetic relationships of the species and varieties of the Cryptococcus species complex to test the hypothesis that each haploid molecular type is monophyletic. We excluded the molecular type VNIII, which contains hybrid individuals of serotype AD. The phylogenetic analysis is based on comparing patterns of sequence variation across four unlinked genes involved in housekeeping, production of secreted enzymes and virulence: actin (ACT1) [17], orotate-phosphoribosyl transferase (URA5) [3], phospholipase B (PLB1) [18] and the gene encoding a 110-kDa neutral metalloendopeptidase (IDE) involved in degradation of insulin in humans and mammals [19]. The first three genes were chosen because they are polymorphic among various groups of the Cryptococcus species complex [3], [9], [10], [17]. IDE is novel for this investigation. This gene was chosen because it is polymorphic between both Cryptococcus species [19]. In addition, this gene is conserved across a wide range of organisms [20], [21], [22], which makes it a candidate locus to be used for studying inter-species phylogenetic relationships [23]. The herein presented multigene sequencing data revealed seven major haploid lineages within the Cryptococcus species complex and provide further evidence to consider these major molecular types as individual varieties, if not species.
Results
Strains
From a collection of 2000 cryptococcal strains previously analyzed, we selected ten strains each of the haploid molecular types VNI, VNII, VNIV, VGI, VGIII and VGIV, and 13 strains of VGII, representing six continents [9], [15], [24], [25], [26, Meyer et al. unpublished data] (Table 1 and 2). Only South America contained strains of every molecular type. In addition, four VNB strains, which until now had only been reported from Africa [15], were used to represent this new molecular type.
Table 1. List of strains used in this study, including general strain information, serotype (ST), mating type (MAT), molecular type (MT) and the allele assignment for the four genes used in the multigene analysis.
| Isolates | WM No. | Country | Source | ST | MAT | MT | References | Allele Assignment | |||
| ACT1 | URA5 | PLB1 | IDE | ||||||||
| Cryptococcus neoformans var. grubii | |||||||||||
| ATCC 90112 | WM 419 | USA | CLIN | A | alpha | VNI | [74] | 1 | 1 | 1 | 1 |
| M27049 | WM 2573 | South Africa | CLIN | - | alpha | VNI | This study | 2 | 1 | 2 | 1 |
| WM 721 | WM 721 | India | ENV | A | alpha | VNI | This study | 3 | 1 | 2 | 1 |
| WM 148R | WM 148 | Australia | CLIN | A | alpha | VNI | [9] | 3 | 2 | 16 | 1 |
| RV 59369 | WM 1416 | Belgium | ENV | A | alpha | VNI | This study | 1 | 1 | 1 | 2 |
| NIH 193 | WM 1421 | USA | ENV | A | alpha | VNI | This study | 2 | 1 | 1 | 1 |
| LA 26 | WM 1641 | Mexico | ENV | A | alpha | VNI | [9] | 2 | 3 | 15 | 1 |
| LA 182 | WM 1897 | Spain | CLIN | A | alpha | VNI | [9] | 2 | 1 | 15 | 1 |
| LA 264 | WM 1742 | Chile | CLIN | A | alpha | VNI | [9] | 2 | 1 | 1 | 1 |
| LA 473 | WM 1948 | Colombia | CLIN | A | alpha | VNI | [9] | 1 | 1 | 1 | 1 |
| H99 | WM 846 | USA | CLIN | A | alpha | VNI | [75] | - | - | - | - |
| JG-02 | WM 2529 | USA | CLIN | - | alpha | VNII | This study | 5 | 5 | 5 | 5 |
| M27053 | WM 2577 | South Africa | CLIN | A | alpha | VNII | This study | 4 | 4 | 3 | 18 |
| PR-101 | WM 1352 | India | CLIN | - | alpha | VNII | This study | 6 | 5 | 5 | 19 |
| UON 11536 | WM 1462 | South Africa | CLIN | - | alpha | VNII | This study | 6 | 5 | 5 | 5 |
| WM626R | WM 626 | Australia | CLIN | A | alpha | VNII | [9] | 6 | 7 | 5 | 6 |
| Hamden C3-1 | WM 1408 | Brazil | ENV | A | alpha | VNII | [8] | 7 | 21 | 4 | 3 |
| RV 58146 | WM 1412 | Zaire | ENV | A | alpha | VNII | [12] | 5 | 6 | 5 | 4 |
| LA 146 | WM 553 | Brazil | ENV | A | alpha | VNII | [9] | 6 | 5 | 5 | 5 |
| LA 404 | WM 1816 | Mexico | CLIN | A | alpha | VNII | [9] | 6 | 5 | 5 | 5 |
| LA 511 | WM 1986 | Colombia | CLIN | A | alpha | VNII | [9] | 7 | 21 | 4 | 3 |
| bt1 | Botswana | CLIN | - | - | VNB | [15] | - | - | - | - | |
| bt22 | Botswana | CLIN | - | - | VNB | [15] | - | - | - | - | |
| bt31 | Botswana | CLIN | - | - | VNB | [15] | - | - | - | - | |
| bt131 | Botswana | CLIN | - | - | VNB | [15] | - | - | - | - | |
| Cryptococcus neoformans var. Neoformans | |||||||||||
| WM 629R | WM 629 | Australia | CLIN | D | alpha | VNIV | [9] | 13 | 10 | 6 | 8 |
| RKI-M186/99 | WM 04.174 | Germany | CLIN | D | alpha | VNIV | This study | 9 | 11 | 17 | 7 |
| RKI-M318/90 | WM 04.172 | Germany | CLIN | D | alpha | VNIV | This study | 9 | 11 | 17 | 7 |
| B-3501 | WM 2242 | USA | CLIN | D | alpha | VNIV | [76] | 12 | 23 | 8 | 20 |
| CBS 7816 | Thailand | ENV | D | a | VNIV | [12] | 12 | 22 | 8 | 7 | |
| LA268 | WM 04.168 | Chile | CLIN | D | alpha | VNIV | [9] | 12 | 9 | 7 | 9 |
| JEC 20 | WM 01.126 | USA | NA | D | a | VNIV | [77] | 12 | 22 | 8 | 7 |
| JEC 21 | WM 01.127 | USA | NA | D | alpha | VNIV | [77] | 11 | 22 | 8 | 7 |
| KRIMM 2 | WM 02.142 | Russia | CLIN | - | alpha | VNIV | This study | 10 | 8 | 6 | 21 |
| LA262 | WM 1740 | Chile | CLIN | D | alpha | VNIV | [9] | 8 | 10 | 6 | 8 |
| Cryptococcus gattii | |||||||||||
| LA1 | WM 1616 | Mexico | CLIN | B | alpha | VGI | [9] | 15 | 16 | 9 | 15 |
| 503 2738 | WM 1251 | Papua New Guinea | CLIN | B | alpha | VGI | [78] | 16 | 16 | 11 | 14 |
| WM 179R | WM 179 | Australia | CLIN | B | alpha | VGI | [9] | 15 | 17 | 9 | 14 |
| Joe | WM 1243 | Papua New Guinea | CLIN | B | alpha | VGI | [78] | 15 | 16 | 11 | 14 |
| MC-S-022 | WM 2634 | Thailand | CLIN | B | alpha | VGI | [79] | 17 | 31 | 23 | 16 |
| TP 0688 | WM 727 | USA | ENV | B | alpha | VGI | This study | 15 | 16 | 11 | 14 |
| TP 1414 | WM 2540 | New Zealand | VET | B | alpha | VGI | This study | 15 | 16 | 9 | 14 |
| LA175 | WM 1899 | Spain | CLIN | B | alpha | VGI | [9] | 14 | 19 | 24 | 15 |
| LA 564 | WM 2039 | Colombia | CLIN | B | alpha | VGI | [9] | 15 | 16 | 18 | 14 |
| F 2863 | WM 02.204 | Canada | VET | B | alpha | VGI | [6] | 14 | 18 | 25 | 24 |
| WM 1008 | WM 1008 | Australia | ENV | - | alpha | VGII | [78] | 20 | 12 | 13 | 12 |
| WM 178R | WM 178 | Australia | CLIN | B | alpha | VGII | [9] | 21 | 25 | 33 | 13 |
| MC-S-239 | WM 06.7 | Thailand | CLIN | B | alpha | VGII | [79] | 18 | 13 | 31 | 12 |
| RAM 002 | WM 03.27 | Australia | ENV | - | alpha | VGII | [78] | 19 | 12 | 13 | 12 |
| CBS 7750 | WM 06.13 | USA | ENV | B | alpha | VGII | [6] | 19 | 13 | 31 | 12 |
| LA 43 | WM 04.191 | Uruguay | ENV | B | alpha | VGII | [9] | 21 | 12 | 30 | 12 |
| LA 84 | WM 477 | Brazil | CLIN | - | alpha | VGII | [9] | 18 | 24 | 28 | 12 |
| CDC R369 | WM 02.46 | Canada | CLIN | B | alpha | VGII | [6] | 18 | 13 | 31 | 12 |
| NIH 444 | WM 02.81 | USA | CLIN | B | alpha | VGII | [80] | 18 | 13 | 31 | 12 |
| RB52 | WM 02.317 | Canada | ENV | B | alpha | VGII | [6] | 18 | 12 | 13 | 12 |
| AV 55 | WM 05.77 | Greece | CLIN | B | a | VGII | [81] | 21 | 14 | 14 | 12 |
| AV 54W | WM 05.75 | Greece | CLIN | B | alpha | VGII | [81] | 21 | 15 | 13 | 12 |
| AV 54S | WM 05.76 | Greece | CLIN | B | alpha | VGII | [81] | 21 | 15 | 13 | 12 |
| WM 175R | WM 175 | USA | ENV | B | alpha | VGIII | [9] | 22 | 26 | 26 | 11 |
| CN043 | WM 2423 | New Zealand | CLIN | - | a | VGIII | [78] | 23 | 29 | 19 | 22 |
| TP 0686 | WM 728 | USA | ENV | B | alpha | VGIII | This study | 22 | 26 | 26 | 11 |
| TP 0689 | WM 161 | USA | ENV | B | alpha | VGIII | This study | 22 | 28 | 26 | 11 |
| TP 0696 | WM 726 | USA | ENV | B | alpha | VGIII | This study | 22 | 26 | 26 | 11 |
| UCLA 380C | WM 1665 | USA | N | C | alpha | VGIII | [76] | 23 | 30 | 22 | 11 |
| LA 290 | WM 1699 | Paraguay | CLIN | - | alpha | VGIII | [9] | 23 | 30 | 10 | 11 |
| LA 382 | WM 1846 | Venezuela | CLIN | C | alpha | VGIII | [9] | 23 | 30 | 19 | 11 |
| LA 622 | WM 2176 | Colombia | CLIN | B | a | VGIII | [9] | 22 | 26 | 21 | 11 |
| LA 644 | WM 2158 | Colombia | ENV | C | alpha | VGIII | [9] | 23 | 30 | 19 | 11 |
| WM 779R | WM 779 | South Africa | VET | C | alpha | VGIV | [9] | 24 | 32 | 27 | 23 |
| B-5748 | WM 2364 | India | CLIN | B | alpha | VGIV | This study | 24 | 32 | 27 | 23 |
| B-5742 | WM 2363 | India | CLIN | B | alpha | VGIV | This study | 24 | 32 | 27 | 23 |
| M27055 | WM 04.20 | South Africa | CLIN | C | alpha | VGIV | This study | 24 | 32 | 12 | 23 |
| M27056 | WM 2579 | South Africa | CLIN | - | alpha | VGIV | This study | 24 | 32 | 12 | 23 |
| V00709 | WM 780 | South Africa | CLIN | C | alpha | VGIV | This study | 24 | 32 | 12 | 23 |
| V00869 | WM 2876 | South Africa | CLIN | C | alpha | VGIV | This study | 24 | 32 | 29 | 23 |
| LA 390 | WM 1802 | Mexico | CLIN | - | a | VGIV | [9] | 25 | 20 | 32 | 17 |
| LA 392 | WM 1804 | Mexico | CLIN | - | a | VGIV | [9] | 25 | 20 | 32 | 17 |
| LA 568 | WM 2041 | Colombia | CLIN | B | alpha | VGIV | [9] | 26 | 27 | 20 | 10 |
| Outgroups | |||||||||||
| Filobasidiella depauperata | |||||||||||
| CBS7841 | Canada | ENV | NA | NA | NA | [82] | - | - | - | - | |
| Cryptococcus albidus | |||||||||||
| CBS142 | WM 773 | Japan | ENV | NA | NA | NA | [83] | - | - | - | - |
Table 2. Geographic distribution and mating type data of all studied isolates.
| Molecular type | Continent | Total* | |||||
| Africa | Asia | Australia | Europe | North America | South America | ||
| VNI | 1 | 1 | 1 | 2 | 2 | 3 | 10 (0) |
| VNII | 3 | 1 | 1 | 0 | 1 | 4 | 10 (0) |
| VNIV | 0 | 1 (1) | 3 | 3 (1) | 2 | 10 (2) | |
| VGI | 0 | 1 | 4 | 1 | 2 | 2 | 10 (0) |
| VGII | 0 | 1 | 3 | 3 (1) | 4 | 2 | 13 (1) |
| VGIII | 0 | 0 | 1 (1) | 0 | 5 | 4 (1) | 10 (2) |
| VGIV | 5 | 2 | 0 | 0 | 0 | 3 (2) | 10 (2) |
| Total | 8 | 9 | 14 | 15 | 16 | 21 | 83 (7) |
Mating type analysis
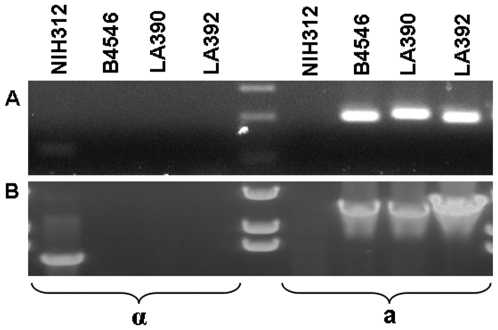
Primers specific for the MFα and MFa pheromone confirmed that MATα was predominant (66 out of 73 strains). Seven strains possessed the MAT a allele: 2 VNIV, 1 VGI, 2 VGIII and 2 VGIV strains (Table 1). Non-specific amplicons were produced by MAT a strains of VGIII and VGIV (data not shown). Therefore, bands of a length corresponding to the MAT a amplicon were sequenced, and BlastN searches revealed 98% sequence similarity with the mating pheromone a 2 (MFa2) gene of the C. gattii strain E566 (GenBank accession No. AY710429). The two VGIII strains CN043 and LA 622 as well as the two VGIV strains LA 390 and LA 392 were accordingly designated as mating type a (GenBank Accession No. EU408654 – EU408657) (Table 1 and 2). To our knowledge, this is the first report of the MAT a mating type in strains of the molecular type VGIV. To confirm this finding we used primers specific for the SXI1α and SXI2 a genes and obtained the same result as above (Figure 1).
Figure 1. Mating type specific PCR amplification of the two mating type a VGIV strains LA390 and LA392.
A) primers MFαU and MFαL: MFa2U and MFa2L; B) primers SXI1αF and SXI1αR: SXI2aF and SXI2aR.
Mating
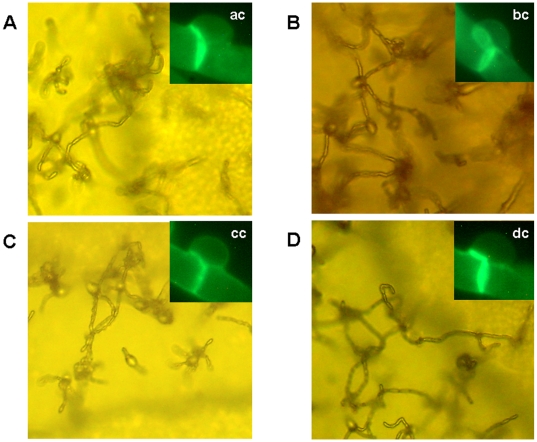
Mating of the two VGIV strains, LA 390 and LA 392, with the MATα reference strain of C. gattii, serotype C, NIH312 [27] and the crg1α mutant derivatives MATα supermater tester strain, JF101 [28] produced dikaryotic hyphae with fused clamp connections, basidia and bacilli-shaped basidiospores (Figure 2). However, the strains LA 390 and LA 392 failed to mate with the MAT a reference tester strains B4546 [29] and JF109 [28] (data not shown). Therefore, both strains, LA 390 and LA 392, were confirmed to possess the MAT a mating type allele. No haploid fruiting was observed when the samples were incubated alone on V8 juice agar.
Figure 2. Mating reaction on V8 media of the two mating type a VGIV strains.
A) LA390×NIH312; B) LA390×JF101; C) LA392×NIH312; D) LA392×JF101. All strains revealed typical bacilli-shape basidiospores and clamp connections (ac, bc, cc and dc).
Sequencing data
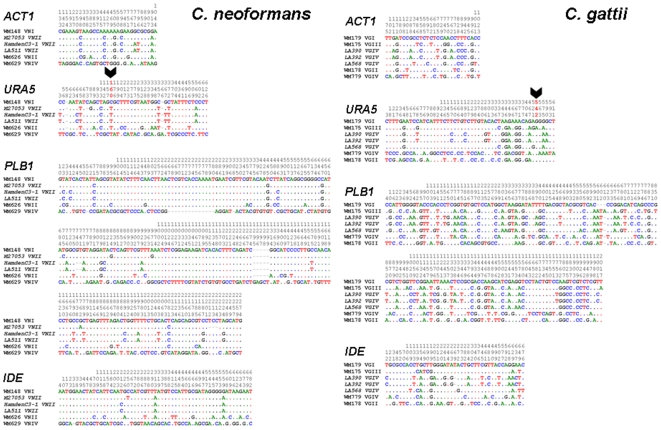
Four independent genetic loci, ACT1, URA5, PLB1 and IDE, were sequenced and all sequences were deposited in GenBank under the following accession numbers: ACT1 (EU408478–408550); URA5 (EU399554–EU399626); PLB1 (EU408624–EU408653, EU408658–408700) and IDE (EU408551–EU408623). The outgroup sequences have the following GenBank accession numbers: F. depauperata ACT1 (EU399627) and URA5 (EU399628); C. albidus ACT1 (EU399629).
Phylogenetic analyses of individual and combined loci
Maximum parsimony and Bayesian methods were used to analyze phylogenetic relationships among the 73 cryptococcal strains selected, using four independent genetic loci, ACT1, URA5, PLB1 and IDE.
The four loci of the intron-excluded and intron-included datasets contained a total of 564 and 834 parsimoniously informative characters, respectively. A hypervariable region containing poly-T was found in the intron of the PLB1 gene of some VGII strains. This region was excluded from the analysis due to sequencing problems. A heuristic search of the ACT1 gene sequences resulted in two maximum parsimony trees (Length 351, CI 0.855, RI 0.957); the URA5 gene produced 1152 maximum parsimony trees (Length 208, CI 0.897, RI 0.973); the PLB1 gene produced six maximum parsimony trees (Length 395, CI 0.922, RI 0.994); and the IDE gene produced 24 maximum parsimony trees (Length 113, CI 0.912, RI 0.993). Character information and substitution models of each locus are presented in Table 3. Bayesian analyses revealed topologies very similar or identical to those obtained using maximum parsimony. The topologies of each gene in both intron-excluded and intron-included datasets were identical or very similar (data not shown).
Table 3. Phylogenetic characters of the intron-excluded and intron-included data sets.
| Locus | Character (intron-excluded) | Character (intron-included) | substitution model | ||||
| total | constant | parsimony informative | total | constant | parsimony informative | ||
| ACT1 | 1124 | 848 | 177 | 1321 | 808 | 187 | GTR+G |
| URA5 | 621 | 457 | 71 | 724 | 509 | 102 | SYM+G |
| PLB1 | 1877 | 1549 | 309 | 2265 | 1819 | 423 | HKY+G |
| IDE | 581 | 483 | 85 | 684 | 549 | 122 | K80+G* |
| Combined | 4203 | 3337 | 564 | 4994 | 3685 | 834 | N/A |
Note: N/A = not applicable (partition of the dataset was used).
The second best model was chosen by the ModelTest program since the best (TrN+I and TrN+G) could not be operated in the MrBayes program due to the limitation of this software.
To focus on the relationships among species and subgroups, and to avoid detecting incongruence among recombining strains within the lineages, a subset of strains was used for the analysis of combined data from four loci. This subset included a single representative strain from each of the major lineages identified by the single locus analyses (see Material and Methods for the strain numbers). Prior to the analysis, the genes were tested using incongruence length difference/partition homogeneity test (ILD/PHT), which revealed no significant incongruence among the loci, when a conservative threshold of P<0.0001 was used. Because ILD test is prone to type I errors of incorrectly rejecting the null hypothesis of congruence among the datasets, a conservative threshold of P<0.0001 is recommended for interpreting the results of this test [30]. In our case, the P value of the ILD test of the combined dataset was 0.002, indicating that the null hypothesis of congruence cannot be rejected [30].
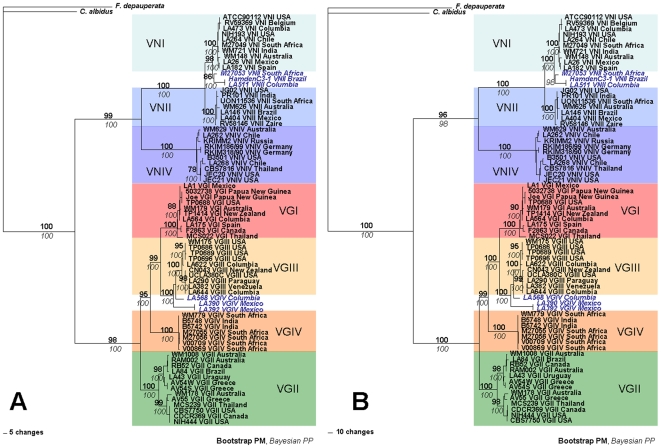
A heuristic search of the combined loci found 110 maximum parsimony trees (Length 1089, CI 0.864, RI 0.984) without introns and 1023 maximum parsimony trees (Length 1706, CI 0.866, RI 0.984) with introns. Bayesian analyses of the combined dataset revealed that topologies were very similar or identical to those obtained using maximum parsimony. Bayesian analyses of the data, including both exons and introns as well as excluding the introns generated phylograms with identical topologies and comparable statistical support. Both analyses strongly support the monophyly of C. neoformans and C. gattii (Figure 3). As expected, the VNI and VNII molecular types, which represent C. neoformans var. grubii have a sibling relationship with the VNIV molecular type, representing C. neoformans var. neoformans. In addition VNIV is basal to VNI and VNII (Figure 3). Clades representing molecular types VGI, VGIII and VGIV are more closely related to one another than to the sibling VGII clade, which is positioned basal to them (Figure 3).
Figure 3. Combined genealogies of (A) the intron-excluded and (B) the intron-included datasets with separate substitution models for each partition.
Parsimony bootstrap support above 75 is indicated in bold. Bayesian posterior probability above 95 is indicated italicized. The phylogenetic tree is rooted using Filobasidiella depauperata and Cryptococcus albidus as outgroups. The blue bold italic letters represent VNII-1 and VGIV-1 clades of C. neoformans and C. gattii, respectively.
Unexpectedly, three MATα strains with the URA5-RFLP pattern of molecular type VNII, one each from South Africa (M27053), Brazil (HamdenC3-1) and Colombia (LA 511), clustered with the VNI clade and not with the expected VNII clade. Similarly, two MAT a strains from Mexico (LA 390 and LA 392) and one MATα strain form Colombia (LA 568) with the URA5-RFLP pattern of the molecular type VGIV, clustered with the VGIII clade. For convenience, these groups will be referred to as the “VNII-1 group” and the “VGIV-1 group”, respectively (Figure 3).
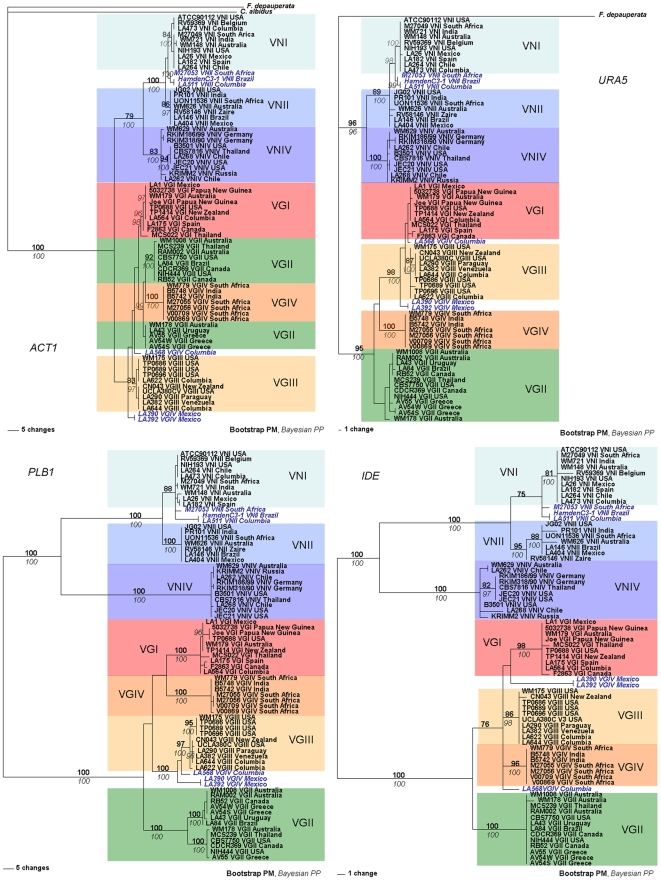
For the C. neoformans taxa, the topology of the individual loci never conflicted with the phylogram of the combined loci. The analysis of the two C. neoformans var. grubii clades (VNI and VNII) confirmed their sibling relationship with the C. neoformans var. neoformans clade (VNIV) (Figure 4). Conversely, the topologies of the individual loci of C. gattii conflicted with the phylogram of the combined loci. On the PLB1, URA5 and IDE phylogenies, the VGI, VGIII and VGIV clades are clustered together and formed a sibling group with VGII. However, the phylogram generated from ACT1 has a different topology in which, VGII strains are subdivided into two subclades. The deep branches of the ACT1 phylogram did not achieve strong statistical support in the C. gattii clade, unlike the combined data set. However, both analyses generated high statistical support for the major clades, with the exception of the VGI and VGII clades of the URA5 phylogram (Figure 4).
Figure 4. Gene genealogies of the four individual loci generated by Maximum Parsimony analysis.
Parsimony bootstrap support above 75 is indicated in bold. Bayesian posterior probability above 95 is indicated italicized. Phylogenetic trees are unrooted. The blue bold italic letters represent VNII-1 and VGIV-1 clades of C. neoformans and C. gattii, respectively.
The VNII-1 strains formed a monophyletic group, which was positioned between the VNI and VNII clades. Although Figure 3 and 4 suggest that the VNI-1 clade is more closely related to VNI, Figure 5 indicates that it has partial VNII sequence characteristics. The URA5 sequence analysis of the VNII-1 isolates revealed that these isolates are missing the recognition site for Sau96I at position 160, which is a specific, identifying marker of VNI isolates (Figure 5), and the overall sequence similarity of the combined four loci showed that those isolates were more similar to VNI (99.41%) than VNII (98.91%) (Table 4).
Figure 5. Variable sequence positions of all alignments of the VNII-1 and VGIV-1 strains revealed sequence similarity to VNI (ATCC90112 and WM148) and VGIII (WM175 and CN043) respectively.
However, some parts of the sequence were similar to that of VNII (WM626 and RV58146) for the VNII-1 strains and VGIV (WM779 and M27056) for the VGIV-1 strains. Black thick arrows reveal one of the cutting sites for URA5 RFLP giving VNII and VGIV patterns for the VNII-1 and VGIV-1 strains respectively.
Table 4. Sequence similarity matrix among the haploid molecular type clades of the Cryptococcus species complex.
| VNI | VNII | VNII-1 | VNII | VNIV | VGI | VGII | VGIII | VGIV | VGIV-1 | VGIV | |
| VNI | 0.16 | 0.11 | 0.13 | 0.35 | 0.49 | 0.45 | 0.43 | 0.44 | 0.43 | 0.43 | |
| VNII | 98.98 | 0.15 | 0.05 | 0.36 | 0.51 | 0.45 | 0.46 | 0.46 | 0.46 | 0.45 | |
| VNII-1 | 99.41 | 98.91 | 0.11 | 0.34 | 0.49 | 0.45 | 0.44 | 0.45 | 0.44 | 0.43 | |
| VNII | 99.11 | 99.63 | 99.16 | 0.35 | 0.5 | 0.45 | 0.45 | 0.45 | 0.45 | 0.44 | |
| VNIV | 94.74 | 94.71 | 94.68 | 94.7 | 0.45 | 0.42 | 0.44 | 0.43 | 0.45 | 0.42 | |
| VGI | 90.92 | 90.81 | 90.72 | 90.78 | 91.16 | 0.24 | 0.19 | 0.23 | 0.19 | 0.19 | |
| VGII | 91.37 | 91.22 | 91.23 | 91.23 | 91.68 | 97.28 | 0.24 | 0.19 | 0.24 | 0.18 | |
| VGIII | 91.18 | 91.15 | 91.00 | 91.11 | 91.40 | 98.07 | 97.40 | 0.24 | 0.12 | 0.18 | |
| VGIV | 91.02 | 90.82 | 90.82 | 90.82 | 91.22 | 97.21 | 97.20 | 97.32 | 0.22 | 0.07 | |
| VGIV-1 | 90.91 | 90.84 | 90.73 | 90.81 | 91.13 | 97.75 | 97.16 | 98.87 | 97.06 | 0.17 | |
| VGIV | 90.99 | 90.82 | 90.80 | 90.82 | 91.2 | 97.37 | 97.19 | 97.79 | 99.11 | 97.76 |
VNII = VNII+VNII-1; VGIV = VGIV+VGIV-1; numbers in italics designated values of standard errors.
Similarly, strains from the VGIV-1 group did not consistently form a monophyletic relationship with any of the C. gattii molecular type clades and were not supported by either analysis, except for the PLB1 gene, whose topology was similar to that of the combined loci (Figure 3 and 4). However, the sequence analysis of the four genes revealed VGIII strains that contained integrated parts of VGIV sequences, which resemble the intermediate position of the VNII-1 strains of C. neoformans (Figure 5). The URA5 sequence analysis of the VGIV-1 isolates revealed that those isolates share the recognition site for Sau96I with VGIV at position 542, which lead to their identification as VGIV (Figure 5), but the sequence similarity varied specifically for each of the four loci we investigated. The overall similarity to VGIII was 98.87% and to VGIV, 97.06%, resulting in an exchange of the position of those isolates in the individual gene trees (Table 4).
Recombination and clonality
To determine the extent of clonality and recombination in populations of different molecular types, we used three different tests of linkage disequilibrium, (i) the incongruence length difference/partition homogeneity test (ILD/PHT), (ii) two measures of index of association (IA and rBarD), and (iii) the phylogenetic incompatibility test. Since clonal reproduction can mask the effect of recombination, we prepared two different databases: one included all strains of each molecular type and the other included the clone corrected data of each molecular type from which identical genotypes were removed.
The ILD/PHT test showed similar results for each dataset, including and excluding individual genotypes (clone corrected). With the exception of VGII, the null hypothesis of clonality was not rejected. VGII showed incongruence in the phylogenies that allowed the rejection of clonality (P<0.0001).
In the second test we used two indexes of association measures, IA and rBarD. These tests are expected to be zero if populations are freely recombining and greater than zero if there is association between alleles (clonality). In the IA and rBarD tests including all isolates, the null hypothesis of recombination was rejected in all molecular types with the exception of VNI, which had the lowest value (Table 5). However, when the clonally corrected dataset was used, the IA and rBarD showed clear evidence of recombination in the molecular types VNI, VGII and VGIII (Table 5) and confirmed the predominance of clonal reproduction among the other molecular types (VNII, VNIV, VGI and VGIV).
Table 5. Multilocus linkage disequilibrium analyses in each of the haploid molecular types of the Cryptococcus species complex.
| Population | No. of isolates | No. of haplotypes | All isolates | Haplotypes only | ||||
| IA | rBarD | PhI | IA | rBarD | PhI | |||
| VNI | 10 | 8 | 0.1595 | 0.0535 | 1 | 0.0551 | 0.0185 | 1 |
| VNII | 10 | 7 | 1.5655*** | 0.5307*** | 1 | 0.8809** | 0.3257** | 1 |
| VNIV | 10 | 8 | 1.0722*** | 0.3643*** | 1 | 0.6535* | 0.018* | 1 |
| VGI | 10 | 9 | 1.0818*** | 0.3692*** | 1 | 0.9189** | 0.3151** | 1 |
| VGII | 13 | 10 | 0.4573** | 0.1538** | 0.6666 | 0.1194 | 0.0399 | 0.6666 |
| VGIII | 10 | 7 | 0.9015*** | 0.3038*** | 1** | 0.4605 | 0.16059 | 1 |
| VGIV | 10 | 5 | 2.2432*** | 0.7641*** | 1** | 2** | 1** | 1 |
Note: PhI, Phylogenetic incompatibility.
P<0.05.
P<0.01.
P<0.001
The third analysis, the phylogenetic incompatibility test only rejected the null hypothesis of random mating for the inclusive datasets of VGIII and VGIV (Table 5).
Genetic variation of each locus
Overall, the ACT1 gene was the most conserved locus (Table 6). Analysis of the sequence variation of each molecular type revealed that sequences of the VNIV and VGII clades were the most variable in C. neoformans and C. gattii, respectively (Table 6). In contrast to the overall comparable genetic variation of the Cryptococcus species complex, the IDE locus was exceptionally conserved among individual molecular types (Table 6). The overall genetic diversities of C. neoformans and C. gattii were similar (P<0.376), but the clades of C. gattii were more diverse than those of C. neoformans (Table 7). The genetic diversity between clades was higher for C. gattii (VGI-VGIV) than C. neoformans var. grubii (VNI and VNII) (P<0.001) and C. neoformans var. neoformans (VNIV) (P<0.038) (Table 7).
Table 6. Percent similarity representing the genetic variation among the haploid molecular types of the Cryptococcus species complex.
| Locus | ACT1 | URA5 | PLB1 | IDE | |||||
| Total Characters | 1062 | 525 | 1877 | 581 | |||||
| Constant Characters | %Total | %PUI | %Total | %PUI | %Total | %PUI | %Total | %PUI | |
| C. neoformans | VNI | 99.72 | 99.72 | 99.81 | 99.81 | 99.68 | 99.79 | 99.66 | 100 |
| VNII | 100 | 100 | 99.43 | 100 | 100 | 100 | 98.97 | 100 | |
| VNII-1 | 100 | N/A | 100 | N/A | 99.99 | N/A | 100 | N/A | |
| VNIV | 99.25 | 99.53 | 99.24 | 99.24 | 99.84 | 99.89 | 98.80 | 99.48 | |
| C. gattii | VGI | 99.53 | 99.91 | 99.05 | 99.43 | 99.36 | 99.68 | 100 | 100 |
| VGII | 99.34 | 99.44 | 98.29 | 99.43 | 98.88 | 99.04 | 99.83 | 100 | |
| VGIII | 99.81 | 99.81 | 99.24 | 99.62 | 99.63 | 99.79 | 99.83 | 100 | |
| VGIV | 100 | 100 | 100 | 100 | 99.89 | 99.95 | 100 | 100 | |
| VGIV-1 | 99.99 | N/A | 99.99 | N/A | 99.99 | N/A | 99.98 | N/A | |
| Mean* | 99.66 | 99.77 | 99.29 | 99.65 | 99.61 | 99.73 | 99.58 | 99.93 | |
| C.s. complex** | 92.47 | 93.03 | 84.57 | 86.67 | 82.53 | 83.54 | 83.13 | 85.37 | |
Note: The VNII-1 and VGIV-1 PI were excluded due to limitation of the PAUP program. (*Mean = mean of variable percentage of all original molecular types. ** C.s. complex = variable percentage calculated from the whole datasets. PUI = parsimony un-informative. N/A = not applicable).
Table 7. Number of polymorphic sites of the intron-excluded combined dataset among different clades of the Cryptococcus species complex.
| Species and clades | Molecular type* | No. of polymorphic characters | No. of parsimony informative characters |
| C. neoformans | 255 | 233 | |
| C. n. var. grubii clade | 73 | 57 | |
| VNI | 12 | 7 | |
| VNII | 9 | 0 | |
| C. n. var. neoformans clade | VNIV | 22 | 14 |
| C. gattii | 236 | 201 | |
| VGI+VGIII+VGIV clade | 181 | 153 | |
| VGI | 24 | 10 | |
| VGIII | 14 | 8 | |
| VGIV | 2 | 1 | |
| VGII clade | VGII | 38 | 27 |
Notes: *VNII-1 and VGIV-1 were excluded. Genetic diversity (polymorphic characters) of C. gattii and C. neoformans is comparable (P<0.376). More genetic variation in the VGI+VGIII+VGIV and VGII clades, compared to the C.n. var. grubii (P<0.001) and C.n. var. neoformans clades (P<0.038), was observed.
Evolutionary Divergence
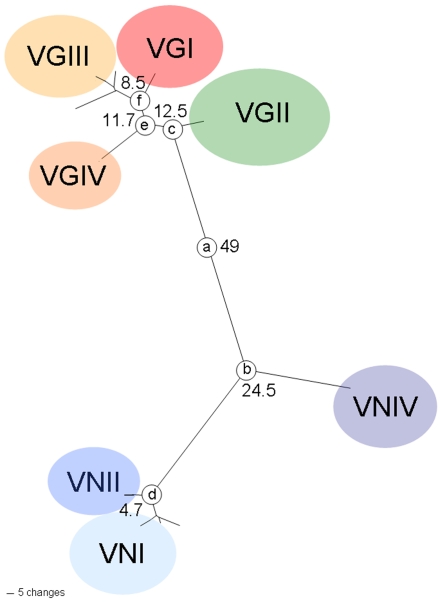
Maximum-likelihood estimations with and without a molecular clock demonstrated that every gene did not differ significantly from molecular clock expectations (P = 0.99). Comparing the two species, the C. gattii lineages (12.5 million years ago) evolved later than the major C. neoformans lineages, C. neoformans var. grubii and C. neoformans var. neoformans (24.5 million years ago), suggesting more recent recombination events (Table 8 and Figure 6). However, the most recent speciation event took place around 4.7 million years ago splitting the two monophyletic lineages within C. neoformans var. grubii, VNI and VNII (Table 8 and Figure 6).
Table 8. Genetic distance and estimates of the times since divergence among gene lineages of the Cryptococcus species complex.
| Between taxon/major molecular type group | Node | Genes | Time since divergence in million years (95% confidence interval) | ||||
| ACT1 | URA5 | PLB1 | IDE | Averages** (95% confidence interval) | |||
| C.n. - C.g. | a | 0.0399 | 0.1042 | 0.1315 | 0.0910 | 0.0980 (0.0890–0.1071) | 49.0 (44.5–53.5) |
| C.n. var. grubii – C.n. var. neoformans | b | 0.0132 | 0.0426 | 0.0705 | 0.0506 | 0.0490 (0.0401–0.0579) | 24.5 (20.0–28.9) |
| VNI – VNII* | d | 0.0058 | 0.0088 | 0.0089 | 0.0185 | 0.0094 (0.0076–0.0113) | 4.7 (3.8–5.7) |
| VGI – VGIII* | f | 0.0083 | 0.0071 | 0.0262 | 0.0129 | 0.0171 (0.0133–0.0209) | 8.5 (6.7–10.4) |
| VGI,VGIII – VGIV* | e | 0.0145 | 0.0447 | 0.0262 | 0.0117 | 0.0235 (0.0196–0.0273) | 11.7 (9.8–13.6) |
| VGI,VGIII,VGIV – VGII* | c | 0.0099 | 0.0296 | 0.0326 | 0.0242 | 0.0221 (0.0221–0.0280) | 12.5 (11.1–14.0) |
Notes: * VNII-1 and VNIV-1 was not included in this calculation due to ambiguous placements in the molecular type lineages.
weighted averages of the genetic distance of all genes combined.
Figure 6. Genealogy of the intron-excluded combined dataset revealed the time since divergence among lineages of the Cryptococcus species complex.
The number stated beside each node represents the ages of divergence in million years. Unlabelled branches are corresponding to VGIV-1 or VNII-1 groups, respectively.
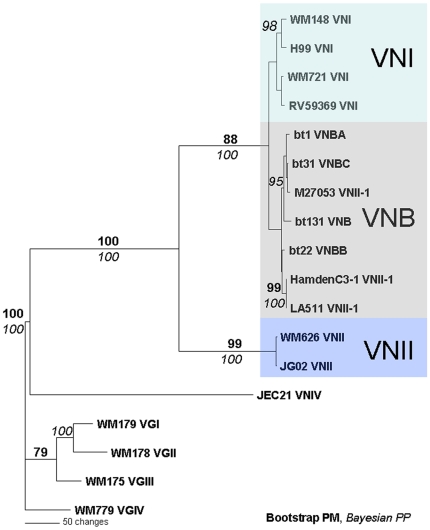
VNII-1 group is identical to the previously identified VNB molecular type according to parsimony and Bayesian analysis
To determine whether VNII-1 isolates cluster with molecular type VNB, we performed a phylogenetic analysis with representative isolates of each molecular type including sequences of VNB isolates previously identified (bt1, bt31, bt131, bt22) [15]. The ILD test showed congruence for all pairs of the genes except IGS1 when pairing the data with URA5 and GPD1 (P<0.0001). However, since the IGS1 was used to describe the VNB molecular type [15], we included IGS1 in the combined dataset. A heuristic search of the ACT1 gene sequences found five maximum parsimony trees (Length 796, CI 0.884, RI 0.928). The tree topologies from the Bayesian analysis were similar to that found in the parsimony analysis. Despite the lack of significant support (<75 for parsimony bootstrap and <95 for Bayesian posterior probability), the VNII-1 strains were related to the VNB strains (Figure 7). Strain M27053 was closely related to strain bt31 with 100% support from the Bayesian posterior probability and 72% support from maximum parsimony bootstrap. The clade containing strain M27053 was significantly supported with a Bayesian posterior probability of 95% (Figure 7). The sequences were deposited in GenBank under the following accession numbers: URA5 (EU929059–62), GPD1 (FM180481–92), IGS1 (FM180493–504).
Figure 7. Genealogy of the combined dataset (PLB1, URA5, GPD1, IGS1) showing that the VNII-1 group isolates cluster with the VNB isolates previously described [15].
Discussion
The multigene phylogeny based on the ACT1, URA5, PLB1 and IDE sequences provides an additional support for the currently accepted two species concept for the pathogenic Cryptococcus species complex (C. neoformans and C. gattii). The seven clades in the phylogenetic trees reflect the haploid molecular types recognized previously by M13 fingerprint, AFLP and RFLP analyses [8], [9]. For both species the obtained phylogenetic trees correspond to these molecular types and are highly supported by both maximum parsimony and Bayesian analyses. Clinical and environmental isolates clustered together in the respective major molecular types as previously determined in an analysis of six genetic loci (ITS1/2, IGS1, CNLAC1, RPB1, RPB2 and TEF1) [14]. Here we found that the sequence diversity was comparable among strains of C. neoformans and C. gattii strains. Within the sample of C. neoformans, strains of C. neoformans var. grubii (VNI, VNII-1 = VNB and VNII) were more than twice as variable as strains of C. neoformans var. neoformans, confirming data reported in previous studies using different genetic loci and different sets of strains [8], [13]. MLST analysis of C. neoformans strains from sub-Saharan Africa had also revealed extensive genetic diversity among C. neoformans var. grubii strains [31].
Based on the four loci analyzed, C. neoformans and C. gattii share an equivalent level of molecular variation. Extensive surveys of the Cryptococcus species complex in South America have revealed extensive genetic diversity among strains of C. gattii, as well as the coexistence of both mating types in nature and evidence of recombination [9], [26], [32]. These points, taken together, suggest that the evolutionary origin of C. gattii may be in South America.
Occasionally, molecular typing based on M13 fingerprinting, AFLP and RFLP analysis is misleading. We obtained conflicting results when comparing strains that were typed using PCR-based (PCR fingerprinting, RFLP and AFLP) analyses and sequence-based methods. For example, strains of “VNII-1” were genotyped as VNII according to the URA5 RFLP analysis but following multigene sequence analysis, they clustered with the VNI clade. Similarly, “VGIV-1” strains were identified as VGIV by URA5 RFLP analysis but clustered with the VGIII clade on multigene sequence analysis. Moreover, URA5-RFLP analysis of the VNII-1 and VGIV-1 clades gave different molecular types from those obtained by PLB1-RFLP analysis (data not shown). These discrepancies may be attributable to insufficient markers, genetic drift or recombination events (Figure 5).
Similar to results presented by others [11], [12], our data indicate a lack of geographic concordance with the phylogeny, which suggest recent global dispersal of the Cryptococcus species complex. In addition, the apparent bias of MATα over MAT a, as reported in previous surveys [33], [34], would limit the ability of the yeast to undergo sexual reproduction and recombination, which would retard speciation. Thus, clonal populations have been documented in several cryptococcal habitats, including Australia [35], [36], Thailand [37] and Canada [6] where the MAT a is scarce.
To investigate the question of recombination within the Cryptococcus species complex, we analyzed the congruence or incongruence of individual gene genealogies (ILD/PHT test) [38], [39] and the linkage disequilibrium (IA, rBarD and phylogenetic incompatibility) [38], [40] to infer the extent of clonality. Though clonal, asexual reproduction is the main reproductive mode in the Cryptococcus species complex, the existence of recombination and putative sexual reproduction has been demonstrated in several natural populations [31], [35], [41], [42]. In this investigation, the VGII clade was the only molecular type displaying unequivocal evidence of recombination in all the tests. Recent studies have provided evidence of sexual reproduction among strains of VGII on a global scale [43] and in local populations of Australia [35], [44]. Frequent recombination in the global population of VGII could explain the capacity of this genotype to expand its ecological range, as has been seen in the case of the Vancouver Island outbreak [6]. In contrast, the global populations of VGI, VGIV and VNII seem to be predominantly clonal. Previous analysis of AFLP and MLST data also found limited evidence for recombination for these molecular types [15], [35]. However, recent studies of selected populations found evidence for genetic re-assortment among environmental strains of VGI [45] and from veterinary strains of VNI [46]. For VNI and VGIII, we observed evidence of linkage equilibrium, but the ILD/PHT analysis data showed the presence of clonality. Among global isolates, our results reflect clonality for most molecular types within the Cryptococcus species complex except VGII, but they do not reject the possibility of sexual reproduction within any molecular type if sampled within a circumscribed population [e.g. 47]. The ILD/PHT P value for each gene combined individually (P values ranged from 0.010 for ACT∶PLB1 to 0.835 for URA5∶IDE) and all genes combined (P = 0.002) showed no significant incongruence using a cut off point of 0.0001 [30], indicating a low degree or absence of recombination between the major molecular types, when only strains representative of the main lineages were used.
The multigene parsimony and Bayesian trees of the ACT1 gene of C. gattii showed some incongruences in the topologies between the molecular types, indicating that more recent recombinational events may have occurred. This observation is consistent with the estimation that the clades of C. gattii diverged prior to those of C. neoformans, as indicated in Figure 6. This divergence time is approximately in agreement with previous studies, which used a different set of genetic loci [11], but closer to the recent estimations based on the whole genome, which estimated a split between the two species at 80 million years [48]. The time since divergence between each molecular type clade of C. gattii (11.7 million years between VGIV and VGI/VGIII and 8.5 million years between VGI and VGIII) is much more than between VNI and VNII (4.7 million years ago) (Figure 6). In addition, the divergence between VGII and VGI/VGIII/VGIV estimated to be 12.5 million years is in a similar range as the previously obtained divergence between the two varieties of C. neoformans, C. neoformans var. grubii and C. neoformans var. neoformans [11]. Hence, changing each clade to at least varietal status should be considered.
The VNB clade is recognized as a unique and highly diverse subpopulation of haploid isolates found in southern Africa [15]. Since the current study identified a special clade for the VNII-1 strains in the phylogenetic trees, which was in a similar position as the previously described VNB strains [15], [49], we conducted a preliminary phylogenetic analysis of the VNII-1 strains using four genetic loci, PLB1, URA5, GPD1 and IGS1, and included a representative strain from each VNB sub-group: bt1 (VNB-A), bt22 (VNB-B), bt131 (VNB) and bt31 (VNB-C) [15]. The sequence analysis showed that the VNII-1 strains are closely related to the VNB clade (Figure 7). One of the strains, bt31, was almost identical to the strain M27053, which originated from South Africa. The other two isolates, Hamden C3-1 and LA511, which originated from South America (Brazil and Colombia, respectively), had related sequences, but without significant support from both analyses. However, these results suggest that VNB strains may not be unique to southern Africa. Although the clade containing strains Hamden C3-1 and LA511 did not receive significant support here, a previous study showed that strain Hamden C3-1 clustered with a VNB strain [14]. Considering their sequence similarities (Figure 5), VNB and VNII-1 strains may represent a link between the VNI and VNII clades. Whether VNB merits status as a distinct molecular type remains an open question, which needs to be addressed specifically in further studies with more VNB strains to be collected from Africa, South America and elsewhere. Bovers et al., 2008 have established a link, using MLST and AFLP analysis, between VNB isolates and their AFLP group 1A, where their AFLP group 1B clusters with isolates of the molecular type VNII [14]. URA5-RFLP analysis is not able to make this differentiation, where both AFLP groups, AFLP1A and AFLP1B, are associated with VNII, which includes the VNII-1/VNB isolates. Perhaps more importantly, DNA sequence markers are more reliable and discriminatory than AFLP genotypes. However, this report has also demonstrated that additional markers as well as additional strains are needed to resolve the number of legitimate clades of both C. neoformans and C. gattii.
A group of strains (VGIV-1) with an unusual clustering was also found in the C. gattii clade. The VGIV-1 strains clustered between VGIII and VGIV. To date, all the strains of VGIV-1 have been isolated in South America. If this geographic origin is substantiated, they perhaps represent a recombining population. Similar to VNB strains in southern Africa [31], there is a large population of fertile MAT a strains in South America [26], [50].
Two of those unusual clustering C. gattii strains (LA 390 and LA 392, both from Mexico) are the first VGIV strains identified as being MAT a . The mating type of the two strains has been identified by two independent mating type specific PCR, which amplified only the MF a pheromone gene and the SXI2 a gene from both strains. The mating type was confirmed by mating experiments with the C. gattii, serotype C, MATα reference tester strain NIH312 [27] as well as with the MATα supermater strain JF101 [28] (Figure 2).
This study provided further information on the genetic diversity among members of the Cryptococcus species complex showing that it consists of at least seven monophyletic lineages, excluding the AD hybrid (VNIII) strains. The present study highly supports the two species concept recognized currently. The genetic variation found between the major molecular types of C. neoformans var. grubii is in the same range as the genetic variation found between C. gattii types. There are three monophyletic phylogenetic clades within the species C. neoformans two closely related lineages (VNI and VNII) observed in C. neoformans var. grubii and a basal lineage for C. neoformans var. neoformans (VNIV). The four molecular types of C. gattii (VGI, VGII, VGIII and VGIV) are highly supported by both maximum parsimony and Bayesian analyses. A similar topology was obtained previously investigating the intergenic spacer regions [12] and PRP8 inteins [13], with high support (over 80%) from parsimony and Neighbor Joining analyses, respectively. The molecular type VGII forms the basal clade within the C. gattii branch of the phylogenetic tree and is phylogenetically more distant than the other three sibling clades, VGI, VGIII and VGIV. This result correlates with the finding of a specific mating characteristic for the VGII Vancouver Island outbreak isolates, which produced a basidium on stunted filaments close to the surface of the colonies when mated with other VGII strains [28]. The same morphology was also obtained by matings between other pairs of VGII isolates from different parts of the world [51]. This study revealed a level of genetic variability among the different molecular types/monophyletic lineages, which are comparable to, or in fact slightly greater than that found for C. neoformans var. grubii and C. neoformans var. neoformans.
The molecular clock analysis of the genetic diversity among the seven haploid major molecular lineages support the phylogenetic species concept, in which strains that form consistently the same monophyletic groups should be considered to be independent species [52], [53]. Especially the molecular types within C. gattii should be considered as varieties. However, the occasional hybrid strains that form between either of the two currently recognized species ([16], Trilles and Meyer pers. communication) or between the different monophyletic groups [54], [55] question the biological species concept [56], which defines a species as consisting of members that are able to mate and undergo sexual recombination. Therefore the question: whether there are more than two species within the Cryptococcus species complex or whether the molecular types/monophyletic lineages within each species deserve varietal status, remains open. A combination between the six recently published loci [14] and the four reported here would be desirable together with further morphological and mating studies to draw final conclusions.
Materials and Methods
Studied strains
Seventy three strains composed of at least ten representative strains of each haploid molecular type of the Cryptococcus species complex from different parts of the world, and the outgroup species Cryptococcus albidus were retrieved from the Molecular Mycology Research Laboratory culture collection at Westmead Hospital, University of Sydney, Westmead, NSW, Australia (Table 1). The four VNB strains were retrieved from the Culture Collection of the Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, NC, USA. The hybrid molecular type, VNIII (serotype AD), was excluded from the study due to ambiguity of phylogenetic resolution in terms of variation in numbers of copies of each locus, and deserves to be studied separately [42], [49]. The strains were grown on Saboraud Dextose Agar (2% glucose, 2% peptone and 2% agar) for 72 hr before extracting DNA.
DNA extraction
DNA extractions were performed by the liquid nitrogen grinding method as previously described [9]. Genomic DNA of the other outgroup Filobasidiella depauperata was kindly provided by Ferry Hagen (CBS – Fungal Biodiversity Centre, Utrecht, the Netherlands).
Serotyping
The serotypes of the studied isolates were determined by the slide agglutination test using the Crypto Check Kit according to the manufactures instructions (Iatron Labs. Tokyo, Japan).
Identification of the molecular type
URA5-RFLP analysis using the enzymes HhaI and Sau96I was performed initially to verify the molecular type of each studied strain as previously described [9].
Determination of the mating type by PCR
Two pairs of mating-type specific primers were used. MFαU and MFαL primers identified the α-mating type (MATα) [57]. MFa2U and MFa2L recognized the a-mating type (MAT a) [28] (Table 9). The MAT a strains of VGIII and VGIV were confirmed by sequencing the mating type specific amplicons and comparing them with the appropriate homologous sequences in GenBank uisng BlastN searches. MAT a of VGIV were additionally verified using a second set of mating type specific primers, SXI1αF and SXI1αR identifying MATα and SXI2aF and SXI2aR recognizing MAT a [44] (Table 9). The used PCR conditions have been published previously [28], [44], [57].
Table 9. List of primers used in this study.
| Primers names | Primers sequences | Note | Source |
| CNACT1 | 5′ AATCTCGCCCAACATGT 3′ | Amplify ACT1 | This study |
| CNACT1R | 5′ TTAGAAACACTTTCGGTGGACG3′ | Amplify ACT1 | This study |
| CNACT1F2 | 5′ CCAAGCAGAACCGAGAGAAG 3′ | Internal primers of ACT1 | This study |
| URA5 | 5′ ATGTCCTCCCAAGCCCTCGACTCCG 3′ | Amplify URA5 | [8] |
| SJ101 | 5′ TTAAGACCTCTGAACACCGTACTC 3′ | Amplify URA5 | [8] |
| PLBCNAF | 5′ TAAAGTGCTTGGTGGGAACC 3′ | Amplify PLB1 from VN | This study |
| PLBCNAR | 5′ TCTCGCGAGGATTACAGGAT 3′ | Amplify PLB1 from VN | This study |
| PLBCG2F | 5′ TCCCCTTCAACACAGCTCTT 3′ | Amplify PLB1 from VG | This study |
| PLBCG2R2 | 5′ CACCTATCTTCGCTGCATCA 3′ | Amplify PLB1 from VG | This study |
| PLBCNIF1 | 5′ GGTTACCGTGCAATGCTGT 3′ | Internal primers of PLB1 | This study |
| PLBCNIF2 | 5′ GGTGCTTTCACCCCTATTGA 3′ | Internal primers of PLB1 | This study |
| PLBCNIR1 | 5′ CGGGAAATATCAGCTTGGTC 3′ | Internal primers of PLB1 | This study |
| IDEF | 5′ CCAAGGCGGACAAGGCTGCGG 3′ | Amplify IDE | [19] |
| IDER | 5′ GTAGAGGTGATCCATGTCGGG 3′ | Amplify IDE | [19] |
| ACT1CAF1 | 5′ GGTGTCATGGTCGGTATGG 3′ | Amplify ACT1 from CA | This study |
| ACT1CAR1 | 5′ GTACTTTCGCTCGGGAGGAG 3′ | Amplify ACT1 from CA | This study |
| ACT1CAR2 | 5′ AGCTTCTCCTTGATGTCTC 3′ | Amplify ACT1 from CA | This study |
| URA5DF1 | 5′ CCWTACTTCTTCAAYGCYGG 3′ | Amplify URA5 from FD | This study |
| MFLL | 5′ CTTCACTGCCATCTTCACCA 3′ | Mating type α determination of VN | [69] |
| MFLR | 5′ GACACAAAGGGTCATGCCA 3′ | Mating type α determination of VN | [69] |
| MFAL | 5′ CGCCTTCACTGCTACCTTCT 3′ | Mating type a determination of VN | [69] |
| MFAR | 5′ AACGCAAGAGTAAGTCGGGC 3′ | Mating type a determination of VN | [69] |
| MFαU | 5′ TTCACTGCCATCTTCACCACC 3′ | Mating type α determination of VG | [47] |
| MFαL | 5′ TCTAGGCGATGACACAAAGGG 3′ | Mating type α determination of VG | [47] |
| MFa2U | 5′ ACACCGCCTGTTACAATGGAC 3′ | Mating type a determination of VG | [38] |
| MFa2L | 5′ CAGCGTTTGAAGATGGACTTT 3′ | Mating type a determination of VG | [38] |
| SXI1αF | 5′ TACATCACCGGTCATATCTGC 3′ | Mating type α determination of VGIV | [68] |
| SXI1αR | 5′ CTGGAGAAGCGCCTCACTGGA 3′ | Mating type α determination of VGIV | [68] |
| SXI2aF | 5′ TGATCGCACGAGCCAAATCCC 3′ | Mating type a determination of VGIV | [68] |
| SXI2aR | 5′ GGCTTCCTGACAACACTTCTA 3′ | Mating type a determination of VGIV | [68] |
| GPD1F | 5′ CCACCGAACCCTTCTAGGATA 3′ | Amplify GPD1 | [70] |
| GPD1R | 5′ CTTCTTGGCACCTCCCTTGAG 3′ | Amplify GPD1 | [70] |
| IGS1F | 5′ ATCCTTTGCAGACGACTTGA 3′ | Amplify IGS1 | [14] |
| IGS1R | 5′ GTGATCAGTGCATTGCATGA 3′ | Amplify IGS1 | [14] |
Notes: VN = molecular type VNI, VNII and VNIV; VG = molecular type VGI, VGII, VGIII and IV, FD = Filobasidiella depauperata, CA = Cryptococcus albidus.
Mating
The fertility of MAT a VGIV strains was investigated by mating them with the following MATα and MAT a reference tester strains: C. gattii clinical, serotype C, strains NIH312 (MATα) [27] and B4546 (MAT a) [29], and crg1α mutant derivatives (“supermater” tester strains), JF101 (MATα) and JF109 (MAT a) [28]. The MAT a VGIV strains (LA390 and LA392) reported herein were co-cultured with each reference tester strain. Cells from each mating partner were mixed on V8 juice agar (5% [vol/vol], 3 mM KH2PO4, 4% [wt/vol] agar, adjusted to pH 5 [58]) and incubated in darkness for up to four weeks. Sexual reproduction was confirmed by the presence of chains of basidiospores and fused clamp connections. All strains were individually incubated on V8 juice agar as described above to differentiate haploid fruiting from mating.
Chromosomal location of the gene loci studied
Four independent genetic loci were used in the multigene sequence analysis. The chromosomal location for each of those genes was determined via BlastN searches against the genome of the serotype D strains JEC21 [59] in GenBank. The ACT1 locus is located on chromosome 1, the PLB1 locus is located on chromosome 13, the URA5 locus is located on chromosome 7 and the IDE locus is located on chromosome 12.
Gene amplification and sequencing
The ACT1 [17], URA5 [3], PLB1 [18] and IDE [19], [22] genes were amplified from each strain using specific primers (see Table 9). Each PCR contained 50 ng of genomic DNA, 50 ng of each primers, 0.2 mM dNTP, 3 mM MgCl2 and 0.5 unit of either AmpliTaq® (Applied biosystems, CA, USA), BioTaq® (Bioline, NSW, Australia) or ExTaq® (Takara, Shiga, Japan) DNA polymerase with 1X of compatible buffer in a total volume of 50 µl. PCR conditions for the ACT1 gene amplification were as follows: 3 min of initial denaturation at 94°C, followed by 35 cycles of 45 seconds at 94°C, 45 seconds at 56°C, 1 min at 72°C and terminating with 7 min of final extension at 72°C. Amplification conditions of the other genes were similar to the protocol for ACT1, but the annealing temperatures and extension times differed, as follows: URA5 62°C and 2 min; PLB1 for strains of VNI, VNII and VNIV 60°C and 2 min; PLB1 for strains of VGI, VGII, VGIII and VGIV 58°C and 2 min; and IDE 62°C and 1 min. ACT1 of F. depauperata was amplified directly using the primers CNACT1 and CNACT1R whereas and the ACT1 of C. albidus was amplified using the primers ACT1CAF1 and ACT1CAR1 (Table 9). URA5 of F. depauperata was amplified using the primers URA5DF and SJ101 (Table 9). To resolve non-specific bands generated by several URA5 amplifications of F. depauperata we cloned the amplicons using the pGEM®-T Easy Vector System I according to the manufacturer's protocol (Promega®, NSW, Australia). PCR amplicons and the URA5 plasmid were purified and sequenced by the ABI Big dye Terminator method (Macrogen Inc., Korea) using the amplification and additional internal primers (Table 9). Sequences were assembled using either the program Sequencher version 4.6 (Gene Codes, MI, USA) or Bioedit version 7.0.5.3 [60]. Intron and exon positions were determined by aligning the sequences with the reference sequences of each gene as follows: ACT1 (GenBank Accession No. U10867), URA5 (GenBank Accession No. AF032436), PLB1 (GenBank Accession No. AF223383), IDE (GenBank Accession No. XM568105). Sequences including and excluding introns were aligned with Bioedit version 7.0.5.3 [60] using Clustal W [61]. Sequences were deposited in Genbank (see below). Combined datasets were created by a combination of the ACT1, URA5, PLB1 and IDE sequences. Sequences from the closet siblings of the pathogenic Cryptococcus species complex: F. depauperata and C. albidus were used as outgroups [1], [62].
Phylogenetic analyses
The phylogenetic relationships within the Cryptococcus species complex were inferred by parsimony and likelihood methods. Parsimony analysis was conducted with the program PAUP* 4.0b10, using the heuristic search option [63]. For the maximum parsimony analysis starting trees were obtained by stepwise addition with 100 random sequence additions. Tree bisection-reconnection (TBR) was used for branch-swapping. Maximum parsimony phylograms were generated for each of the four loci, as well as the combined datasets, which included every locus of all the isolates. Bootstrap analysis using 500 heuristic replicates was used to estimate support for the clades of each locus with MaxTree set to 100. Gaps in the sequences were treated as missing data and all characters were equally weighted.
Bayesian phylogenetic analysis was used to estimate the probability of the taxonomic structure given the data at each individual locus, as well as the combined data from all four loci using MrBayes 3.1.2 [64]. First, the model of nucleotide substitution that best fit the data of each locus was determined by a likelihood ratio test using the program PAUP* 4.0b10. The likelihood score files, created in PAUP, were then analyzed with the program ModelTest 3.7 on the ModelTest server 1.0 (http://darwin.uvigo.es/software/modeltest_server.html) using the Akaike information criterion (AIC) [65]. In the analysis of the combined loci, parameter estimates of each locus were unlinked, allowing independent substitution models for each locus. Two analyses were performed simultaneously and used to calculate the posterior probabilities, as estimated from uniform priors, of the clades of each locus and the combined loci. Each analysis included four simultaneous and incrementally heated Markov chains; each replicate used default heating values. Markov chains were initiated from a random tree and were run for 1,000,000 generations. Samples were taken every 100th generation. Standard deviations of split frequencies of the two runs were monitored until they converged. The last 5,000 samples of each analysis containing the standard deviations ≥0.02 were used to generate consensus trees using PAUP under the 50% majority rule. The clade posterior probabilities of each clade and the overall topology of each replicate were compared to verify that each consensus tree converged on a similar phylogeny.
Sequence similarity determination
The sequence similarity matrix among the major haploid molecular type clades was created by the program MEGA version 4.1 [66] based on the intron-included dataset. P-distances were generated and converted to similarity percentage using Microsoft Excel program. Standard errors were computed using 500 bootstrap replicates.
Combinability assessment
Prior to combined analyses the combinability of the data were explored using incongruence length difference/partition homogeneity test (ILD/PHT) [67], [68] implanted in PAUP. To avoid detecting incongruence that is expected within lineages (see below), IDL/PHT was restricted to datasets containing only single reference strains representing the major haploid molecular types and the major lineages obtained by the phylogenetic analyses of the Cryptococcus species complex (strains: ATCC90112, WM148, M27053, WM626, HamdenC3-1, RV58146, WM629, JEC21, WM179, MCS022, WM178, RB52, WM175, LA644, WM779, M27055, LA390 and LA568). The ILD/PHT analysis used only informative characters and entailed a random stepwise-addition maximum parsimony heuristic searches with 10000 replicates (TBR; maxtrees = 500). Since a significance threshold of 0.05 forces the ILD/PHT computation to be too conservative [30], the null hypothesis of congruence was rejected only if P<0.0001.
Recombination assessment
To determine the extent of recombination, if any, within each molecular type, we implemented three complementary tests: the ILD/PHT test, the Index of Association (IA) and the phylogenetic incompatibility.
In the first test, we used the ILD/PHT with the objective to find signals of recombination in our dataset. In the absence of recombination, all the loci should provide the same phylogenetic result and therefore all the gene trees should be compatible and alleles at different regions should be associated. In contrast, in a recombining population each locus is expected to have a unique evolutionary history resulting in a different phylogeny for each locus thus this phylogenies are incongruent and incompatible. This test was conducted using PAUP including all the isolates under the same conditions explained previously.
In the second test we used the IA, that is the most common measure of multi-locus linkage. The IA uses an estimate of multi-locus association within a haploid population through assessment of the number of loci that are different when individuals are compared. The expected difference between a recombinant population and a clonal one is that a clonal population will have a skewed distribution with a statistically significant excess of extreme distances, whereas a recombinant population will have a genetic distance with a normal distribution [47], [69]. In view of the fact that the number of loci analyzed can influence the expected value of the IA we complemented these analysis using the statistic rBarD that is a modification of IA, which removes this dependency on number of loci. For IA and rBarD analysis, we used the program MultiLocus 1.3 [70] (http://www.agapow.net/software/multilocus/) using 1000 artificially recombining datasets. The null hypothesis of recombination is rejected if the IA or rBarD of the observed dataset is significantly (P<0.05) different than the range of values produced from the artificially recombined datasets.
In the third test, we calculated the proportion of pairwise loci that were phylogenetically incompatible. Two loci are compatible if all the observed genotypes are explainable by mutation rather than recombination or homoplasy [71]. For example, if there are two loci with two alleles each, the two loci are incompatible if all four possible genotypes can be found in the population, showing a possible genetic exchange between strains. This test was performed using the program Multilocus 1.3 using 1000 artificially recombining datasets. P values of less than 0.05 indicate that the hypothesis of random recombination should be rejected.
Genetic variability analysis
Genetic variations of each locus were determined by using the program PAUP* 4.0b10. The variation among all sequences, as well as the parsimoniously informative characters, was calculated from each reference molecular type strain and from all cryptococcal strains.
Numbers of polymorphic characters of the combined loci from the intron-excluded dataset of each clade were compared using the Chi-Square test. The statistical analysis was performed with the program SPSS 15.0.0 (LEAD Technologies, Inc., IL, USA) and significance was defined when P value<0.05.
Estimation of genetic divergence among lineages
Genetic divergence among each lineages was estimated as previously describe [11] with modification. A maximum-likelihood tree for each gene was created based on the selected evolutionary models with and without molecular clock and the Likelihood Ratio Test statistic was calculated. Chi-square test was performed to estimate whether the four genes evolved according to the molecular clock model (P value>0.05). Distances according to the selected evolutionary model for each gene were calculated from the maximum-likelihood tree [63]. Estimates of the time of divergence and hybridization assumed the consensus mutation rate of 2×10−9 per nucleotide per year for protein coding genes [11], [72], [73]. The distance was calculated between the most recent nodes of the hybrid to the tip of the phylogeny. Weighted average and standard deviation of the distance were calculated using the program SPSS 15.0.0 (LEAD Technologies, Inc., IL, USA). Among the four genes, the estimates of age show that the earliest hybridization event in each isolate is the maximum age of hybridization. The 95% confidence intervals were calculated using the program Excel (Microsoft Corporation, USA).
Investigation of the relationship of VNII-1 group and the VNB molecular type
A recent study in Botswana has proposed a new molecular type, VNB for the Cryptococcus species complex [15]. We determined whether our VNII-1 isolates are in fact resided in this new molecular type. Parsimony and Bayesian analysis were performed as described above, using a combined dataset of PLB1, URA5, GPD1, and IGS1 with representatives of each molecular type including: VNI: WM148, H99, WM721, RV59369; VNII: WM626, JG02; VNII-1: M27053, HamdenC3-1, LA511; VNB: bt1, bt31, bt131, bt22; VNIV: JEC21 and the standard molecular type strains of C. gattii. Bayesian analysis was performed based on HKY evolutionary model with the gamma distribution according to the program ModelTest 3.7 using the combined dataset obtained as described above. The combinability as tested as described above. Sequences of PLB1, GPD1 and IGS1 of the VNB strains were obtained from the online database (http://www.mlst.net/). Gene sequences of strain H99 were obtained from the genome project database (http://www.broad.mit.edu/) as well as GPD1 and IGS1 sequences of JEC21 (http://www.ncbi.nlm.nih.gov/). URA5 of the VNB strains were amplified and sequenced as described above. GPD1 and IGS1 of the other strains were amplified using the primers GPD1F/GPD1R [68] and IGSF/IGSR [15] under the following conditions: 3 min of initial denaturation at 94°C, followed by 35 cycles of 45 seconds at 94°C, 45 seconds at 63°C (GPD1) or 60°C (IGS1), 1 min at 72°C and lastly, 7 min of final extension at 72°C.
Acknowledgments
The authors would like to thank Teun Boekhout (Centralalbureau voor Schimmelcultures, Utrecht, The Netherlands), Katrin Tintelnot (Robert Koch Institute, Berlin, Germany), Marcia Lazera and Bodo Wanke (Servico de Micologia, Instituto de Pesquisa Clinica Evandro Chagas, Av. Brasil 4365, Rio de Janeiro, Brazil), Elizabeth Castaneda (Grupo de Microbiologia, Instituto Nacional de Salud, Bogota, DC, Columbia), John Graybill (Faculty of Medicine, University of Texas, San Antonio, Texus, USA), Aristea Velegraki (Department of Microbiology, University of Athens, Athens, Greece), Dinah Parr (Department of Microbiology, Auckland Hospital, Auckland, New Zealand), Nikola Yelinov (Kashkin Research Institute of Medical Mycology, Saint-Petersburg, Russia), David Ellis (Women's and Children's Hospital, University of Adelaide, Adelaide, Australia), Mark Krockenberger and Richard Malik (Department of Veterinary Sciences, University of Sydney, NSW, Australia), Murray Fyfe (British Columbia Centre for Disease Control, Vancouver, British Columbia, Canada), Melanie-Ann John, Medical School, University of Natal, Durban, South Africa), Valerie Davis (South African Institute for Medical Research, Johannesburg, South Africa) and Samaniya Sukroongreung (Faculty of Medical Technology, Mahidol University, Bangkok, Thailand) for generously contributing strains to the current study. The authors like to thank David Ellis (Women's and Children's Hospital, University of Adelaide, Adelaide, Australia) for the serotyping and John Taylor (UC Berkeley, USA) for his helpful comments to the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by an NH&MRC research grant ID 352303 to WM and CNPq research grant 476612 and MS/CNPq/FAPERGS-PPSUSII 0700845 to MHV. The funding supported the general research project and had no impact on study design or any other aspect of the research reported herein.
References
- 1.Casadevall A, Perfect JR. Cryptococcus neoformans. Washington, DC: ASM press; 1998. [Google Scholar]
- 2.Kwon-Chung KJ, Bennett JE. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984;120:123–130. doi: 10.1093/oxfordjournals.aje.a113861. [DOI] [PubMed] [Google Scholar]
- 3.Franzot SP, Fries BC, Cleare W, Casadevall A. Genetic relationship between Cryptococcus neoformans var. neoformans strains of serotypes A and D. J Clin Microbiol. 1998;36:2200–2204. doi: 10.1128/jcm.36.8.2200-2204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M. (1557) Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. basillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon. 2002;51:804–806. [Google Scholar]
- 5.Sorrell TC. Cryptococcus neoformans variety gattii. Med Mycol. 2001;39:155–168. [PubMed] [Google Scholar]
- 6.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–1799. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1790::AID-ELPS1790>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Boekhout T, Theelen B, Diaz M, Fell JW, Hop WC, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- 9.Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latouche GN, Huynh M, Sorrell TC, Meyer W. PCR-restriction fragment length polymorphism analysis of the phospholipase B (PLB1) gene for subtyping of Cryptococcus neoformans isolates. Appl Environ Microbiol. 2003;69:2080–2086. doi: 10.1128/AEM.69.4.2080-2086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Vilgalys R, Mitchell TG. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol Ecol. 2000;9:1471–1481. doi: 10.1046/j.1365-294x.2000.01021.x. [DOI] [PubMed] [Google Scholar]
- 12.Diaz MR, Boekhout T, Kiesling T, Fell JW. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Res. 2005;5:1129–1140. doi: 10.1016/j.femsyr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Butler MI, Poulter RT. The PRP8 inteins in Cryptococcus are a source of phylogenetic and epidemiological information. Fungal Genet Biol. 2005;42:452–463. doi: 10.1016/j.fgb.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Bovers M, Hagen F, Kuramae EE, Boekhout T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet Biol. 2008;45:400–421. doi: 10.1016/j.fgb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172:2223–2238. doi: 10.1534/genetics.105.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607. doi: 10.1111/j.1567-1364.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- 17.Cox GM, Rude TH, Dykstra CC, Perfect JR. The actin gene from Cryptococcus neoformans: structure and phylogenetic analysis. J Med Vet Mycol. 1995;33:261–266. doi: 10.1080/02681219580000521. [DOI] [PubMed] [Google Scholar]
- 18.Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- 19.Faganello J, Dutra V, Schrank A, Meyer W, Schrank IS, et al. Identification of genomic differences between Cryptococcus neoformans and Cryptococcus gattii by Representational Difference Analysis (RDA). Med Mycol. 2008:1–8. doi: 10.1080/13693780802272148. [DOI] [PubMed] [Google Scholar]
- 20.Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19:608–624. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- 21.Chesneau V, Rosner MR. Functional human insulin-degrading enzyme can be expressed in bacteria. Protein Expr Purif. 2000;19:91–98. doi: 10.1006/prep.2000.1217. [DOI] [PubMed] [Google Scholar]
- 22.Garcia JV, Gehm BD, Rosner MR. An evolutionarily conserved enzyme degrades transforming growth factor-alpha as well as insulin. J Cell Biol. 1989;109:1301–1307. doi: 10.1083/jcb.109.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Authier F, Bergeron JJ, Ou WJ, Rachubinski RA, Posner BI, et al. Degradation of the cleaved leader peptide of thiolase by a peroxisomal proteinase. Proc Natl Acad Sci U S A. 1995;92:3859–3863. doi: 10.1073/pnas.92.9.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igreja RP, Lazera Mdos S, Wanke B, Galhardo MC, Kidd SE, et al. Molecular epidemiology of Cryptococcus neoformans isolates from AIDS patients of the Brazilian city, Rio de Janeiro. Med Mycol. 2004;42:229–238. doi: 10.1080/13693780310001644743. [DOI] [PubMed] [Google Scholar]
- 25.Katsu M, Kidd S, Ando A, Moretti-Branchini ML, Mikami Y, et al. The internal transcribed spacers and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans species complex. FEMS Yeast Res. 2004;4:377–388. doi: 10.1016/S1567-1356(03)00176-4. [DOI] [PubMed] [Google Scholar]
- 26.Escandon P, Sanchez A, Martinez M, Meyer W, Castaneda E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 2006;6:625–635. doi: 10.1111/j.1567-1364.2006.00055.x. [DOI] [PubMed] [Google Scholar]
- 27.Wickes BL, Moore TD, Kwon-Chung KJ. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology. 1994;140 (Pt 3):543–550. doi: 10.1099/00221287-140-3-543. [DOI] [PubMed] [Google Scholar]
- 28.Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varma A, Kwon-Chung KJ. Construction of stable episomes in Cryptococcus neoformans. Curr Genet. 1998;34:60–66. doi: 10.1007/s002940050366. [DOI] [PubMed] [Google Scholar]
- 30.Cunningham CW. Can three incongruence tests predict when data should be combined? Mol Biol Evol. 1997;14:733–740. doi: 10.1093/oxfordjournals.molbev.a025813. [DOI] [PubMed] [Google Scholar]
- 31.Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, et al. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot Cell. 2003;2:1162–1168. doi: 10.1128/EC.2.6.1162-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trilles L, Lazera M, Wanke B, Theelen B, Boekhout T. Genetic characterization of environmental isolates of the Cryptococcus neoformans species complex from Brazil. Med Mycol. 2003;41:383–390. doi: 10.1080/1369378031000137206. [DOI] [PubMed] [Google Scholar]
- 33.Kwon-Chung KJ, Bennett JE. Distribution of alpha and alpha mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol. 1978;108:337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- 34.McClelland CM, Chang YC, Varma A, Kwon-Chung KJ. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 2004;12:208–212. doi: 10.1016/j.tim.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, et al. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot Cell. 2005;4:1403–1409. doi: 10.1128/EC.4.8.1403-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halliday CL, Carter DA. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var gattii isolates from Australia. J Clin Microbiol. 2003;41:703–711. doi: 10.1128/JCM.41.2.703-711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sriburee P, Khayhan S, Khamwan C, Panjaisee S, Tharavichitkul P. Serotype and PCR-fingerprints of clinical and environmental isolates of Cryptococcus neoformans in Chiang Mai, Thailand. Mycopathologia. 2004;158:25–31. doi: 10.1023/b:myco.0000038435.14281.f4. [DOI] [PubMed] [Google Scholar]
- 38.Taylor J, Jacobson D, Fisher M. The evolution of asexual fungi: Reproduction, Speciation and Classification. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 39.Pringle A, Baker DM, Platt JL, Wares JP, Latge JP, et al. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution. 2005;59:1886–1899. [PubMed] [Google Scholar]
- 40.Maynard-Smith JM, Smith NH, O'Rourke M, Spratt BG. 1993 How clonal are bacteria? [Google Scholar]
- 41.Boekhout T, van Belkum A. Variability of karyotypes and RAPD types in genetically related strains of Cryptococcus neoformans. Curr Genet. 1997;32:203–208. doi: 10.1007/s002940050267. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Mitchell TG. Comparative gene genealogical analyses of strains of serotype AD identify recombination in populations of serotypes A and D in the human pathogenic yeast Cryptococcus neoformans. Microbiology. 2003;149:2147–2154. doi: 10.1099/mic.0.26180-0. [DOI] [PubMed] [Google Scholar]
- 43.Kidd SE, Guo H, Bartlett KH, Xu J, Kronstad JW. Comparative gene genealogies indicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryot Cell. 2005;4:1629–1638. doi: 10.1128/EC.4.10.1629-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell LT, Fraser JA, Nichols CB, Dietrich FS, Carter D, et al. Clinical and environmental isolates of Cryptococcus gattii from Australia that retain sexual fecundity. Eukaryot Cell. 2005;4:1410–1419. doi: 10.1128/EC.4.8.1410-1419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saul N, Krockenberger M, Carter D. Evidence of recombination in mixed-mating-type and alpha-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryot Cell. 2008;7:69. doi: 10.1128/EC.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bui T, Lin X, Malik R, Heitman J, Carter D. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, {alpha} mating type populations. Eukaryot Cell. 2008;7:1771–1780. doi: 10.1128/EC.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell LT, Carter DA. Looking for sex in the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:588–598. doi: 10.1111/j.1567-1364.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- 48.Sharpton TJ, Neafsey DE, Galagan JE, Taylor JW. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol. 2008;9:R24. doi: 10.1186/gb-2008-9-1-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Litvintseva AP, Lin X, Templeton I, Heitman J, Mitchell TG. Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa. PLoS Pathog. 2007;3:e114. doi: 10.1371/journal.ppat.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escandon P, Ngamskulrungroj P, Meyer W, Castaneda E. In vitro mating of Colombian isolates of the Cryptococcus neoformans species complex. Biomedica. 2007;27:308–314. [PubMed] [Google Scholar]
- 51.Ngamskulrungroj P, Sorrell TC, Chindamporn A, Chaiprasert A, Poonwan N, et al. Association between fertility and molecular sub-type of global isolates of Cryptococcus gattii molecular type VGII. Med Mycol. 2008;46:665–673. doi: 10.1080/13693780802210734. [DOI] [PubMed] [Google Scholar]
- 52.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 53.Mayden RL. A hierarchy of species concepts: the denouncement in the saga of the species problem. In: Claridge MF, Dawah HA, Wilson MR, editors. Species: the units of biodiversity. London: Chapman and Hall; 1997. pp. 381–424. [Google Scholar]
- 54.Cogliati M, Esposto MC, Tortorano AM, Viviani MA. Cryptococcus neoformans population includes hybrid strains homozygous at mating-type locus. FEMS Yeast Res. 2006;6:608–613. doi: 10.1111/j.1567-1364.2006.00085.x. [DOI] [PubMed] [Google Scholar]
- 55.Lengeler KB, Cox GM, Heitman J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun. 2001;69:115–122. doi: 10.1128/IAI.69.1.115-122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayr E. Systematics and the origin of species: from the viewpoint of a zoologist. New York: Columbia University Press; 1942. p. 334. [Google Scholar]
- 57.Halliday CL, Bui T, Krockenberger M, Malik R, Ellis DH, et al. Presence of alpha and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J Clin Microbiol. 1999;37:2920–2926. doi: 10.1128/jcm.37.9.2920-2926.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon-Chung KJ, Bennett JE, Rhodes JC. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Van Leeuwenhoek. 1982;48:25–38. doi: 10.1007/BF00399484. [DOI] [PubMed] [Google Scholar]
- 59.Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 61.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kidd S. Molecular Epidemiology and Characterisation of Genetic Structure to Assess Speciation within the Cryptococcus neoformans Complex. Westmead: The University of Sydney; 2003. p. 429. [Google Scholar]
- 63.Swofford D. PAUP*. Phylogenetic using parsimony (* and other methods). 4 ed. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 64.Huelsenbeck J, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Oxford. 2001:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 65.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 66.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 67.Farris JS, Kallersjo M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- 68.Huelsenbeck JP, Bull JJ, Cunningham CW. Combining data in phylogenetic analysis. Trends Ecol Evol. 1996;11:152–158. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 69.Taylor JW, Geiser DM, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12(1):126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agapow PM, Burt A. Indices of multilocus linkage disequilibrium. Mol Ecol Notes. 2001;1:101–102. [Google Scholar]
- 71.Estabrook GF, Landrum L. A simple test for the possible simultaneous evolutionary divergence of two amino acid positions. Taxon. 1975;24:609–613. [Google Scholar]
- 72.Stephens JC, Nei M. Phylogenetic analysis of polymorphic DNA sequences at the Adh locus in Drosophila melanogaster and its sibling species. J Mol Evol. 1985;22:289–300. doi: 10.1007/BF02115684. [DOI] [PubMed] [Google Scholar]
- 73.Li WH, Tanimura M, Sharp PM. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25:330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- 74.Espinel-Ingroff A, Kish CW, Jr, Kerkering TM, Fromtling RA, Bartizal K, et al. Collaborative comparison of broth macrodilution and microdilution antifungal susceptibility tests. J Clin Microbiol. 1992;30:3138–3145. doi: 10.1128/jcm.30.12.3138-3145.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perfect JR, Ketabchi N, Cox GM, Ingram CW, Beiser CL. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol. 1993;31:3305–3309. doi: 10.1128/jcm.31.12.3305-3309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer W, Lieckfeldt E, Kuhls K, Freedman EZ, Borner T, et al. DNA- and PCR-fingerprinting in fungi. Exs. 1993;67:311–320. doi: 10.1007/978-3-0348-8583-6_28. [DOI] [PubMed] [Google Scholar]
- 77.Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen S, Sorrell T, Nimmo G, Speed B, Currie B, et al. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin Infect Dis. 2000;31:499–508. doi: 10.1086/313992. [DOI] [PubMed] [Google Scholar]
- 79.Sukroongreung S, Nilakul C, Ruangsomboon O, Chuakul W, Eampokalap B. Serotypes of Cryptococcus neoformans isolated from patients prior to and during the AIDS era in Thailand. Mycopathologia. 1996;135:75–78. doi: 10.1007/BF00436454. [DOI] [PubMed] [Google Scholar]
- 80.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 81.Velegraki A, Kiosses VG, Pitsouni H, Toukas D, Daniilidis VD, et al. First report of Cryptococcus neoformans var. gattii serotype B from Greece. Med Mycol. 2001;39:419–422. doi: 10.1080/mmy.39.5.419.422. [DOI] [PubMed] [Google Scholar]
- 82.Samson RA, Stalpers JA, Weijman AC. On the taxonomy of the entomogenous fungus Filobasidiella arachnophila. Antonie Van Leeuwenhoek. 1983;49:447–456. doi: 10.1007/BF00399323. [DOI] [PubMed] [Google Scholar]
- 83.Kooiman P. The Chemical Structure of the Extracellular “Starch” Produced by Cryptococcus Albidus and C. Laurentii Var. Flavescens. Antonie Van Leeuwenhoek. 1963;29:169–176. doi: 10.1007/BF02046049. [DOI] [PubMed] [Google Scholar]