Abstract
CIP75 is a member of the UbL(ubiquitin-like)-UBA (ubiquitin-associated) domain containing protein family, which has a variety of functions. One specific role described for several members of the UbL-UBA family is the involvement in the proteasomal degradation of target proteins. We have reported that CIP75 interacts with the gap junction protein, connexin43 (Cx43), and that CIP75 may modulate the proteasomal degradation of Cx43. Thus, CIP75 may have a critical role in regulating Cx43 levels, and thus intercellular gap junctional communication. This study reports the development of monoclonal antibodies (MAbs) against CIP75 and the characterization of these antibodies through immunoblotting, immunoprecipitation, and immunofluorescence microscopy analyses. These MAbs will be useful tools in future studies to elucidate the role of CIP75 in Cx43 proteasomal degradation as well as other potential activities.
Introduction
Regulation of connexins is critical in maintaining normal cell function. Connexins compose gap junctions, plasma membrane channels that mediate the direct cell-to-cell exchange of small molecules such as ions, second messengers, and small metabolites.(1) Cx43 is the most widely expressed connexin and is critical in various physiological events, such as cell growth, differentiation, and certain developmental processes.(2–4) Therefore, proper regulation of Cx43 levels, and thus Cx43 gap junction channels, is essential in maintaining normal cellular functions.
Cx43 has been demonstrated to have a high turnover rate for a plasma membrane protein with a half-life of 1.5–5 h.(5–7) While it has been shown that Cx43 degradation occurs via the lysosomal and proteasomal proteolytic pathways,(8–12) how this happens and what controls the degradation is not clear. We have previously demonstrated a novel interaction between Cx43 and CIP75, which is dependent upon the UBA domain of CIP75.(13) Our initial studies have indicated that CIP75 affects Cx43 turnover, and that this occurs through proteasomal degradation. Further studies are required to elucidate the mechanism by which CIP75 is involved in Cx43 proteasomal degradation.
Members of the UbL-UBA protein family have been implicated in the proteasomal degradation pathway. Rad23 and PLIC2 have been reported to interact with subunits of the proteasome, specifically the S2 and S5a proteins of the 19S subunit, through their UbL domain.(14–16) We have reported that CIP75 is also able to interact with Rpn1/S2 and Rpn10/ S5a through its UbL domain.(13) The UBA domain has been shown by other groups to interact with ubiquitin and ubiquitinated proteins,(16–21) suggesting a role for UbL-UBA proteins as adaptors or shuttles to bring proteins marked for degradation to the proteasome.
In this study, we have generated and characterized a series of MAbs to the various regions (UBA and UbL domains) of CIP75 that can function in immunoblotting, immunoprecipitation, and immunofluorescence microscopy experiments. These MAbs will be invaluable to further elucidate the nature of the CIP75 interaction with Cx43 and perhaps lead to a more general characterization of the role of CIP75 in proteasomal degradation as well as the identification of other CIP75 interaction partners. In doing so, this may allow a better understanding of how Cx43 and gap junction communication is regulated, as well as how proteins may be transported to the proteasome for degradation.
Methods
Protein expression in bacteria and purification
Full-length CIP75 was subcloned into the pTrcHis vector and expressed as a His-tagged fusion protein in BL21 Escherichia coli following induction with 0.1 mM IPTG for 2 h at 37°C. Bacteria were harvested, washed once with PBS, and lysed by sonication. Cell lysates were incubated with Ni+ Sepharose Fast Flow (GE Healthcare, Piscataway, NJ) for 3 h at 4°C to bind His proteins. The Sepharose was washed with 2 column volumes of PBS and then eluted with 500 mM imidazole. Purified CIP75 was concentrated to 300 μg/mL in Centricon columns (Millipore, Billerica, MA) and imidazole was diluted down to 150 mM with PBS. CIP75 with a deletion of the UbL domain at the N-terminus (CIP75ΔUbL) or the UBA domain at the C-terminus (CIP75ΔUBA) and the Src tyrosine kinase negative control proteins were also expressed as His-tagged fusion proteins and purified as described above.
The UBA and UbL domains were subcloned into the pGEX-6P2 vector and expressed as a glutathione S-transferase (GST) fusion protein in BL21 E. coli following induction with 0.1 mM IPTG for 2 h at 37°C. Bacteria were harvested, washed once with PBS, and lysed by sonication. Cell lysates were incubated with glutathione agarose (Sigma, St. Louis, MO) for 1 h at 4°C to bind GST proteins. The glutathione agarose was washed with 2 column volumes of PBS and then eluted with 20 mM glutathione in 50 mM Tris-HCl (pH 9.5). The GST only protein was expressed in, and purified from, bacteria in a similar manner.
Immunization of mice and generation of CIP75 hybridomas
BALB/c mice were immunized with 10–15 μg of purified CIP75 protein in either Freund's complete or alum adjuvants. Booster immunizations were given at 3-week intervals in either Freund's incomplete or alum adjuvants. Test bleeds were assayed for positive reactions to CIP75 by indirect enzyme-linked immunoabsorbant assay (ELISA). Spleen cells from each immunized group of mice were fused to P3x63Ag8.653 mouse myeloma cells in the presence of polyethylene glycol (PEG) to produce monoclonal antibodies according to established techniques.(22,23) Hybridomas were then selected with hypoxanthine, aminopterin, and thymidine (HAT) supplemented medium and allowed to grow on macrophage plates in preparation for ELISA. Positive wells were subcloned two to three times and tested by indirect ELISA. Selected hybridoma lines were later grown in BD serum-free cell medium, and concentrated monoclonal antibody supernatants were generated using the CELLine flask system (BD Bioscience, San Jose, CA).
Monoclonal antibody screening by ELISA
Round-bottom 96-well plates (Corning, Lowell, MA) were coated with 1 μg of antigen diluted in PBS overnight at 4°C. Plates were washed with PBS thrice and then blocked with 5% nonfat milk in borate buffer (167 mM boric acid, 134 mM NaCl [pH 8.0]). Supernatants from hybridoma cultures were added and incubated for 1 h. Plates were washed with 0.5x borate buffer thrice before incubation with secondary goat anti-mouse IgG antibody conjugated to alkaline phosphatase (Southern Biotech, Birmingham, AL) for 1 h. Plates were washed with 0.5x borate buffer thrice, then incubated with p-nitrophenyl phosphate (PnPP, Thermo Scientific, Waltham, MA) diluted to 1 mg/mL in diethanolamine substrate (DEA) buffer (Thermo Scientific) for 30 min. The OD at 405 nm was detected on a Victor3 plate reader (Perkin Elmer, Waltham, MA).
Isotypes of the monoclonal antibodies were determined using the Mouse Mono-Ab-ID kit (Invitrogen, Carlsbad, CA) through the capture assay.
Cell line and culture conditions
Human cervical carcinoma cells (HeLa) lacking endogenous Cx43 expression were cultured in high glucose Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum, 20 mM L-glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin.
Western blot and immunoprecipitation
Purified proteins were separated by 10% SDS-PAGE, transferred to PVDF membranes, and immunoblotted for CIP75 using monoclonal supernatants. Antibody binding was detected using ECL. For immunoprecipitation experiments, cells were lysed in 0.2% NP-40 lysis buffer (0.2% NP-40, 150 mM NaCl, 20 mM Tris-HCl [pH 8.0], 10 mg/mL leupeptin, 10 mg/mL aprotinin, 1 mM benzamidine, and 2 mM PMSF) and lysates were clarified by centrifugation at 14,000 rpm for 30 min at 4°C. The BCA assay reagent kit (Biorad, Hercules, CA) was used to determine protein concentrations. Clarified lysates were incubated with monoclonal CIP75 antibodies bound to protein G agarose (Thermo Scientific) for 1 h at 4°C. The agarose was washed five times with lysis buffer and then proteins were released from the agarose by boiling for 5 min in SDS-PAGE sample buffer. The proteins were analyzed by immunoblotting as described above.
Immunofluorescence microscopy
HeLa cells were transiently transfected with pcDNA-Flag-CIP75 using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were labeled with either mouse anti-Flag M2 antibody (Sigma) or different CIP75 monoclonal antibodies, as previously described.(13) The cells were co-labeled with rabbit anti-calnexin (Stressgen, Ann Arbor, MI). Secondary antibodies used were anti-mouse Alexa 488 and anti-rabbit Alexa 594 (Molecular Probes, Eugene, OR). Subcellular localization of CIP75 and calnexin was examined using a TCS SP5 AOBS confocal microscope (Leica, Wetzlar, Germany).
Results
Preparation of mouse monoclonal antibodies to CIP75
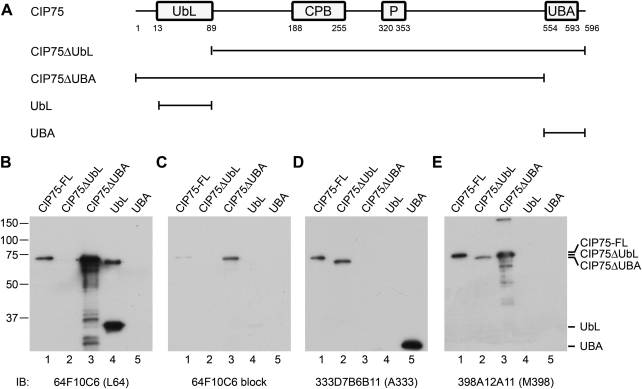
Recombinant CIP75 was expressed as a His fusion protein in bacteria, purified, and used as the antigen to immunize mice. Initial ELISA assays were conducted on test bleeds and showed a positive immune reaction. Hybridomas were generated from these mice and clones were tested for reactivity to different regions of the CIP75 protein through ELISA. As negative controls, a GST only and His-Src fusion proteins were used to screen against His-reactive clones. Full-length CIP75, CIP75ΔUbL (deleted for the N-terminal region including the UbL domain), CIP75ΔUBA (deleted for the C-terminal region including the UBA domain), and the UbL or UBA domains alone were used to establish the domain specificity for each antibody. Clones were generated that demonstrated reactivity specifically to the N-terminal UbL domain (clones 64F10C6 and 68B7D11E8), the C-terminal UBA domain (clone 333D7B6B11), or to the middle region of CIP75 (clone 398A12A11).
Recognition of monoclonal antibodies to CIP75 in Western blot and immunoprecipitation analysis
Monoclonal antibodies were further tested for CIP75 reactivity by immunoblotting (Fig. 1). Purified full-length CIP75, CIP75 deletion mutants, and UbL or UBA domains alone were used to confirm MAb specificities. Similar to the ELISA results, clones 64F10C6 (L64, Fig. 1B) and 68B7D11E8 (L68, data not shown) recognized full-length CIP75 (lane 1), the C-terminal deletion mutant (lane 3), and the UbL only domain (lane 4), but not the N-terminal deletion mutant (lane 2) or the UBA only domain (lane 5), indicating that these antibodies specifically recognized the UbL domain. Clone 333D7B6B11 (A333, Fig. 1D) recognized full-length CIP75 (lane 1), the N-terminal deletion mutant (lane 2), and the UBA only domain (lane 5), but not the C-terminal deletion mutant (lane 3) or the UbL only domain (lane 4), indicating specificity to the UBA domain. Clone 398A12A11 (M398, Fig. 1E) recognized full-length CIP75 (lane 1), and the N- and C-terminal deletion mutants (lanes 2 and 3), but neither of the individual domains (lanes 4 and 5), indicating that this clone is specific to the middle region of the protein. These results confirm the initial ELISA data. The specificity of the immunoblotting recognition of CIP75 was confirmed in protein-blocking experiments. Pre-incubation of clone 64F10C6 (L64) supernatant with the purified UbL domain prior to immunoblotting resulted in the markedly decreased recognition of the CIP75 proteins (Fig. 1C). For future use, the MAb nomenclature will be L64 for 64F10C6; L68 for 68B7D11E8; A333 for 333D7B6B11; and M398 for 398A12A11.
FIG. 1.
Detection of purified CIP75 proteins by immunoblot. Purified full-length CIP75, CIP75 mutants lacking the UbL or UBA domains, and UbL or UBA only domains were used to test MAb specificity. (A) Schematic of full-length CIP75, the CIP75ΔUbL or CIP75ΔUBA deletion mutants, and the UbL and UBA only domains. Purified full-length CIP75, CIP75ΔUbL, CIP75ΔUBA, and either the UbL- or UBA-only domains were immunoblotted with: (B) L64 MAb; (C) L64 MAb preblocked with the UbL domain; (D) A333 MAb; and (E) M398 MAb. Migration positions of the molecular mass markers are indicated at left (B–E), and migration positions of the purified proteins are indicated at right (B–E).
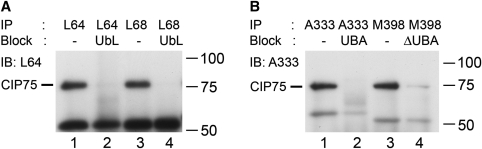
To further assess the functional capabilities of these MAbs, endogenously expressed CIP75 immunoprecipitated from HeLa cell lysates was resolved by SDS-PAGE and detected by immunoblotting (Fig. 2). All four MAbs were able to immunoprecipitate endogenous CIP75 from the cell lysates (Fig. 2A, lanes 1 and 3, and Fig. 2B, lanes 1 and 3). As in the immunoblot assays, purified domains were used to block interaction between the MAbs and endogenously expressed CIP75 in the HeLa cell lysate during the immunoprecipitation. Preblocking the antibodies resulted in little or no immunoprecipitated CIP75 (Fig. 2A, lanes 2 and 4, and Fig. 2B, lanes 2 and 4). Additionally, since purified CIP75 domains were used in the blocking experiments (UbL domain for L64 and L68, UBA domain for A333, and the C-terminal deletion mutant for M398), the regional specificity of each antibody was also confirmed.
FIG. 2.
Detection of endogenous CIP75 by immunoprecipitation. CIP75 endogenously expressed in HeLa cells was immunoprecipitated with: L64 MAb (A, lane 1); L64 MAb preblocked with the UbL domain (A, lane 2); L68 MAb (A, lane 3); L68 MAb preblocked with UbL (A, lane 4), A333 MAb (B, lane 1); A333 MAb preblocked with UBA (B, lane 2); M398 MAb (B, lane 3); and M398 MAb preblocked with the CIP75ΔUBA mutant (B, lane 4). The immunoprecipitated proteins were detected by immunoblotting using the L64 MAb (A) or A333 MAb (B). Molecular mass markers are indicated at the right of each panel.
Recognition of monoclonal antibodies to CIP75 in immunofluorescence microscopy analysis
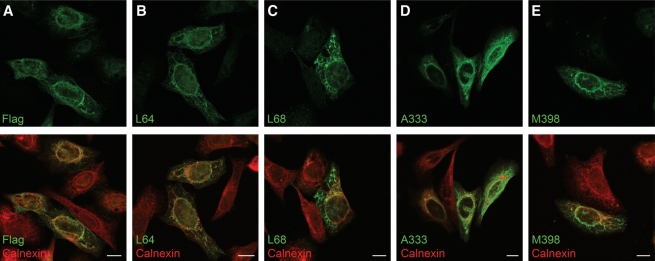
Lastly, MAbs were tested for their ability to recognize CIP75 cells by immunofluorescence microscopy. HeLa cells were transiently transfected with Flag-tagged CIP75 and labeled with the various MAbs (Fig. 3). The HeLa cells were co-labeled with calnexin, a marker for the ER, as we have previously shown that CIP75 co-localizes to the ER.(13) The cells were also labeled with the Flag antibody as a positive control. All CIP75 MAbs showed a similar staining pattern as the Flag antibody (Fig. 3A–E, top panels). In addition, all CIP75 MAbs demonstrated a similar pattern of staining as calnexin (Fig. 3, bottom panels), supporting our previously published results.
FIG. 3.
Immunofluorescence confocal microscopy analysis of Flag-CIP75-transfected HeLa cells. The subcellular localization of CIP75 (green) with the ER marker calnexin (red) was visualized by laser confocal microscopy. Top row, CIP75 localized by Flag antibody (A), L64 MAb (B), L68 MAb (C), A333 MAb (D), and M398 MAb (E). Bottom row, Flag or CIP75 MAb images merged with the calnexin antibody images.
These combined results, along with isotyping data, are summarized in Table 1.
Table 1.
Comparison of CIP75 MAb Specificities and Activities
| CIP75 MAb | Region specificity | Immunoblotting | Immunoprecipitationa | Immunofluorescenceb | Isotype |
|---|---|---|---|---|---|
| L64 | UbL domain | Yes | Yes | Yes | IgG2a, kappa |
| L68 | UbL domain | Yes | Yes | Yes | IgG2a, kappa |
| A333 | UBA domain | Yes | Yes | Yes | IgG1, kappa |
| M398 | Middle region | Yes | Yes | Yes | IgG1, kappa |
Detection of endogenous protein.
Detection of exogenous protein.
Discussion
The proper regulation of gap junctional communication between neighboring cells is essential for normal physiological functions. Changes in the levels of Cx43 can lead to significant changes of Cx43 at the plasma membrane in gap junction channels, and thus the level of gap junctional intercellular communication.(24) Therefore, understanding how Cx43 levels are regulated can aid in understanding the mechanisms that control the communication between neighboring cells. One mechanism that regulates the intracellular levels of Cx43 is protein turnover, through either the lysosomal or proteasomal degradation pathways. It has been demonstrated previously by a number of investigators that both pathways play a role in Cx43 turnover,(8–12) and we have shown that CIP75 is one Cx43-interacting partner that affects Cx43 proteasomal degradation.(13) However, the precise mechanism by which CIP75 acts to facilitate the proteasomal turnover of Cx43 is still unclear.
In this study, we have successfully generated a panel of mouse MAbs to the CIP75 protein. Most importantly, we have generated MAbs with specificities to three different domains/regions of CIP75: UbL at the N-terminus, UBA at the C-terminus, and the middle region (Table 1). In addition, these CIP75 MAbs were found to be useful in immunoblotting, immunoprecipitation, and immunofluorescence microscopy experiments, and they consisted of IgG1 or IgG2a heavy chain and kappa light chain isotypes (Table 1). As we have previously demonstrated that the CIP75 UbL domain binds to the S2 and S5a components of the 19S proteasomal subunit and the UBA domain binds to Cx43,(13) MAbs that bind to different regions of CIP75 will allow us to investigate the importance of CIP75 and its domains in the proteasome-mediated degradation of Cx43 and hence its ability to establish gap junctional communication.
Acknowledgments
This work was supported by grant CA052098 (AFL) and a P30 core grant CURE award (AK) from the National Cancer Institute, National Institutes of Health (Bethesda, MD).
References
- 1.Goodenough DA. Goliger JA. Paul DL. Connexins, connexons, and intercellular communication. Annu Rev Biochem. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- 2.Vinken M. Vanhaecke T. Papeleu P. Snykers S. Henkens T. Rogiers V. Connexins and their channels in cell growth and cell death. Cell Signal. 2006;18:592–600. doi: 10.1016/j.cellsig.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Wei CJ. Xu X. Lo CW. Connexins and cell signaling in development and disease. Annu Rev Cell Dev Biol. 2004;20:811–838. doi: 10.1146/annurev.cellbio.19.111301.144309. [DOI] [PubMed] [Google Scholar]
- 4.White TW. Paul DL. Genetic diseases and gene knockouts reveal diverse connexin functions. Annu Rev Physiol. 1999;61:283–310. doi: 10.1146/annurev.physiol.61.1.283. [DOI] [PubMed] [Google Scholar]
- 5.Darrow BJ. Laing JG. Lampe PD. Saffitz JE. Beyer EC. Expression of multiple connexins in cultured neonatal rat ventricular myocytes. Circ Res. 1995;76:381–387. doi: 10.1161/01.res.76.3.381. [DOI] [PubMed] [Google Scholar]
- 6.Laird DW. Puranam KL. Revel JP. Turnover and phosphorylation dynamics of connexin43 gap junction protein in cultured cardiac myocytes. Biochem J. 1991;273(Pt 1):67–72. doi: 10.1042/bj2730067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musil LS. Cunningham BA. Edelman GM. Goodenough DA. Differential phosphorylation of the gap junction protein connexin43 in junctional communication-competent and -deficient cell lines. J Cell Biol. 1990;111:2077–2088. doi: 10.1083/jcb.111.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beardslee MA. Lerner DL. Tadros PN. Laing JG. Beyer EC. Yamada KA. Kleber AG. Schuessler RB. Saffitz JE. Dephosphorylation and intracellular redistribution of ventricular connexin43 during electrical uncoupling induced by ischemia. Circ Res. 2000;87:656–662. doi: 10.1161/01.res.87.8.656. [DOI] [PubMed] [Google Scholar]
- 9.Girao H. Pereira P. Phosphorylation of connexin 43 acts as a stimuli for proteasome-dependent degradation of the protein in lens epithelial cells. Mol Vis. 2003;9:24–30. [PubMed] [Google Scholar]
- 10.Laing JG. Beyer EC. The gap junction protein connexin43 is degraded via the ubiquitin proteasome pathway. J Biol Chem. 1995;270:26399–26403. doi: 10.1074/jbc.270.44.26399. [DOI] [PubMed] [Google Scholar]
- 11.Laing JG. Tadros PN. Westphale EM. Beyer EC. Degradation of connexin43 gap junctions involves both the proteasome and the lysosome. Exp Cell Res. 1997;236:482–492. doi: 10.1006/excr.1997.3747. [DOI] [PubMed] [Google Scholar]
- 12.Musil LS. Le AC. VanSlyke JK. Roberts LM. Regulation of connexin degradation as a mechanism to increase gap junction assembly and function. J Biol Chem. 2000;275:25207–25215. doi: 10.1074/jbc.275.33.25207. [DOI] [PubMed] [Google Scholar]
- 13.Li X. Su V. Kurata WE. Jin C. Lau AF. A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. J Biol Chem. 2008;283:5748–5759. doi: 10.1074/jbc.M709288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elsasser S. Gali RR. Schwickart M. Larsen CN. Leggett DS. Muller B. Feng MT. Tubing F. Dittmar GA. Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 15.Schauber C. Chen L. Tongaonkar P. Vega I. Lambertson D. Potts W. Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson CR. Seeger M. Hartmann-Petersen R. Stone M. Wallace M. Semple C. Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 17.Bertolaet BL. Clarke DJ. Wolff M. Watson MH. Henze M. Divita G. Reed SI. UBA domains mediate protein-protein interactions between two DNA damage-inducible proteins. J Mol Biol. 2001;313:955–963. doi: 10.1006/jmbi.2001.5105. [DOI] [PubMed] [Google Scholar]
- 18.Chen L. Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L. Shinde U. Ortolan TG. Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funakoshi M. Sasaki T. Nishimoto T. Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci USA. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao H. Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 22.Goding JW. London: Academic Press; 1983. Monoclonal Antibodies: Principles, Practice. [Google Scholar]
- 23.Loo LW. Berestecky JM. Kanemitsu MY. Lau AF. pp60src-mediated phosphorylation of connexin 43, a gap junction protein. J Biol Chem. 1995;270:12751–12761. doi: 10.1074/jbc.270.21.12751. [DOI] [PubMed] [Google Scholar]
- 24.VanSlyke JK. Musil LS. Dislocation and degradation from the ER are regulated by cytosolic stress. J Cell Biol. 2002;157:381–394. doi: 10.1083/jcb.200111045. [DOI] [PMC free article] [PubMed] [Google Scholar]