Abstract
Interest in anthocyanin-pigmented potato tuber flesh is increasing. To genetically map and characterize loci that influence this trait, diploid potato clone 10618-01, which has partially pigmented flesh, was crossed with diploid 320-02, which has white flesh. Almost all progeny exhibited purple coloration in the flesh, with some clones having only a small percentage of tissue pigmented, other clones having most tissue pigmented, and the majority of clones showing intermediate color phenotypes. The two parents and 228 progeny were genotyped with 493 AFLP, 8 CAPS, and 13 SSR markers. QTLs influencing extent of flesh pigmentation were detected on chromosomes 5, 8, and 9. The potato homolog of Petuniaan1, a basic helix-loop-helix (bHLH) transcriptional regulator of anthocyanin biosynthesis, was found to co-localize with the QTL on chromosome 9. A CAPS marker based on this gene was used to evaluate a collection of 21 tetraploid potato clones with highly or fully pigmented red or purple flesh, as well as 53 cultivars with white or yellow flesh. All 21 pigmented-flesh clones shared a marker allele that was present in only 21 of the 53 white and yellow clones, suggesting that a common bHLH allele contributes toward, although it is clearly not sufficient for, highly or fully pigmented tuber flesh in cultivated potato.
Introduction
Consumer interest in potatoes with red or purple flesh has been increasing over the past decade, in part because of novel appearance, and in part because of the perceived benefits of higher antioxidant content (Tsuda et al. 2000; Ross and Kasum 2002; Brown et al. 2003, 2005, 2007; Scalbert et al. 2005). Red and purple tuber flesh color results from the accumulation of anthocyanin pigments (Lewis et al. 1998; Rodriguez-Saona et al. 1998; Naito et al. 1998; Eichhorn and Winterhalter 2005).
Pigmented tuber flesh is conferred by the Pf locus (De Jong 1987). Pf is tightly linked with I, which is required for pigmentation of tuber skin and maps to chromosome 10 (De Jong 1987; Dodds and Long 1955; van Eck et al. 1994). The I locus is also known as D in tetraploid potatoes (Salaman 1910). Pf alone is not sufficient for tuber flesh to be completely pigmented; potatoes with this gene may exhibit a small, intermediate, or large degree of flesh coloration. In many plants, tissue-specific accumulation of anthocyanins is mediated by R2R3MYB genes and/or bHLH regulators (Cone et al. 1986; Ludwig et al. 1989; Ludwig and Wessler 1990; Quattrocchio et al. 1998, 1999; Selinger et al. 1998). The potato ortholog of Petunia an2, an R2R3MYB regulator of anthocyanin production, maps to the same region of the genome as Pf and I (De Jong et al. 2004).
Several other potato genes are also known to influence potato color. The R locus, which co-segregates with dihydroflavonol 4-reductase (De Jong et al. 2003), is required for the production of red anthocyanins, while the P locus, which codes for flavonoid 3′,5′-hydroxylase (Jung et al. 2005), is required for production of purple pigments.
Several recent studies have reported on potential health benefits of consuming potatoes with anthocyanin-pigmented flesh. For example, rats fed with purple potato flakes have significantly higher serum antioxidant potential and hepatic Cu/Zn-superoxide dismutase in the liver (Han et al. 2006). Potato anthocyanin may also help combat both prostate cancer (Reddivari et al. 2007) and breast cancer (Thompson et al. 2009). To more efficiently manipulate tuber flesh color in our applied breeding program, we would like to better understand the genetic basis for flesh pigmentation. Toward this end, we constructed a diploid population that segregates for degree of tuber flesh coloration, and report here on a QTL analysis of the pigmented flesh trait, as well as a follow-up marker analysis of tetraploid potato cultivars with and without pigmented tuber flesh.
Materials and methods
Plant materials
Diploid potato clone 10618-01, which has purple, partially pigmented tuber flesh, was crossed as a female with white-fleshed diploid 320-02 to form an F1 mapping population consisting of 228 clones. Both diploid parents were kindly provided by H. De Jong (AAFC, Fredericton, NB). WIS clones with pigmented tuber flesh were kindly provided by S. Janksy (USDA-ARS, Madison, WI). POR04PG01-2 was kindly provided by C. Brown (USDA-ARS, Prosser, WA). NY clones were provided by the Cornell University potato breeding program.
Phenotyping
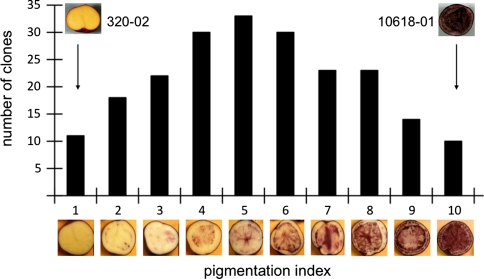
The extent of tuber flesh coloration was evaluated with tubers produced in the greenhouse. Each clone was grown in two pots, and the largest tuber from each pot was scored. The average score of the two tubers was used for QTL analysis. All 228 progeny, as well as both parents, were genotyped. After genotyping, two daughter clones appeared to be identical; one of the two was excluded from subsequent analysis. Thirteen of the remaining 227 clones did not tuberize well or died in 2006, and were not included in any QTL analyses. A further 15 clones did not tuberize well or died in 2007, and were not used for QTL analyses that year. Flesh coloration was scored on an arbitrary 1–10 scale (1 = no flesh pigmentation, 10 = almost completely pigmented), where the distribution of pigment was assessed in equatorial cross sections of mature tubers. Flesh coloration was scored in both 2006 and 2007.
Marker analysis
Genomic DNA was extracted from plants grown in the greenhouse using a Qiagen DNA kit, following the manufacturer’s instructions. AFLP markers were generated with 14 Pst+2/Mse+3 and 10 Eco+2/Mse+3 AFLP primer combinations according to Vos et al. (1995), using 33P-labeled PstI or EcoR1 primers. AFLP amplification products were separated on a 5% denaturing polyacrylamide gel. Sizes of amplification products were estimated by comparison to a Sequamark 10 base ladder (Research Genetics, Huntsville, AL). Images were visualized by exposing film against dried acrylamide sequencing gels. Eight CAPS markers (Konieczny and Ausubel 1993) (SbeII—Chen et al. 2001; F35H-4F/4R—Jung et al. 2005; 21BA, bch6, CT203, GP24, UGPase, and zep are described in Table 1) and 13 polymorphic SSR markers of known chromosomal location (STM0003, STM1104, STM1106, STM1053, STM2020, STM2022, STM3009, STM3010, STM3016, StI011, StI014, StI041, and StI049) (Milbourne et al. 1998; Feingold et al. 2005) were used to identify chromosomes.
Table 1.
CAPS markers developed in this study
| Marker | Approx. product size (bp) | Restriction enzyme | Chromosome | Primer sequences |
|---|---|---|---|---|
| 21BA | 470/400 | None | 10 | F: GTGATTATGTCATCCAAAAGTTTATAG R: GAATTTCTGAGGTTGAGGTCTTA |
| ans | 700 | HaeIII | 8 | F: TATTGCTTGTACTTCTATTTTTCGAGATAG R: CTTGGCATATTCACTTGTTGCT |
| bch6 | 400 | BcrI | 6 | F: AACAACCTCACATGTTTCTCCAA R: CAAATGTACCCAACATTTCGGTTA |
| chi | 800 | MseI | 5 | F: ATAGAGGTTTGGAGATTGAAGG R: ACTACACTTTGCTGCAGGGGA |
| chs | 1600 | AluI | 5 | F: GCGACTCCTTCGAACTGTG R: TGAAGTTTTTCGGGCTTTAGGC |
| CT203 | 760 | AluI | 10 | F: AGTGACGATGATGACAGAGGAGAA R: AAATGGACTAAAGCATATAGCCGG |
| GP24 | 900 | AluI | 6 | F: CTGCAGTCAAGGGATACATTT R: GCGTCTCTGCAATCTATTTCT |
| jaf13 | 1480 | RsaI | 8 | F: GAAGATCCTAACCTCATTCAGCAAATAAAA R: GTTGCTTAAAATTATGGAGGCACTGA |
| Stan1 | 1600 | TaqI | 9 | F: CGGCCCTAGTTATGATGAATTATCACA R: ACCTCCACTTTAAGTTCCCTTAGC |
| UGPase | 600 | RsaI | 11 | F: CACCTTGACTGATGAGGGCTAT R: TGGCACCAGCAGCTACTCTA |
| zep | 1000 | BfuCI | 2 | F: AGAGGGATTTAAGTGCTATCAGAG R: CCAGTATAACAAGTGTAGCCAGAG |
Mapping
Marker data were analyzed with JoinMap 3.0 (Van Ooijen and Voorrips 2001). Linkage groups were assembled using the Kosambi function (Kosambi 1943). Twenty-two linkage groups were assembled at LOD thresholds of eight or greater. Chromosomes 2 and 7 of female parent 10618-01 were assembled at LOD 6 and LOD 5, respectively. Each linkage group was labeled with at least one anchor marker of known location.
QTL analysis
QTL analysis was performed with the program MapQTL 5 (Van Ooijen 2004). Two analysis models (Kruskal–Wallis and Interval Mapping) were used. A LOD threshold of 2.85 for declaring significance (P < 0.05) for interval mapping was established by empirically permuting the data 1000 times. Linkage map and QTL locations were visualized using MapChart 2.1 (Voorrips 2002). QTL analysis was repeated with phenotypic data from 2006 and 2007.
Association of QTLs with anthocyanin pathway genes
CAPS markers were designed against five potato anthocyanin pathway genes (ans, Stan1, chi, chs, jaf13; Table 1). For each CAPS marker, genomic DNA was amplified using the following thermal profile: 94°C for 2 min, then 35 cycles of [94°C, 20 s; 72°C, 60 s; 56°C, 30 s]. PCR products were digested with the corresponding restriction enzyme for 3 h, then visualized on a 2% agarose gel.
Results
The progeny of a cross between diploid 10618-01, which has purple skin and partially colored (purple and white) tuber flesh, and diploid 320-02, which has red skin and white flesh, segregated extensively for extent of purple color in tuber flesh. After harvest in 2006, the flesh of 11 progeny did not appear to be pigmented at all, the flesh of 10 progeny were heavily pigmented, while the remaining 193 progeny displayed intermediate degrees of purple flesh coloration (Fig. 1). All progeny had purple tuber skin. The extent of tuber flesh coloration was scored on a 1–10 scale (Fig. 1).
Fig. 1.
Distribution of flesh color phenotypes observed in 2006 in the F1 progeny of a cross between diploid clones 10618-01 and 320-02
To identify loci influencing extent of flesh coloration, the progeny and both parents were evaluated with 514 molecular markers including 493 AFLP, 13 SSR, and 8 CAPS markers. Analysis with JoinMap 3.0 readily separated markers into 12 maternal and 12 paternal linkage groups. A total of 496 markers could be placed in linkage groups with a LOD score of 5 or higher. The map of 10618-01 totaled 753 cM in length and comprised 212 markers, while the map of 320-02 totaled 907 cM in length and was made up of 284 markers. All 24 linkage groups included at least one anchor marker of known chromosomal location.
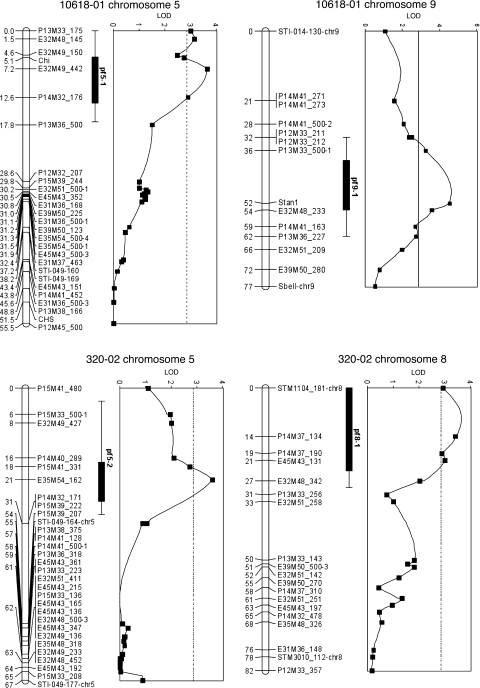
Marker and year 2006 trait data were then analyzed using both nonparametric (Kruskal–Wallis) and parametric (interval mapping) approaches. Kruskal–Wallis analysis revealed significant (P < 0.001) loci on chromosome 5 of both parents: for 10618–01, at AFLP marker E32M49-442, and for 320–02, at marker E53M54-162 (Table 2). In addition, highly significant loci were detected on chromosome 8 of 320-02 at marker P14M37-134 (P < 0.0005) and on chromosome 9 of 10618-01 at marker E32M48-233 (P < 0.0001) (Table 2). QTLs at comparable locations were identified by interval mapping (Fig. 2, Table 2). The same loci were detected when phenotypic data for year 2007 was analyzed separately. Tuber pigmentation scores were not identical in 2006 and 2007, but were highly correlated (r2 = 0.61). Two additional loci were detected only in 2007, on chromosome 8 of 10618-01 at marker E32M48-318 (LOD 3.1) and chromosome 3 of 320-02 at marker E39M50-249 (also with LOD 3.1) (data not shown).
Table 2.
QTLs detected by Kruskal–Wallis (KW) and Interval Mapping (IM) using phenotype data from 2006
| QTL model | Parent | QTL parameters | |||
|---|---|---|---|---|---|
| Chromosome | Markera | Significance value | Percent variation (r2) | ||
| KW | 10618-01 | 9 | E32M48-233 | 0.0001 | N/A |
| 5 | E32M49-442 | 0.001 | N/A | ||
| 320-02 | 8 | P14M37-134 | 0.0005 | N/A | |
| 5 | E35M54-162 | 0.001 | N/A | ||
| IM | 10618-01 | 9 | E32M48-233 | LOD 3.6 | 8.1 |
| 5 | E32M49-442 | LOD 3.6 | 8.1 | ||
| 320-02 | 8 | P14M37-134 | LOD 2.9 | 6.5 | |
| 5 | E35M54-162 | LOD 3.7 | 8.1 | ||
aMarker with highest significance score
Fig. 2.
Location of QTLs that influenced extent of tuber flesh coloration in 2006. Map locations for anthocyanin-related genes chi, chs, and Stan1 are also shown
We subsequently tested whether any known anthocyanin biosynthetic or regulatory genes co-localize with these QTLs. Potato chromosome 5 is known to code for at least two anthocyanin biosynthesis genes, chalcone isomerase (chi) and chalcone synthase (chs); chromosome 8 is known to harbor a basic helix-loop-helix (bHLH) anthocyanin regulatory gene (homolog of Petunia jaf13), as well as anthocyanidin synthase (ans); and chromosome 9 is known to code for a bHLH gene homologous to petunia anthocyanin 1 (an1) (Spelt et al. 2000; De Jong et al. 2004). CAPS markers were developed for all these genes (Table 1). Two of the five genes mapped under QTLs detected in 10618-01: chi on chromosome 5 and Stan1, the potato homolog of an1, on chromosome 9 (Fig. 2). Stan1 explained more phenotypic variation—11%—than AFLP marker E32M48-233, which explained 8.1% (Table 2). The relationship between the potato homologs of jaf13 and ans with the QTL on chromosome 8 could not be evaluated, as neither CAPS marker was polymorphic in 320-02. The fifth gene, chs, mapped far from the QTL on chromosome 5 of 10618-01 (Fig. 2).
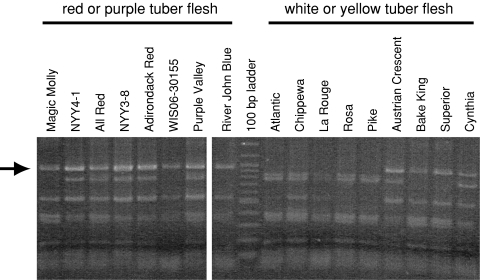
CAPS markers based on ans, chi, Stan1, and jaf13 were tested for possible relationship with pigmented flesh in a panel of diverse potato germplasm consisting of 21 tetraploid potato clones with red or purple flesh and 53 clones with white or yellow tuber flesh. The Stan1 CAPS marker revealed a common digestion product, about 980 bp in size, in all 21 of the clones with pigmented flesh (Fig. 3 and Table 3). This same digestion product was present in only 21 of 53 white- and yellow-fleshed clones (Fig. 3 and Table 3), suggesting that a common bHLH allele contributes toward, but is not sufficient, for the ability to accumulate anthocyanin in potato tuber flesh. No association with flesh color was observed with CAPS markers based on jaf13, ans or chi in the same panel.
Fig. 3.
Association between colored tuber flesh and a CAPS marker allele based on the potato homolog of Petunia an1. Genomic DNA was amplified with Stan1 primers (Table 1), restricted with Taq I, and electrophoresed through a 2% agarose gel. An arrow denotes the approximately 980 bp band present in all clones tested with red or purple tuber flesh
Table 3.
Presence/absence of ≈980 bp Stan1 marker allele in a panel of potato clones with and without anthocyanin-pigmented tuber flesh
| Potato clone | Flesh color | Stan1 980 bp fragment present (1 = yes, 0 = no) |
|---|---|---|
| Adirondack Blue | Purple | 1 |
| Adirondack Red | Red | 1 |
| All Red | Red | 1 |
| Huckleberry | Red | 1 |
| Magic Molly | Purple | 1 |
| NYH52-1 | Purple | 1 |
| NYS48-6 | Purple | 1 |
| NYY3-8 | Red | 1 |
| NYY4-1 | Red | 1 |
| POR04PG01-2 | Purple | 1 |
| Purple Peruvian | Purple | 1 |
| Purple Valley | Purple | 1 |
| River John Blue | Purple | 1 |
| WIS00-4252-1 | Purple | 1 |
| WIS01-1131-1 | Purple | 1 |
| WIS01-1131-5 | Red | 1 |
| WIS06-3124 | Purple | 1 |
| WIS06-30155 | Purple | 1 |
| WIS06-30244 | Purple | 1 |
| WIS06-30340 | Purple | 1 |
| WIS99-2743 | Purple | 1 |
| Allegany | White | 0 |
| Amandine | Yellow | 1 |
| Andover | White | 0 |
| Atlantic | White | 0 |
| Austrian Crescent | Yellow | 1 |
| Bake King | White | 1 |
| Bintje | Yellow | 1 |
| Carola | Yellow | 0 |
| Chieftain | White | 0 |
| Chippewa | White | 0 |
| Cynthia | Yellow | 0 |
| Desiree | Yellow | 1 |
| Eva | White | 0 |
| German Butterball | Yellow | 1 |
| Idarose | White | 1 |
| Katahdin | White | 0 |
| Kennebec | White | 0 |
| Keuka Gold | Yellow | 1 |
| La Rouge | White | 0 |
| Lehigh | Yellow | 0 |
| Lenape | White | 0 |
| Monona | White | 0 |
| Nordonna | White | 1 |
| Norland | White | 1 |
| NY97 | White | 1 |
| NY99 | White | 0 |
| NY115 | White | 0 |
| NY118 | White | 1 |
| NY120 | White | 1 |
| NY121 | White | 0 |
| NY123 | White | 0 |
| NY127 | White | 0 |
| NY128 | White | 0 |
| NY129 | White | 0 |
| NY130 | White | 0 |
| NY132 | White | 0 |
| NYT15-1 | White | 0 |
| Pike | White | 0 |
| Prince Hairy | White | 1 |
| Reba | White | 0 |
| Red La Soda | White | 0 |
| Redsen | White | 1 |
| Rideau | White | 1 |
| Rosa | White | 0 |
| Salem | White | 0 |
| Sandy | Yellow | 0 |
| Serrana Inta | Yellow | 1 |
| Snowden | White | 0 |
| Stirling | White | 1 |
| Superior | White | 1 |
| Sylvia | Yellow | 0 |
| Yagana | Yellow | 1 |
| Yukon Gold | Yellow | 1 |
Discussion
This study detected loci on three chromosomes—5, 8, and 9—that mediate degree of tuber flesh pigmentation. Alleles influencing this trait descended from both the white- and purple-fleshed parents, with the white-fleshed parent contributing alleles from chromosome 5 and 8, and the purple-fleshed parent contributing alleles from chromosomes 5 and 9.
The only locus that has previously been implicated in pigmentation of tuber flesh, Pf, is presumably located on chromosome 10, as Pf is tightly linked to I (De Jong 1987), and I has been mapped to chromosome 10 (Van Eck et al. 1994). If Pf segregated in this cross, we would have expected half the progeny to exhibit white flesh. Instead, only 11 of 214 progeny had unpigmented flesh, suggesting that 10618-01 is homozygous for Pf, and that Pf is necessary, but not sufficient, for anthocyanin-pigmented tuber flesh. No polymorphic markers from chromosome 10 segregated aberrantly, so Pf must have been transmitted to either half or all progeny. As tuber skin color did not segregate in this cross—all progeny had purple-skinned tubers—the genes required for anthocyanin production per se were present in all progeny. Thus, the relatively few white-fleshed progeny must have been white for a reason other than lacking a necessary biosynthetic gene.
Although the genes underpinning flesh coloration QTLs were not conclusively established in this study, two promising candidates—chi (for a QTL on chromosome 5) and a bHLH transcription factor similar to Petunia an1 (for a QTL on chromosome 9)—were identified. Both of these genes mapped close to, or under, the peak of the respective QTLs. It is not obvious how chi, an anthocyanin biosynthetic gene, might influence degree of flesh pigmentation; perhaps this gene exhibits functional variation in its promoter region, leading to differences in the range of tissues in which it can be expressed. That the potato homolog of Petunia an1 may play a role in tissue-specific expression was not surprising, as bHLH regulators of anthocyanin biosynthesis, such as delila in Antirrhinum majus (Goodrich et al. 1992), B in Zea mays (Selinger et al. 1998), ivs in Ipomoea tricolor (Park et al. 2004), tt8 of Arabidopsis thaliana (Nesi et al. 2000; Baudry et al. 2006), the rice Purple leaf (Pl) locus (Sakamoto et al. 2001), and the rice red grain locus Rc (Sweeney et al. 2006) are all known to mediate tissue-specific expression of anthocyanins.
Further evidence that the potato homolog of Petunia an1 (or a gene tightly linked to it) is associated with pigmented tuber flesh came from a comparison of varieties with and without pigmented flesh. All 21 pigmented flesh clones tested to date share an approximately 980 bp Stan1 CAPS marker allele. Eight of the pigmented flesh clones evaluated were developed in Wisconsin (WIS clones), six were developed in New York (NY and Adirondack clones), one was developed in Alaska (Magic Molly), one was developed in Washington (POR clone), one was developed in Korea (Purple Valley), and the remaining four are of unknown origin. Though potato clones that accumulate anthocyanin in tuber flesh are not uncommon in Andean landraces, this trait has generally been selected against in modern potato breeding, just as pigmented kernels were selected against in maize (Johannessen et al. 1970) and pigmented grains were selected against in rice (Sweeney et al. 2007). Nevertheless, as understanding of the potential health benefits conferred by anthocyanins has increased over the past decade, interest in consuming anthocyanin-rich plant tissues has also increased dramatically. Markers based on Stan1 may thus prove useful for those seeking to more efficiently manipulate the nutritionally important trait of pigmented tuber flesh in applied potato breeding programs.
Acknowledgments
We thank Shuping Cheng for helping with SSR data collection, and the field crew of the Cornell potato breeding program for maintaining the diploid population.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46:768–779 [DOI] [PubMed]
- Brown CR, Wrolstad R, Durst R, Yang CP, Clevidence B (2003) Breeding studies in potatoes containing high concentrations of anthocyanins. Am J Potato Res 80:241–250 [DOI]
- Brown CR, Culley D, Yang CP, Durst R, Wrolstad R (2005) Variation of anthocyanin and carotenoid contents and associated antioxidant values in potato breeding lines. J Am Soc Hort Sci 130:174–180
- Brown CR, Culley D, Bonierbale M, Amoros W (2007) Anthocyanin, carotenoid content, and antioxidant values in Native South American Potato Cultivars. Hortscience 42:1733–1736
- Chen X, Salamini F, Gebhardt C (2001) A potato molecular-function map for carbohydrate metabolism and transport. Theor Appl Genet 102:284–295 [DOI]
- Cone KC, Burr FA, Burr B (1986) Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA 83:9631–9635 [DOI] [PMC free article] [PubMed]
- De Jong H (1987) Inheritance of pigmented tuber flesh in cultivated diploid potatoes. Am Potato J 64:337–343 [DOI]
- De Jong WS, De Jong DM, De Jong H, Kalazich J, Bodis M (2003) An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.). Theor Appl Genet 107:1375–1383 [DOI] [PubMed]
- De Jong WS, Eannetta NT, De Jong DM, Bodis M (2004) Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet 108:423–432 [DOI] [PubMed]
- Dodds KS, Long DH (1955) The inheritance of colour in diploid potatoes: types of anthocyanidins and their genetic loci. J Genet 53:136–149 [DOI]
- Eichhorn S, Winterhalter P (2005) Anthocyanins from pigmented potato (Solanum tuberosum L.) varieties. Food Res Int 38:943–948 [DOI]
- Feingold S, Lloyd J, Norero N, Bonierbale M, Lorenzen J (2005) Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). Theor Appl Genet 111:456–466 [DOI] [PubMed]
- Goodrich J, Carpenter R, Coen ES (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68:955–964 [DOI] [PubMed]
- Han KH, Sekikawa M, Shimada K, Hashimoto M, Hashimoto N, Noda T, Tanaka H, Fukushima M (2006) Anthocyanin-rich purple potato flake extract has antioxidant capacity and improves antioxidant potential in rats. Br J Nutr 96:1125–1133 [DOI] [PubMed]
- Johannessen CL, Wilson MR, Davenport WA (1970) The domestication of maize: process or event? Geogr Rev 60:393–413 [DOI]
- Jung CS, Griffiths HM, De Jong DM, Cheng S, Bodis M, De Jong WS (2005) The potato P locus codes for flavonoid 3′, 5′-hydroxylase. Theor Appl Genet 110:269–275 [DOI] [PubMed]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4:403–410 [DOI] [PubMed]
- Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
- Lewis CE, Walker JRL, Lancaster JE, Sutton KH (1998) Determination of anthocyanins, flavonoids and phenolic acids in potatoes. I: coloured cultivars of Solanum tuberosum L. J Sci Food Agric 77:45–57 [DOI]
- Ludwig SR, Wessler SR (1990) Maize R gene family: tissue-specific helix-loop-helix proteins. Cell 62:849–851 [DOI] [PubMed]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA 86:7092–7096 [DOI] [PMC free article] [PubMed]
- Milbourne D, Meyer RC, Collins AJ, Ramsay LD, Gebhardt C, Waugh R (1998) Isolation, characterisation and mapping of simple sequence repeat loci in potato. Mol Gen Genet 259:233–245 [DOI] [PubMed]
- Naito K, Umemura Y, Mori M, Sumida T, Okada T, Takamatsu N, Okawa Y, Hayashi K, Saito N, Honda T (1998) Acylated pelargonidin glycosides from a red potato. Phytochemistry 47:109–112 [DOI]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12:1863–1878 [DOI] [PMC free article] [PubMed]
- Park KI, Choi JD, Hoshino A, Morita Y, Iida S (2004) An intragenic tandem duplication in a transcriptional regulatory gene for anthocyanin biosynthesis confers pale-colored flowers and seeds with fine spots in Ipomoea tricolor. Plant J 38:840–849 [DOI] [PubMed]
- Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R (1998) Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J 13:475–488 [DOI] [PubMed]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11:1144–1433 [DOI] [PMC free article] [PubMed]
- Reddivari L, Vanamala J, Chintharlapalli S, Safe SH, Miller JC (2007) Anthocyanin fraction from potato extracts is cytotoxic to prostate cancer cells through activation of caspase-dependent and caspase-independent pathways. Carcinogenesis 28:2227–2235 [DOI] [PubMed]
- Rodriguez-Saona LE, Giusti MM, Wrolstad RE (1998) Anthocyanin pigment composition of red-fleshed potatoes. J Food Sci 63:458–465 [DOI]
- Ross JA, Kasum CM (2002) Dietary flavonoids: bioavailability, metabolic effects, and safety. Annu Rev Nutr 22:19–34 [DOI] [PubMed]
- Sakamoto W, Ohmori T, Kageyama K, Miyazaki C, Saito A, Murata M, Noda K, Maekawa M (2001) The Purple leaf (Pl) locus of rice: the Pl(w) allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol 42:982–991 [DOI] [PubMed]
- Salaman RN (1910) The inheritance of colour and other characters in the potato. J Genet 1:7–46 [DOI]
- Scalbert A, Johnson IT, Saltmarsh M (2005) Polyphenols: antioxidants and beyond. Am J Clin Nutr 81(Suppl 1):215S–217S [DOI] [PubMed]
- Selinger DA, Lisch D, Chandler VL (1998) The maize regulatory gene B-Peru contains a DNA rearrangement that specifies tissue-specific expression through both positive and negative promoter elements. Genetics 149:1125–1138 [DOI] [PMC free article] [PubMed]
- Spelt C, Quattrocchio F, Mol JN, Koes R (2000) Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12:1619–1632 [DOI] [PMC free article] [PubMed]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18:283–294 [DOI] [PMC free article] [PubMed]
- Sweeney MT, Thomson MJ, Cho YG, Park YJ, Williamson SH, Bustamante CD, McCouch SR (2007) Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet 3(8):e133 [DOI] [PMC free article] [PubMed]
- Thompson MD, Thompson HJ, McGinley JN, Neil ES, Rush DK, Holm DG, Stushnoff C (2009) Functional food characteristics of potato cultivars (Solanum tuberosum L.): phytochemical composition and inhibition of 1-methyl-1-nitrosourea induced breast cancer in rats. J Food Compos Anal, in press
- Tsuda T, Horio F, Osawa T (2000) The role of anthocyanins as an antioxidant under oxidative stress in rats. Biofactors 13:133–139 [DOI] [PubMed]
- Van Eck HJ, Jacobs JME, Van Den Berg PMMM, Stiekema WJ, Jacobsen E (1994) The inheritance of anthocyanin pigmentation in potato (Solanum tuberosum L.) and mapping of tuber skin colour loci using RFLPs. Heredity 73:410–421 [DOI]
- Van Ooijen JW (2004) MapQTL ® 5, software for the zapping of quantitative trait loci in experimental populations. Kyazma BV, Wageningen, the Netherlands
- Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0, software for the calculation of genetic linkage maps. Plant Research International. Wageningen, the Netherlands
- Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78 [DOI] [PubMed]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 21:4407–4414 [DOI] [PMC free article] [PubMed]