SUMMARY
Group II introns are hypothesized to share common ancestry with both nuclear spliceosomal introns and retrotransposons, which collectively occupy the majority of genome space in higher eukaryotes. These phylogenetically diverse introns are mobile retroelements that move through an RNA intermediate. Disruption of Escherichia coli genes encoding enzymes that catalyze synthesis of global regulators cAMP and ppGpp inhibits group II intron retromobility. These small molecules program genetic transitions between nutrient excess and starvation. Accordingly, we demonstrated that glucose depletion of wild-type cells and cAMP supplementation of mutants stimulated retromobility. Likewise, amino acid starvation, which induces the alarmone ppGpp, activated retromobility. In both cases, retrotransposition to ectopic sites was favored over retrohoming. Interestingly, these stimulatory effects are mediated at the level of the DNA target, rather than of expression of the retroelement. Thereby, during metabolic stress, cAMP and ppGpp control group II intron movement in concert with the cell’s global genetic circuitry, stimulating genetic diversity.
Keywords: Retroelements, retrotransposons, diversity-generating elements, bacterial stress response
INTRODUCTION
The effect of host factors on retroelement propagation is a nascent but burgeoning field, which considers the relationship between retrotransposon proliferation and host survival (Beauregard et al., 2008). Introns are mobile retroelements that parasitize their hosts, scavenging cellular enzymes, such as polymerases, nucleases and DNA ligase to complete the RNA-based mobility process (Smith et al., 2005). However, little is known of how these selfish elements respond to environmental or nutritional insults to their hosts. There must undoubtedly be a delicate balance whereby these retroelements are normally silenced by the cell to minimize deleterious effects, while being stimulated to spread when their niche is threatened.
The RNA of a mobile group II intron can integrate either site-specifically into a DNA homing site through a process known as retrohoming, or more randomly into ectopic sites through a process termed retrotransposition (Belfort et al., 2002; Lambowitz and Pyle, 2006). Both reactions, of which retrohoming is at least 100-fold more efficient, are catalyzed by a ribonucleoprotein (RNP) complex that consists of an intron-encoded protein (IEP) and the spliced intron lariat RNA. The group II intron from Lactococcus lactis, Ll.LtrB, is mobile in its native host and in Escherichia coli (Mills et al., 1997; Cousineau et al., 1998), reverse splicing into DNA. The Ll.LtrB IEP, called LtrA, has reverse transcriptase, RNA maturase and DNA endonuclease activities (Matsuura et al., 1997), which function in retromobility. In retrohoming, a nick introduced by the endonuclease, on the strand opposite to the integrated intron, generates a primer for cDNA synthesis, with the integrated intron serving as the template for cDNA synthesis. However, in certain cases the endonuclease activity is dispensable, particularly in retrotransposition, when replication forks act as primers for cDNA synthesis (Ichiyanagi et al., 2002; Zhong and Lambowitz, 2003).
Strikingly, in a recent screen of an E. coli mutant library for cellular functions involved in retromobility of the Ll.LtrB intron, one-half of the disruptions that decreased levels of retrohoming into the chromosome were in genes encoding global regulators (Coros et al., 2008). One of the disruptions in the library was in cyaA and four were in spoT, genes responsible for the synthesis of cAMP and ppGpp, respectively. These two small molecules play key roles in sensing the nutritional status of the cell; accordingly cAMP and ppGpp orchestrate switches in global gene expression. The cyaA gene encodes adenylate cyclase, which catalyses the conversion of ATP to its cyclic monophosphate derivative, cAMP. Then, cAMP interacts with the cAMP receptor protein (CRP) to control gene expression, in response to glucose starvation (Kolb et al., 1993). The cAMP-CRP complex is a major global transcription factor in E. coli, regulating approximately 200 genes (Martinez-Antonio and Collado-Vides, 2003). An even more extreme case of transcriptional reprogramming in E. coli occurs during the stringent response to amino acid starvation, when stable RNA synthesis is inhibited while biosynthetic genes are expressed (Cashel et al., 1996; Chatterji and Ojha, 2001). During the stringent response, cellular ppGpp levels increase, under the control of the relA and spoT genes. In this work, we show that retromobility of Ll.LtrB is stimulated by starvation, and that global regulators cAMP and ppGpp act at the level of the DNA target to coordinate intron-mediated genome restructuring.
RESULTS AND DISCUSSION
Retrohoming into the chromosome, but not into plasmids, is depressed in cyaA and spoT mutants
Host mutants that alter retrohoming of the Ll.LtrB intron were isolated in a phenotypic screen of a Tn5 E. coli transposon library, using cells containing the Ll.LtrB homing site in the chromosomal dedE gene (Fig. 1A) (Coros et al., 2008). The retrohoming assay for this screen used the pET-TORF/RIG donor plasmid, which contains a retromobility indicator gene (RIG) in the intron and LtrA expressed downstream of the intron (Fig. 1A) (Coros et al., 2005). The RIG cassette has a kanamycin resistance (kanR) gene in the opposite orientation to the Ll.LtrB intron. The kanR gene contains the group I td self-splicing intron in the same orientation as the Ll.LtrB intron, such that kanR is only expressed when the Ll.LtrB intron integrates into a DNA target via an RNA intermediate, thereby allowing direct selection for KanR (Fig. 1A).
Figure 1. Chromosomal but not plasmid retrohoming is defective in cyaA and spoT mutants.

A. Chromosomal retrohoming assay. With the pET-TORF/RIG donor (top), only the Ll.LtrB intron that has lost the group I intron via retrohoming can confer KanR, allowing direct selection. The small arrow under the group I intron indicates transcription in the same orientation as the Ll.LtrB intron. Wavy lines depict kanR transcripts, and E1 and E2 represent exon 1 and exon 2 of the −30/+15 homing site in the E. coli genome. After retrohoming, cells become KanR. B. Insertions in spoT and cyaA. The site for each transposon insertion is shown relative to E. coli map coordinates. C. Retrohoming levels are different in chromosomal and plasmid targets. Levels of retrohoming into the chromosomal (dedE) and plasmid targets are given for each disruption, relative to wild-type levels, which are ~10−6/chromosome, and ~5×10−5/plasmid. Chromosomal frequencies are from Coros et al. (2008). Strains MC1061-dedE::HS and MC1061 were used for retrohoming and retrotransposition experiments, respectively. D. Plasmid retrohoming assay. The donor plasmid is pLI1td+KR′ and the recipient is pLHS1. The group II intron is shaded gray. Retrohoming products that have lost the td intron are scored as KanR-CamR recombinants per recipient.
Among 10 mutants with reproducible reductions in the number of KanR retrohoming products, and whose phenotype was maintained when the homing site was located in the chromosomal ycjV gene, five had Tn5 disruptions in genes that encode small-molecule regulators, four in spoT and one in cyaA (Fig. 1B). The other five mutants had insertions in genes involved in RNA processing (pnp and trmE), energy metabolism (atpA), replication (rep), and a putative transporter (yidR) (Coros et al., 2008). Here we focus on the discovery that the levels of retrohoming into the chromosome in the cyaA and spoT mutants were depressed 5- to 12-fold relative to levels of ~10−6 in wild-type cells (Fig. 1C). We had previously shown that LtrA, intron RNA, splicing and plasmid copy number levels are similar to wild-type in the cyaA and spoT mutants (Coros et al., 2008).
To determine whether reduction in retrohoming was a replicon-specific effect, we tested retrohoming into a plasmid target using a donor that has LtrA encoded in the intron (Fig. 1D). Retrohoming levels into plasmid targets were not reduced in the cyaA and spoT mutant strains in this assay (Fig. 1C). This was also the case with a plasmid-to-plasmid real-time PCR assay using the same pET-TORF/RIG donor plasmid as for chromosomal retrohoming, and with a genetic assay with a different target replicon (bhr, Supplementary Fig. 1). The ability to support efficient plasmid retrohoming in the cyaA and spoT hosts suggests that the retrohoming machinery, namely the RNP comprising the intron RNA and IEP, is not limiting; rather, some downstream, target DNA-associated step is inhibited in chromosomal retrohoming in the cyaA and spoT mutants.
The cAMP-CRP complex, glucose starvation and retromobility
To determine whether the decrease in retrohoming in the cyaA::Tn5 mutant was due to reduced levels of cAMP in the cell, we supplemented the growth medium with cAMP (Fig. 2A). The addition of 5 mM or 10 mM cAMP to the medium rescued the retrohoming defect of the cyaA mutant. Therefore, the absence of cAMP inhibits retrohoming of Ll.LtrB into the chromosome.
Figure 2. The cAMP effect and glucose starvation.
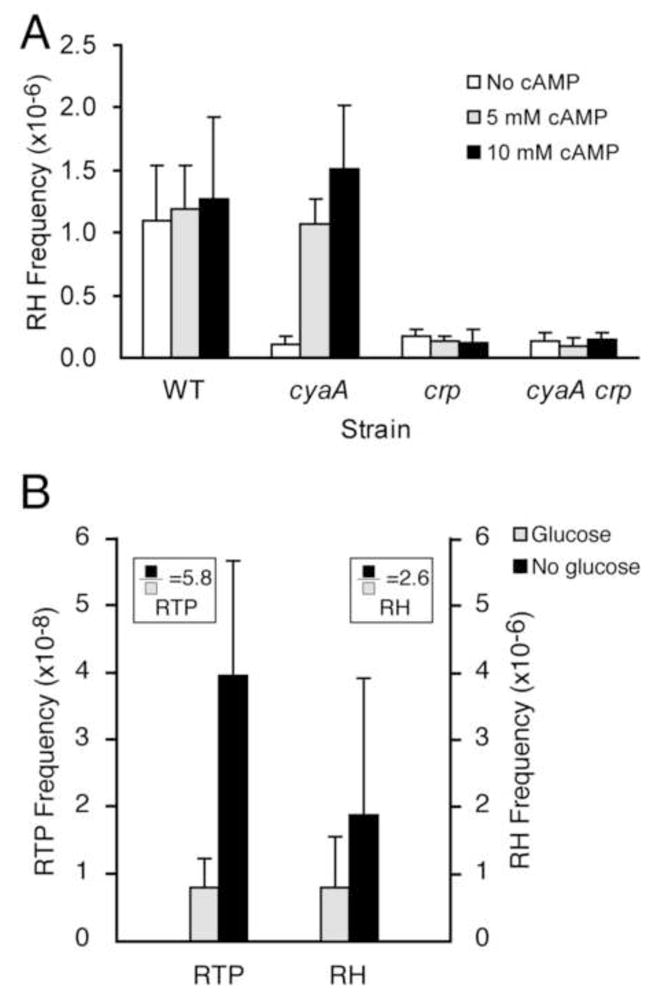
A. Suppression of the cyaA mutant with exogenous cAMP. Retrohoming (RH) frequency of the MC1061-dedE::HS (WT), MC1061-cyaA-dedE::HS (cyaA), MC1061-crp-dedE::HS (crp) and MC1061-cyaA-crp-dedE::HS (cyaA crp) strains was measured in the presence of exogenous cAMP (n ≥ 3; standard deviations shown as error bars). B. Effect of glucose starvation on retromobility. Retrohoming in BL21(λDE3)-dedE::HS and retrotransposition (RTP) in BL21(λDE3) were measured with (+) and without (−) glucose as carbon source and expressed as the ratio of −Glu/+Glu (boxed), with P = 0.0027 for RTP, and P = 0.048 for RH (two-tailed paired T-test); the standard deviations are shown as error bars. The P = 0.036 for the difference between fold-increases of 5.8 for RTP and 2.6 for RH (two-tailed independent T-test).
To establish that the decrease in retrohoming in the absence of cAMP is mediated through cAMP-CRP regulation, we deleted crp from the parental and cyaA mutant hosts. Consistent with a cAMP-CRP-mediated effect, the disruption of crp reduced retrohoming 10-fold (Fig. 2A). Furthermore, the addition of cAMP did not restore retrohoming levels in the cyaA mutant when the crp gene was deleted. These findings show that reduced retrohoming in the absence of cAMP is mediated through the cAMP-CRP global regulatory complex.
Because cAMP-CRP interaction is stimulated by glucose deprivation, we next determined chromosomal retromobility in cells starved for glucose (Fig. 2B). Retrohoming levels were increased at least 2.6-fold in cells starved for glucose. Moreover, in the absence of a chromosomal homing site, retrotransposition was elevated 5.8-fold during glucose starvation. These results confirm that activation of the cAMP-CRP complex by glucose starvation stimulates intron movement, particularly to ectopic sites (Fig. 2B, P = 0.036 for difference between retrohoming and retrotransposition).
The stringent response, ppGpp, and retromobility
SpoT is a bifunctional enzyme, ppGpp synthetase II, which can both synthesize and hydrolyze ppGpp, whereas RelA, ppGpp synthetase I, is involved only in ppGpp synthesis (Cashel et al., 1996; Murray and Bremer, 1996; Chatterji and Ojha, 2001). Interestingly, the host strain MC1061, in which our Tn5 mutagenesis was performed, contains a spoT1 mutation that eliminates the hydrolysis activity of SpoT (Laffler and Gallant, 1974) and relA1, a null mutation. Together these confer low levels of ppGpp (Metzger et al., 1989), but sufficient to support retromobility (Coros et al., 2008). The Tn5 insertions presumably abolish the ppGpp synthetase activity of SpoT, thereby reducing retrohoming frequencies.
Due to the complexity of ppGpp regulation in the MC1061 strain, we constructed single ΔrelA and ΔspoT mutants and a ΔrelA ΔspoT double mutant, to determine how a ppGpp null mutant affects retrohoming (Fig. 3A). Whereas deletion of relA had little effect (Fig. 3A, line 2), deletion of spoT resulted in a 10-fold reduction in chromosomal retrohoming (Fig. 3A, line 3). Furthermore, deletion of both relA and spoT decreased chromosomal retrohoming 100-fold (Fig. 3A, line 4), corroborating the finding that even small amounts of ppGpp facilitate retrohoming. All the mutants checked for plasmid copy number effects, Ll.LtrB RNA and splicing levels, and LtrA protein synthesis; fluctuations in levels were insufficient to account for the dramatic decrease in retrohoming (Supplementary Table S1).
Figure 3. Retromobility, ppGpp levels, and the stringent response.
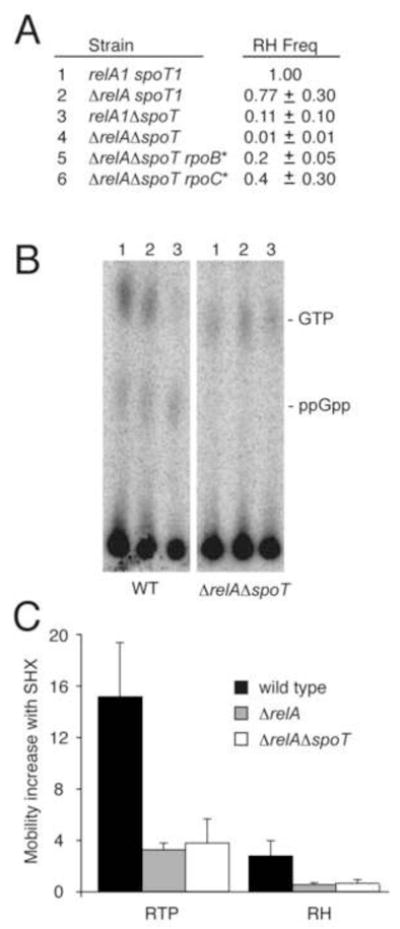
A. Retrohoming in mutants of the stringent response. Retrohoming frequency in mutant hosts was determined relative to “wild-type” MC1061 (relA1 spoT1), using the plasmid-to-chromosome assay (Fig. 1A), with the homing site located in the dedE gene. B. Levels of ppGpp in wild-type BL21(λDE3) and isogenic ΔrelA ΔspoT hosts. Lanes 1, 2 and 3 contain 0, 0.2 and 2 mg/ml SHX, respectively. Levels of ppGpp relative to GTP, expressed as ppGpp (100)/(ppGpp + GTP) were 22, 29 and 67 for WT and 4, 4 and 3 for ΔrelA ΔspoT (lanes 1–3). Results for the ΔrelA were similar to the ΔrelA ΔspoT host (not shown). C. Induction of retrotransposition (RTP) and retrohoming (RH) in mutants. Measurements were in BL21(λDE3) after treatment with 1 mg/ml SHX, and fold-increases were plotted relative to untreated samples (n = 3; standard deviations shown as error bars).
The majority of ppGpp effects in the cell are regulated at the transcriptional level through ppGpp interaction with RNA polymerase (RNAP) (Cashel et al., 1996; Paul et al., 2004). Therefore, to determine whether reduced retrohoming in the ppGpp-deficient strains is mediated through RNAP, we measured retrohoming frequencies in the ΔrelA ΔspoT homing-site strain that contains the suppressor mutations rpoB β RH454 (rpoB* ) and rpoC β′ Δ215–220 (rpoC*); these RNAP suppressors mimic ppGpp-mediated RNAP induction at stringently controlled promoters, enabling ppGpp null mutants to express stringently controlled genes (Bartlett et al., 1998). The rpoB* and rpoC* mutants increased retrohoming 20- and 40-fold, respectively, in the ΔrelA ΔspoT strain (Fig. 3A, lines 5 and 6). These findings suggest that ppGpp regulation of retromobility is being mediated through RNAP.
We next wished to explore group II intron retromobility in cells that are wild type for relA and spoT, and that can therefore induce ppGpp and the stringent response. We therefore induced the stringent response with serine hydroxamate (SHX) in the BL21(λDE3) host, which stimulated both ppGpp levels (Fig. 3B) and retrotransposition (Fig. 3C). Again the effect on retrotransposition was more dramatic than on retrohoming: a 15-fold increase in retrotransposition, and a 3-fold increase for retrohoming at the dedE locus was observed with SHX (Fig. 3C).
To extend the parallel between ppGpp levels and retromobility, we treated the ΔrelA and ΔrelA ΔspoT mutants with SHX. Both retrotransposition and retrohoming levels mirrored ppGpp levels, with < 20% induction in ΔrelA and ΔrelA ΔspoT hosts (Fig. 3B & 3C). Together, these results verify that ppGpp induction during cellular stress stimulates intron movement.
Potential mechanisms for cAMP and ppGpp control of retromobility
It has been proposed that ppGpp is the master molecule that coordinates the genetic transitions between feast and famine (Traxler et al., 2006). Moreover, there is interplay between the ppGpp and cAMP-CRP carbon-scavenging regulons; ppGpp maximizes induction of cAMP-CRP-controlled genes, and ppGpp accumulates in response to carbon limitation (Hardiman et al., 2007). However, the effect of ppGpp on retrohoming appears to be independent of crp, since addition of cAMP did not relieve the phenotype of a ΔrelA ΔspoT mutant (data not shown).
Given that stimulation of retromobility by activation of both the ppGpp and cAMP-CRP regulons is manifest into plasmid targets (Fig. 1C; Supplementary Fig. 1), we argue that RNP formation and the early steps in retrohoming are intact, and rather that a late step, related to the DNA replicon, is affected. We favor two possible explanations to account for the differences in chromosomal and plasmid targeting by the group II intron in the cyaA and spoT mutants. The first interpretation involves the bacterial nucleoid, which includes the chromosome but not small plasmids; the second relates to DNA replication, which is different for the chromosome and plasmids (Fig. 4). We note that the two explanations are not mutually exclusive.
Figure 4. Model for starvation-mediated activation of chromosomal retromobility.

Thick arrows represent increases (upward-directed) or decreases (downward-directed) in an entity. Thin lines represent regulatory flow, with an arrowhead representing positive control, and a bar representing negative control. Carbon-source or amino-acid starvation result in elevated cAMP and ppGpp levels, respectively. The action of cAMP is further potentiated by ppGpp stimulation of CRP. The small molecules are hypothesized to act at the level of the chromosomal target, by increasing the occurrence of replication forks and promoting a favorable nucleoid disposition (boxed), which facilitate retromobility as described in the text. H-NS and StpA also favor retromobility, likely via the nucleoid.
The first evidence for involvement of the nucleoid in retromobility came from experiments in which chromosomal retrotransposition of the Ll.LtrB intron was reduced in the absence of the nucleoid-associated proteins H-NS and StpA (Beauregard et al., 2006), whereas retrohoming into plasmids remained intact (Smith et al., 2005). Consistent with a nucleoid-specific effect, the cAMP and ppGpp mutants were also reduced in retromobility into the chromosome but not into plasmids. Furthermore, since the cAMP-CRP complex is involved in chromosome modeling by repositioning RNAP throughout the nucleoid, and RNAP positioning within the chromosome can be a driving force for nucleoid condensation (Grainger et al., 2005; Jin and Cabrera, 2006), it is likely that the cAMP- and ppGpp-mediated stress responses reduce chromosomal retrohoming through changes in nucleoid structure. Interestingly, there may also be a link between H-NS and cAMP-CRP as well as ppGpp regulons (Johansson et al., 2000). Together these results argue that cAMP- and ppGpp-mediated stress responses, which induce repositioning of RNAP throughout the nucleoid, affect retromobility through changes in nucleoid structure.
Second, group II intron retromobility in L. lactis, E. coli, and Sinorhizobium meliloti can be intimately related to DNA replication (Ichiyanagi et al., 2002; Zhong and Lambowitz, 2003; Martinez-Abarca et al., 2004), which is disrupted in the stress response. In E. coli, ppGpp negatively regulates dnaA, which encodes DnaA, the replication initiator protein, and cAMP binds DnaA, in a complex interplay through which DNA synthesis is inhibited (Cashel et al., 1996; Landoulsi and Kohiyama, 1999; Chatterji and Ojha, 2001). Furthermore, replication elongation is stalled shortly after amino acid starvation-induced ppGpp expression in Bacillus subtilis (Wang et al., 2007). Interestingly, group II introns lacking endonuclease (endo) function, and/or retrotransposing in specific bacterial hosts, target single-stranded DNA at replication forks, rather than double-stranded DNA. The nascent lagging strand is then used as a primer for second-strand cDNA synthesis (Ichiyanagi et al., 2002; Zhong and Lambowitz, 2003). Consistent with a role for replication forks, retrotransposition is increased 9-fold when the forks are stalled in a temperature-sensitive Topoisomerase IV mutant (Beauregard et al., 2006). Likewise, with an Endo− intron donor, which relies on replication forks for second-strand-synthesis, we have observed increases of ~8-fold in retrotransposition over increases with the Endo+ donor when cells are nutritionally stressed by the addition of SHX (DS, VRC and MB, unpublished). Together these observations argue strongly that stress-induced stalled replication forks present an opportunity for mobile introns to proliferate.
Introns as diversity-generating elements
Our report demonstrates that nutritional stress is a stimulus for intron movement, and for the first time indicates that ppGpp promotes retrotransposon activity. Interestingly, polyphosphates, which are also involved in stationary phase and in cellular stress, act in LtrA localization in E. coli (Zhao et al., 2008). Furthermore, cAMP has been implicated in the regulation of a putative LTR-retrotransposon in Schizosaccharomyces pombe (Labudova and Lubec, 1998), suggesting that this small molecule is a universal activator of retrotransposable elements across different domains of life.
Group II introns have evolved two distinct mobility mechanisms, site-specific retrohoming and ectopic retrotransposition, the latter apparently being more stress-sensitive. Similarly, the yeast Ty5 retrotransposon targets heterochromatin, but random integration is stimulated by the host stress response (Dai et al., 2007). It would therefore appear that these different classes of retroelements in phylogenetically diverse hosts, Ty5 retrotransposons in fungi and group II introns in bacteria (reviewed by Beauregard et al., 2008), have evolved a means for replenishing copies of themselves by specific targeting on one hand, and spreading in response to stress by ectopic integration on the other. What sets the cAMP-ppGpp/group II intron response apart is that it alters intron promiscuity at the level of the target, whereas the Ty5 response to stress is controlled by phosphorylation of the integrase. These different mechanisms underscore the evolutionary pressure on these disparate retroelements in times of stress to generate diversity and remodel genomes.
MATERIALS AND METHODS
Bacterial strains and plasmids
The E. coli strains (Supplementary Table 2A) used for retrohoming assays were derived from MC1061(λDE3) or BL21(λDE3). Strains for chromosomal retrohoming have the Ll.LtrB homing site in either the dedE gene (dedE::HS) or ycjV gene (ycjV::HS) (Coros et al., 2008). The ΔrelA and ΔspoT alleles were transduced into BL21(λDE3) using P1 lysates provided by Abbie Coros. The crp gene was disrupted in the MC1061(λDE3) dedE::HS strains by P1 transduction from MG1655 Δcrp (camR) (Kim et al., 1992). The tetR-linked rpoB* RH454 and rpoC* Δ215–220 mutations (Bartlett et al., 1998) were transferred with P1 into MC1061 dedE::HS ΔrelA ΔspoT. TetR colonies were verified by their ability to grow in minimal medium and therefore suppress the ΔrelA ΔspoT phenotype. The PCR primers used for strain construction and verification of site-specific integration are listed in Supplementary Table 3. Plasmids, including donor pLI1td+KR′ (Cousineau et al., 1998), are described in Supplementary Table 2B.
Retromobility assays
Retrotransposition and retrohoming into the chromosome were performed with donor plasmid pET-TORF/RIG-ampR (Fig. 1A & D) (Coros et al., 2005; Coros et al., 2008). Retrotransposition was carried out in the absence of a homing site, whereas retrohoming was performed in cells with the dedE::HS insertion. Briefly, overnight cultures containing the intron donor plasmid in LB-Amp medium were diluted 1/100, grown to an OD600 of 0.2, IPTG was added to 100 μM, the cultures grown for 3 h and plated on LB plates with and without 40 μg/ml kanamycin. Chromosomal retromobility frequencies were determined as KanR/total colonies. Retrohoming into plasmid targets used the pLI1td+KR′ donor (AmpR KanR), and the compatible pLHS1 recipient (CamR) as previously described (Cousineau et al., 1998). Retrohoming frequencies were calcuated, following enzymatic digestion of the donor, as KanR-CamR/CamR colonies.
Glucose and amino acid starvation, and ppGpp measurements
For glucose starvation, overnight cultures grown in M9 minimal medium, were diluted 1/100 in the same medium, minus or plus 0.2% glucose. Cultures were grown, induced and scored as described above. For amino-acid starvation, cells were grown in MOPS EZ rich medium (Teknova) to an OD600 of 0.2. SHX was added at 0.2 to 2.0 mg/ml, and cultures were grown for another 30 min. IPTG was then added at 100 μM and the cultures were grown for 2.5 h; KanR colonies were scored as above. Levels of ppGpp were measured in SHX-treated cultures labeled with H332PO4 (Cashel et al., 1993). At times indicated, 25 μl aliquots were removed into 25 μl of cold formic acid; after two freeze-thaw cycles, the cells were spun down and kept on ice. The samples were then spotted onto PEI-cellulose TLC plates and developed in 1.5 M phosphate buffer. Spots were visualized and quantitated on a Typhoon scanner.
Supplementary Material
Acknowledgments
We thank Melanie Barker, Art Beauregard, Joan Curcio, Keith Derbyshire, Hanna Engelberg-Kulka, Alan Grossman, and Bill Reznikoff for useful discussions and advice, and Maryellen Carl and John Dansereau for their help in preparing the manuscript and figures, respectively. We are also indebted to Richard Gourse for the crp and RNAP suppressor strains and Abbie Coros for the ΔrelA and ΔspoT P1 lysates. DNA sequencing was performed in the Wadsworth Center Molecular Genetics Core. This work was supported by NIH grants GM39422 and GM44844.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartlett MS, Gaal T, Ross W, Gourse RL. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- Beauregard A, Chalamcharla VR, Piazza CL, Belfort M, Coros CJ. Bipolar localization of the group II intron Ll. LtrB is maintained in Escherichia coli deficient in nucleoid condensation, chromosome partitioning and DNA replication. Mol Microbiol. 2006;62:709–722. doi: 10.1111/j.1365-2958.2006.05419.x. [DOI] [PubMed] [Google Scholar]
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Derbyshire V, Parker MM, Cousineau B, Lambowitz AM. Mobile introns: pathways and proteins. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. ASM Press; 2002. pp. 761–783. [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In Escherchia coli and Salmonella. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. cellular and molecular biology. Washington, DC: ASM Press; 1996. [Google Scholar]
- Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Current Opin Microbiol. 2001;4:160–165. doi: 10.1016/s1369-5274(00)00182-x. [DOI] [PubMed] [Google Scholar]
- Coros CJ, Landthaler M, Piazza CL, Beauregard A, Esposito D, Perutka J, Lambowitz AM, Belfort M. Retrotransposition strategies of the Lactococcus lactis Ll. LtrB group II intron are dictated by host identity and cellular environment. Mol Microbiol. 2005;56:509–524. doi: 10.1111/j.1365-2958.2005.04554.x. [DOI] [PubMed] [Google Scholar]
- Coros CJ, Piazza CL, Chalamcharla VR, Belfort M. A mutant screen reveals RNase E as a silencer of group II intron retromobility in Escherichia coli. RNA. 2008;14:2634–2644. doi: 10.1261/rna.1247608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, Lambowitz AM, Belfort M. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol Cell. 2007;27:289–299. doi: 10.1016/j.molcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci USA. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiman T, Lemuth K, Keller MA, Reuss M, Siemann-Herzberg M. Topology of the global regulatory network of carbon limitation in Escherichia coli. J Biotechnol. 2007;132:359–374. doi: 10.1016/j.jbiotec.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Ichiyanagi K, Beauregard A, Lawrence S, Smith D, Cousineau B, Belfort M. Multiple pathways for the Ll. LtrB group II intron include reverse splicing into DNA targets. Mol Microbiol. 2002;46:1259–1271. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- Jin DJ, Cabrera JE. Coupling the distribution of RNA polymerase to global gene regulation and the dynamic structure of the bacterial nucleoid in Escherichia coli. J Struct Biol. 2006;156:2842–2891. doi: 10.1016/j.jsb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Johansson J, Balsalobre C, Wang SY, Urbonaviciene J, Jin DJ, Sonden B, Uhlin BE. Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell. 2000;102:475–485. doi: 10.1016/s0092-8674(00)00052-0. [DOI] [PubMed] [Google Scholar]
- Kim J, Adhya S, Garges S. Allosteric changes in the cAMP receptor protein of Escherichia coli: hinge reorientation. Proc Natl Acad Sci USA. 1992;89:9700–9704. doi: 10.1073/pnas.89.20.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- Labudova O, Lubec G. cAMP upregulates the transposable element mys-1: a possible link between signaling and mobile DNA. Life sciences. 1998;62:431–437. doi: 10.1016/s0024-3205(97)01136-3. [DOI] [PubMed] [Google Scholar]
- Laffler T, Gallant JA. Stringent control of protein synthesis in E. coli. Cell. 1974;3:47–49. doi: 10.1016/0092-8674(74)90036-1. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Pyle AM. Group II introns: Ribozymes that splice RNA and invade DNA. In: Atkins JF, editor. The RNA World. Plainview, NY: Cold Spring Harbor Press; 2006. pp. 469–505. [Google Scholar]
- Landoulsi A, Kohiyama M. Initiation of DNA replication in Δcya mutants of Escherichia coli K12. Biochimie. 1999;81:793–940. doi: 10.1016/s0300-9084(99)00214-x. [DOI] [PubMed] [Google Scholar]
- Martinez-Abarca F, Barrientos-Duran A, Fernandez-Lopez M, Toro N. The RmInt1 group II intron has two different retrohoming pathways for mobility using predominantly the nascent lagging strand at DNA replication forks for priming. Nucleic Acids Res. 2004;32:2880–2888. doi: 10.1093/nar/gkh616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Current Opin Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, Cavanagh S, Dunny GM, Belfort M, Lambowitz AM. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989;264:21146–21152. [PubMed] [Google Scholar]
- Mills DA, Manias DA, McKay LL, Dunny GM. Homing of a group II intron from Lactococcus lactis subsp lactis ML3 . J Bacteriol. 1997;179:6107–6111. doi: 10.1128/jb.179.19.6107-6111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray KD, Bremer H. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J Mol Biol. 1996;259:41–57. doi: 10.1006/jmbi.1996.0300. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Smith D, Zhong J, Matsuura M, Lambowitz AM, Belfort M. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes Dev. 2005;19:2477–2487. doi: 10.1101/gad.1345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Chang DE, Conway T. Guanosine 3′,5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc Natl Acad Sci USA. 2006;103:2374–2379. doi: 10.1073/pnas.0510995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Niu W, Yao J, Mohr S, Marcotte EM, Lambowitz AM. Group II intron protein localization and insertion sites are affected by polyphosphate. PLoS Biol. 2008;6:e150. doi: 10.1371/journal.pbio.0060150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Lambowitz AM. Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription. EMBO Journal. 2003;22:4555–4565. doi: 10.1093/emboj/cdg433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


