Abstract
The aim of this work was to determine the best strategy to display antigens (Ags) on immunochemical devices to improve test selectivity and sensitivity. We comparatively evaluated five Trypanosoma cruzi antigenic recombinant peptides, chose the three more sensitive ones, built up chimeras bearing these selected Ags, and systematically compared by enzyme-linked immunosorbent assay the performance of the assortments of those peptides with that of the multiepitope constructions bearing all those peptides lineally fused. The better-performing Ags that were compared included peptides homologous to the previously described T. cruzi flagellar repetitive Ag (here named RP1), shed acute-phase Ag (RP2), B13 (RP5), and the chimeric recombinant proteins CP1 and CP2, bearing repetitions of RP1-RP2 and RP1-RP2-RP5, respectively. The diagnostic performances of these Ags were assessed for discrimination efficiency by the formula +OD/cutoff value (where +OD is the mean optical density value of the positive serum samples tested), in comparison with each other either alone, in mixtures, or as peptide-fused chimeras and with total parasite homogenate (TPH). The discrimination efficiency values obtained for CP1 and CP2 were 25% and 52% higher, respectively, than those of their individual-Ag mixtures. CP2 was the only Ag that showed enhanced discrimination efficiency between Chagas' disease-positive and -negative samples, compared with TPH. This study highlights the convenience of performing immunochemical assays using hybrid, single-molecule, chimeric Ags instead of peptide mixtures. CP2 preliminary tests rendered 98.6% sensitivity when evaluated with a 141-Chagas' disease-positive serum sample panel and 99.4% specificity when assessed with a 164-Chagas' disease-negative serum sample panel containing 15 samples from individuals infected with Leishmania spp.
Immunological methods are nowadays the elective procedure to diagnose Chagas' disease (http://whqlibdoc.who.int/trs/WHO_TRS_905.pdf). This illness, caused by infection with the parasite Trypanosoma cruzi, has been estimated to affect between 16 and 18 million people in Latin America alone, with a further 100 million considered at risk (http://www.globalhealthprogress.org/issues/ntds_who.php). Chagasic infection is diagnosed mostly when specific antibodies (Abs) against T. cruzi antigens (Ags) are detected in a patient's blood, by use of conventional serological methods, such as enzyme-linked immunosorbent assay (ELISA) and indirect hemmagglutination (IHA). Total homogenate of the parasite at the epimastigote stage provides Ags for serological tests, since it was proved to render the appropriate sensitivity to detect even very low Ab levels (19). However, when using this complex mixture of variable, largely undefined Ags, not only do specificity problems appear but also difficulties in standardizing the method (12, 44, 45). The current trend is to use recombinant proteins as sensitizing elements, since large amounts of them can be obtained in a highly purified form, and additionally, they can be synthesized from DNA sequences engineered to encode peptide fragments in which the specific regions responsible for cross-reactivity have been excised (2, 12, 40, 48). Thus, a number of recombinant peptides have been used for serological diagnosis, based upon their capability to improve the test performance in different aspects compared to the total parasite homogenate (TPH) (http://whqlibdoc.who.int/trs/WHO_TRS_905.pdf) (1-3, 8, 9, 15, 16, 18, 25, 26, 30, 34-38, 41-44, 50-53). Bearing in mind the sensitivity loss when using single recombinant peptides, several authors who have evaluated the performance of recombinant Ags in separate assays have suggested that a peptide mixture would reach a sensitivity equal to the sum of those of the individual Ags (3, 31, 34, 42, 50). Under this assumption, different peptide assortments have been used, improving in different ways the assay performance (3, 5, 8, 16, 31, 42, 51, 52). Alternatively, the use of multiepitope proteins expressing several unrelated antigenic determinants has also been proposed to enhance sensitivity (2, 25, 26). An argument supporting the use of chimeric molecules instead of the assortment of the epitopes expressed separately is that unique molecules facilitate the standardization procedure by lowering purification and immobilization steps and by balancing the number of epitopes on the surface of the immunoassay microplate (4, 12, 13). Recombinant DNA technology to obtain hybrid molecules has been used largely to obtain immunogens for vaccine preparation, but only a small number of authors have taken advantage of this approach in T. cruzi infection diagnosis. To the best of our knowledge, no work reporting on recombinant constructions compares systematically the performance of a multiepitope chimeric Ag with that of the mixture containing all the individual peptides that constitute the chimeric protein under study, and this work aims to cover this aspect.
For this purpose, we focused first on the rational selection of the antigenic peptides from those that have already proved to have diagnostic utility. We evaluated them alone, and we synthesized new multiepitope chimeric constructions by fusing the Ags that rendered better signal-to-noise ratios. Afterward, we analyzed the Ags' utility for T. cruzi infection diagnosis by comparing the ELISA performances displayed by the chosen synthetic peptides alone, their assortments, the new chimeric proteins with the selected peptides fused, and TPH. Finally, considering the permanent need to improve the selectivity and sensitivity displayed by T. cruzi Ags, the new, best-performing chimeric protein was preliminarily evaluated as a diagnostic tool with a 141-Chagas' disease-positive and 164-Chagas' disease-negative serum sample panel, and the results are discussed.
MATERIALS AND METHODS
Reagents.
All reagents were of analytical grade and were purchased from Sigma (St. Louis, MO), unless otherwise indicated. Molecular biology reagents were purchased from Promega (Madison, WI), unless otherwise stated.
TPH.
Epimastigotes of T. cruzi (Tulahuen strain) were grown in liver infusion tryptose medium supplemented with 10% fetal calf serum (7). TPH from epimastigotes was obtained by resuspension of the washed cells in 5 volumes of 1 mM N-p-tosyl-l-lysine chloromethyl ketone and 1 mM phenyl-methylsulfonyl fluoride in distilled water, frozen and thawed (four cycles), and subjected to posterior sonication (20 kHz, 30 W, 2 min).
Serum panel.
Serum samples from T. cruzi-infected patients (n = 141) were obtained from the Regional Hospital of Reconquista (Santa Fe, Argentina). The T. cruzi infection status of the patients was established by using two different conventional tests based on epimastigote TPH Ags, namely, commercial ELISA (Chagatest ELISA) and IHA (Chagatest IHA) from Wiener Lab (Argentina). The serological condition was ascertained when concordant results were obtained while performing both conventional tests, as established by standard technical procedures and acknowledged by the WHO (10) (http://whqlibdoc.who.int/trs/WHO_TRS_905.pdf). Chagas' disease-negative serum samples without other reactivity (n = 164) were obtained from blood donors from the same hospital. Donors of these negative samples were clinically healthy individuals whose serum samples rendered negative results when tests for syphilis, human immunodeficiency virus, and hepatitis A, B, and C were performed. A leishmaniasic panel (n = 15) was kindly provided by M. E. Brito. Samples consisted of sera from patients with clinical manifestations of cutaneous leishmaniasis who inhabited a region of endemicity (Recife PE, Brazil). These patients were studied at the Centro de Pesquisas Ageu Magalhães, Fundação Oswaldo Cruz, Recife, Brazil, and were determined to be negative for T. cruzi infection.
Construction of expression plasmids.
The nucleotide sequences of the T. cruzi genes encoding RP1, RP2, RP3, RP4, and RP5, homologous to Ags previously described (Table 1), were obtained from the GenBank database. As we have already demonstrated that proteins expressed using pET-32a vector show almost no unspecific reactions with the vector fusion protein TRX, we cloned all the sequences of interest using the mentioned vector (39). Genomic DNA from CL Brener strain epimastigotes was kindly provided by Patricio Diosque. This DNA was used as template for amplification of the selected encoded Ags, by means of standard PCR. Sequences of the primers used were as follows: RP1f (5′-GAATTCAAGAAGAAGCTTGCCGAC-3′), RP1r (5′-GAGCTCGCGTGCCAGCTCCTGTGC-3′), RP2f (5′-GAGCTCCTGATTGGCACGGAAGC-3′), RP2r (5′-GTCGACATCGGGCAAAATCAAAACC-3′), RP3f (5′-GAATTCAGCGTGCCTTGCCGCTGGAAG-3′), RP3r (5′-AAGCTTACGCACATCCTTCACAACAGG-3′), RP4f (5′-GAATTCAGGGCAGCTGAAGCCAC-3′), RP4r (5′-GCGGCCGCCTTCTCCGTCTCCACGGCC-3′), RP5f (5′-GAATTCAGCCGACGCCCAAAAAAGC-3′), and RP5r (5′-GTCGACGGCCTGTCCAAATAGTGA-3′). To build up CP2, the RP5 coding sequence was reamplified from the pET32a/RP5 construction using the primers RP5f (5′-GCGGCCGCAGCCGACGCCCAAAAAAGC-3′) and RP5r (5′-CTCGAGGGCCTGTCCAAATAGTGA-3′). The identity of each nucleotide sequence obtained was confirmed by automatic sequencing in each cloning step (Sequencing Service, GAD, Universidad Nacional de La Plata). Plasmidic DNA minipreparations were performed according to the procedure described by Sambrook et al. (46). Escherichia coli cells bearing the plasmids of interest were harvested overnight in LB medium with 0.1 mg ml−1 ampicillin at 37°C. Competent bacteria were transformed by one-pulse electroporation (2.5 kV, 25 μF) using a Bio-Rad Gene Pulser (Bio-Rad Laboratories Inc.), under the conditions specified by the manufacturer.
TABLE 1.
Comparison of recombinant and previously reported homologous Ags and their identity percentages
| Obtained Ag | Reported Ag (reference) | Identity (%) | Reference(s) reporting high diagnostic performance |
|---|---|---|---|
| RP1 | H49 (9) | 98 | 41, 50, 51 |
| Ag1 (27) | 97 | 15, 37, 38, 42 | |
| FRA (33) | 95 | 3, 8, 16, 44, 50 | |
| JL7 (35) | Sequence not indexed | 34, 50 | |
| RP2 | SAPA (27) | 94 | 15, 37, 42 |
| RP3 | Ag-36 (27) | 97 | 27 |
| MAP (30) | 97 | 52 | |
| JL9 (35) | Sequence not indexed | 34 | |
| RP4 | JL8 (35) | 86 | 34, 35, 50, 52 |
| CRA (33) | 88 | 3, 8, 9, 16, 44, 50 | |
| Ag30 (27) | Sequence not indexed | 15, 27, 37, 42 | |
| RP5 | B13 (18) | 93 | 18, 50, 51 |
| Ag2 (27) | 97 | 15, 25, 26, 37, 42, 43, 53 |
Protein expression and purification.
E. coli BL21(DE3) cells bearing the different plasmidic constructions, pET-32a/RP1, pEt-32a/RP2, pEt-32a/RP3, pET-32a/RP4, pET-32a/RP5, pET-32a/CP1, and pET-32a/CP2, were grown overnight in LB medium, supplemented with 0.1 mg ml−1 ampicillin at 37°C, with agitation. Protein expression was induced for 3 h with isopropyl-β-d-thiogalactopyranoside, washed with phosphate-buffered saline (PBS), centrifuged, and resuspended in 50 mM NaH2PO4 (pH 8), 300 mM NaCl, 10 mM imidazole buffer. The respective RP1, RP2, RP3, RP4, RP5, CP1, and CP2 peptides were purified with a Ni-nitrilotriacetic acid column (GE), as described elsewhere (2). Briefly, once supernatants were applied to the columns, they were washed with the same buffer and eluted into different fractions, using the mentioned buffer plus 50, 100, and 250 mM imidazole, consecutively. The Bradford assay was performed for protein quantification, with the absorbance being read at 590 nm (6). Purity of the recombinant proteins was analyzed by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and staining with Coomassie brilliant blue, according to the method described by Laemmli (32).
Protein antigenicity evaluation.
Polystyrene microplates (Costar) were sensitized with 1,000 ng TPH and 500 ng of recombinant protein per well, respectively, in carbonate buffer (pH 9.6). Peptide mixtures were prepared by adding equal amounts of each protein, and wells were then sensitized with 500 ng of the mixture. The microplates were incubated for 1 h at 37°C and washed thrice with 0.01% Tween in PBS, and the free polystyrene spaces were blocked with 5% skimmed milk in PBS for 1 h at 37°C. Microplates thus sensitized were incubated with a 1:100 dilution of human serum in 1% skimmed milk in PBS. After three washes with 0.01% Tween in PBS, microplates were incubated with peroxidase-conjugated goat anti-human immunoglobulin G (IgG), Fcγ (Zymed), and diluted 1:5,000 in 1% skimmed milk in PBS. CP2 evaluation was carried out using the same protocol, except that sensitized wells were dried before use, and serum samples and peroxidase-conjugated IgG were diluted 1:20 and 1:15,000, respectively, in an attempt to reproduce dilutions currently used in commercial kits. The reaction was developed using tetramethyl benzidine (Zymed) in H2O2, using 1 M H2SO4 as stopper.
Data analysis.
ELISA results, recorded as optical density (OD) at 450 nm, were distributed by using a scatter computer graphic software (GraphPad Prism version 2.00). All serum samples were evaluated in duplicate, with the result of the test being the mean OD value of these simultaneous determinations. ELISA cutoff values were calculated as the mean OD of the true negative serum samples plus 3 standard deviations of that mean. ELISA results were compared with the serologic status, previously confirmed by using two commercial kit assays, ELISA and IHA, according to the WHO acknowledged standard procedure (10) (http://whqlibdoc.who.int/trs/WHO_TRS_905.pdf). ELISA results were analyzed by plotting each positive result as the relative OD (+OD/cutoff, where +OD is the OD value of the positive serum sample and cutoff is the cutoff OD value). Inconclusive results were considered those produced by samples whose OD values fell into the undetermined zone, defined as a cutoff value of ±10%. Ag discrimination efficiency was evaluated by the formula +OD/cutoff, where +OD is the mean OD value of the positive serum samples tested. The discrimination limit was assessed by the formula +mOD/cutoff, where +mOD is the minimum OD produced by the least reactive Chagas' disease-positive serum sample. Sensitivity was expressed as 100 times the number of positive samples detected by using the assayed protein divided by the number of true-positive samples evaluated, confirmed as stated above (49). Specificity was expressed as 100× the number of negative samples detected by using the assayed protein divided by the number of true-negative samples evaluated, confirmed as stated above (49). Comparison and degrees of significance were assessed by Student's t test. The GraphPad Prism software was used to perform Student's t test to compare population distributions.
Nucleotide sequence accession numbers.
CP1, CP2, RP1, RP2, RP3, RP4, and RP5 were deposited in GenBank under the accession numbers FJ440556, FJ440557, FJ440558, FJ440559, FJ440560, FJ440561, and FJ440562, respectively.
RESULTS
Primary evaluation of the antigenic constructions.
The encoding nucleotide sequences of five tandemly repeated Ags previously described were cloned. Numerous authors reported appropriate sensitivities and specificities when using these Ags (Table 1), and a common agreement exists on their good ELISA performance. The sequences of the nucleotides obtained were compared with those previously reported. Their identity percentages are shown in Table 1. RP1, RP2, RP3, RP4, and RP5 displayed 1, 4.5, 2, 3, and 1 repetition, respectively.
The immunochemical performance of the obtained peptides was initially tested to evaluate them as candidates to be used alone, in mixtures, and in multiepitope constructions. The ELISA results for 10 positive and 10 negative serum samples showed that RP3 and RP4 led to a higher number of positive results falling into the undetermined zone than RP1, RP2, and RP5. A second purification step (PAGE with subsequent electroelution) was carried out to confirm that no protein contamination was responsible for the high RP3 and RP4 backgrounds. The results for the same Chagas' disease-negative samples reproduced those previously obtained with purified RP3 and RP4 with Ni-nitrilotriacetic acid columns.
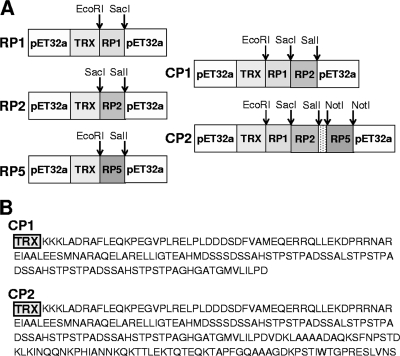
Chimeric proteins were subsequently designed using exclusively the peptides that performed better; CP1 was an assemblage of RP1 and RP2 peptides, whereas CP2 included these peptides plus RP5 (Fig. 1).
FIG. 1.
(A) Schematic representation of the construction and methods used to obtain the plasmids encoding the better-performing recombinant peptides selected, RP1, RP2, and RP5, and the chimeric proteins CP1 and CP2. (B) Amino acid sequences of the multiepitope chimeric proteins CP1 and CP2.
RP1, RP2, RP5, CP1, and CP2 were overexpressed, and after purification, the protein amounts obtained per liter of induced culture were ca. 300 mg, 60 mg, 90 mg, 120 mg, and 90 mg, respectively. All of the Ags were visualized as homogeneous bands when they were subjected to sodium dodecyl sulfate-PAGE and further stained with Coomassie blue.
Protein antigenicity comparison.
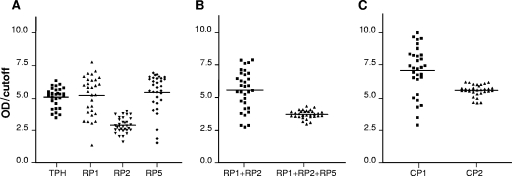
The antigenic performance of the selected peptides was evaluated when the Ags were used either individually (each peptide in a well), in mixtures (all the peptides in the same well), or combined in a unique chimeric construction. The same panel of 32 positive and 32 negative serum specimens was assayed, using as sensitizing Ags in ELISAs TPH, RP1, RP2, RP5, the RP1+RP2 mixture, the RP1+RP2+RP5 mixture, CP1, and CP2. Results for the relative OD distribution obtained for each Ag are shown in Fig. 2.
FIG. 2.
Relative OD distribution obtained for a panel of 32 Chagas' disease-positive serum specimens, using all the Ags studied. Horizontal lines show the discrimination efficiency values (the mean OD values of the positive samples tested divided by the cutoff value). (A) TPH and the isolated peptides RP1, RP2, and RP5. (B) RP1+RP2 mixture and RP1+RP2+RP5 mixture. (C) CP1 (fused RP1-RP2) and CP2 (fused RP1-RP2-RP5).
All positive chagasic serum samples displayed reactivity when assayed with every synthetic Ag. However, different distributions of the relative ODs were observed, rendering discrimination efficiency values of 5.022, 5.140, 2.870, and 5.390 (evaluated as +OD/cutoff) for TPH, RP1, RP2, and RP5, respectively (Fig. 2A). According to the Student t test, RP1 and RP5 behaved alike (P = 0.506), whereas RP2 turned out to be less antigenic than RP1 and RP5 (P = 0.0001, in both cases).
Comparison of the discrimination efficiency value of the RP1+RP2 mixture (Fig. 2B) with that of RP1 alone (Fig. 2A) indicated that they were similar (P = 0.281). However, the discrimination limit, +mOD/cutoff, was higher with the RP1+RP2 mixture than with RP1 alone.
Table 2 lists the cutoff values obtained for the selected Ags assayed, either alone, in mixtures, or as part of chimeric constructions and the discrimination limit (+mOD/cutoff) values produced by the least reactive serum. When analyzing the RP1+RP2+RP5 mixture performance versus the RP1+RP2 mixture performance, the cutoff value obtained for the former mixture was higher than that obtained for the latter (0.480 versus 0.163). This led to a lower discrimination efficiency value for the RP1+RP2+RP5 mixture (P = 0.0001) than for the RP1+RP2 mixture (Fig. 2B).
TABLE 2.
Cutoff values for a panel of 32 Chagas' disease-negative serum specimens and discrimination limits for the selected peptides alone, in mixtures, and in multiepitope chimeric proteins
| Ag | Cutoff (OD) | Discrimination limit (+mOD/cutoff)a |
|---|---|---|
| RP1 | 0.212 | 1.343 |
| RP2 | 0.260 | 1.597 |
| RP5 | 0.270 | 1.520 |
| RP1+RP2 | 0.163 | 2.724 |
| RP1+RP2+RP5 | 0.480 | 2.967 |
| CP1 (RP1-RP2) | 0.162 | 2.874 |
| CP2 (RP1-RP2-RP5) | 0.323 | 4.571 |
The minimum OD values produced by the least reactive serum samples divided by the cutoff value.
CP1 and CP2 antigenic performances were assessed, and the results are depicted in Fig. 2C. The CP1 construction, a unique chimeric molecule bearing both RP1 and RP2 epitopes, showed a discrimination efficiency (7.051) that was significantly higher than that exhibited by the RP1+RP2 mixture (5.564, P < 0.001). The discrimination efficiency shown by CP2, i.e., the chimera that includes the RP1, RP2, and RP5 peptides in a sole protein structure, was 5.543, a value considerably higher than that displayed by the RP1+RP2+RP5 mixture, 3.709, P < 0.0001 (Fig. 2B and C).
The comparison between CP1 and CP2 performances showed that the background produced by CP1, with a cutoff of 0.163, was lower than that obtained when using CP2, with a cutoff of 0.323 (Table 2). Consequently, CP1 discrimination efficiency was higher than that of CP2 (7.051 versus 5.564, respectively, P < 0.0001). However, it was also observed that low-reactivity serum samples produced higher relative ODs with CP2 compared to those with CP1 (Fig. 2C). In Table 2 it can also be seen that CP2 displayed the highest discrimination limit value produced by the least-reactive serum.
Assessment of the best multiepitope chimera sensitivity and specificity.
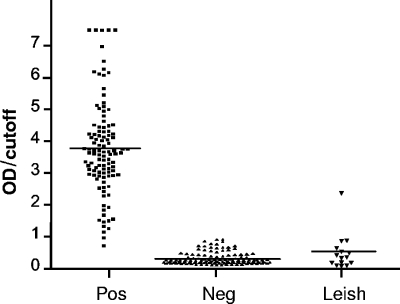
The diagnostic performance of CP2 was evaluated in terms of sensitivity and specificity by using a panel of 109 additional Chagas' disease-positive and 132 additional Chagas' disease-negative serum specimens and 15 serum specimens from patients with clinical manifestations of cutaneous leishmaniasis, without other clinical infection. Results are presented in Fig. 3. The sensitivity and specificity for the whole serum specimen panel studied were 98.6% and 99.4%, respectively, when using CP2.
FIG. 3.
Relative OD distribution obtained using CP2 for a panel of 109 Chagas' disease-positive (Pos) and 132 Chagas' disease-negative (Neg) serum samples, together with 15 serum samples from individuals infected with Leishmania spp. (Leish). Horizontal lines show the discrimination efficiency values (mean OD values of the positive samples tested divided by the cutoff value).
DISCUSSION
Selection of Ags.
Considering that repetitive T. cruzi Ags are immunodominant (17, 24, 33, 35), show high affinity for their specific Abs (11, 12, 24), depict the greatest variability among the different species (14), and display a high signal when an ELISA is performed, we built up peptides homologous to five previously described repetitive Ags, for which a general consensus on their good immunoassay performance exists. The high percentages of identity between these homologous sequences and the sequences we obtained stress the elevated conservation rate of the selected peptides (Table 1).
The primary ELISA results using the Ags independently revealed that RP1, RP2, and RP5 appropriately discriminated between Chagas' disease-positive and -negative samples. Conversely, RP3 and RP4 displayed high ODs for the negative chagasic serum specimen tested. Although these results differ from others previously reported for Ags homologous to RP3 and RP4 (references in Table 1), the discrepancy was attributed to a different antigenic performance originated by the use of another expression system, as used in our case, in agreement with other reports (39).
We therefore excluded RP3 and RP4 from further analysis and selected RP1, RP2, and RP5 to elucidate which of two different Ag-building strategies was more convenient: the use of either recombinant peptide mixtures or single chimeric proteins fusing the DNA sequences all together in the same molecule. To achieve this goal, we compared the ELISA performances of the Ags used separately, in mixtures (RP1+RP2 and RP1+RP2+RP5), or as part of unique fusion proteins CP1 and CP2 carrying RP1-RP2 and RP1-RP2-RP5, respectively.
Performance of single recombinant antigenic peptides versus their mixtures.
In general terms and supporting previous reports, the use of Ag mixtures enhanced the ELISA performance compared with use of the individual Ags (52). Thus, as can be seen in Fig. 2, the discrimination efficiency of the RP1+RP2 mixture was higher than those of isolated RP1 or RP2, whereas the RP1+RP2+RP5 mixture rendered higher relative ODs for low-reactivity serum samples than those displayed by individual RP1, RP2, or RP5 peptides. It should be noted, however, that the discrimination efficiency of the RP1+RP2+RP5 mixture was inferior to those of isolated RP1 and RP5. In point of fact, the RP5 presence notably increased OD values for low-reactivity serum samples exclusively, whereas the OD values for highly reactive samples were similar to the values obtained for the isolated peptides. Additionally, a higher cutoff value was obtained when using the RP1+RP2+RP5 mixture (Table 2) compared with the two-component mixture. Even though this may represent a drawback, the general ELISA performance was indeed improved because conflictive, undetermined results obtained with isolated Ags turned out not to be so when using the three-component mixture, which produced higher OD values that were farther from the cutoff value.
Umezawa et al. reported ELISA results from the use of both isolated antigenic peptides and their mixtures (52). From their results, it can be inferred that the assay sensitivity not only increased for low-reactivity serum samples, as we observed, but also for highly reactive serum samples, when using peptide mixtures. However, their results were obtained with different Ags (MAP, JL8, and TcPo instead of RP1, RP2, and RP5, which were used here). Moreover, they sensitized the microplates with a lower Ag amount (ca. 15 versus 500 ng of peptide mixture per well). Therefore, the reason for the discrepancy is not only the Ag nature but also that small amounts of adsorbed protein lead to the preferential recognition of high-affinity Abs, whereas large amounts of adsorbed proteins allow for the recognition of both low- and high-affinity Abs (28).
Peptide mixtures versus multiepitope chimeras.
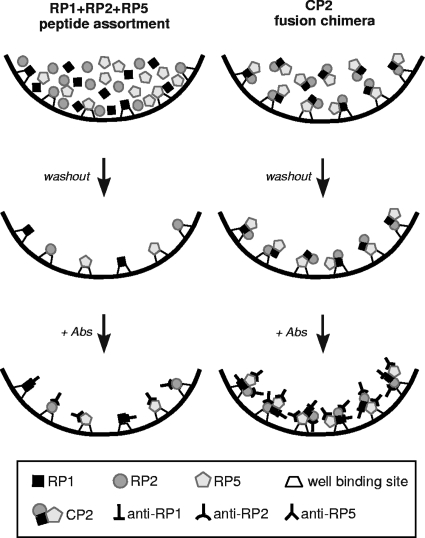
Although the current trend is to use chimeras as sensitizing Ags, the pros and cons regarding the use of chimeric fusion proteins versus recombinant peptide assortments still remain to be proved, since no previous work has carried out a systematic comparison between the two approaches. When analyzing the behavior of chimeras versus the behavior of peptide mixtures, the immunoassay merit figures (e.g., discrimination efficiency and discrimination limit) were boosted again for multiepitope chimeric constructions compared with peptide assortments. Indeed, CP1 discrimination efficiency was remarkably higher (ca. 25%) than that of the RP1+RP2 mixture (7.091 versus 5.614, respectively). Similarly, the CP2 discrimination efficiency was notably higher (ca. 52%) than that displayed by the RP1+RP2+RP5 mixture (5.591 versus 3.682, respectively). It has been proposed that peptides in mixtures may decrease their individual antigenicity once adsorbed on solid phases because of the blockage of essential chains (20-23, 47). Another hypothesis to explain these results is that when antigenic peptides present in the mixture adsorb to the well, they compete for the binding sites, leading to sensitivity loss. This phenomenon has also been proposed to explain the decrease of human IgG attachment to ELISA microplates when the Abs are part of a mixture of diverse molecules (29). On the other hand, when sensitizing microplates with chimeric constructions, the protein may adsorb to the well binding site through certain sites, leaving the rest of the molecule available to freely react without steric constrains. Therefore, even when some epitope blockage may occur, other epitopes could still be exposed appropriately to further interact with their specific Abs (Fig. 4). Hence, multiepitope proteins may render a greater available epitope-to-well active site ratio, which would eventually enhance the sensitivity of the assay.
FIG. 4.
Illustration of the well-sensitizing step, which is followed by the Ag-Ab reaction for the RP1+RP2+RP5 peptide mixture and the CP2 chimeric protein bearing the fused RP1-RP2-RP5 peptides.
From another point of view, our results also point to the less laborious and cheaper strategy, since fewer steps are necessary once the multiepitope chimera has been expressed. Indeed, production of multiepitope chimeras requires purification of only one protein, which substantially diminishes production costs compared to purification of several proteins. Moreover, standardization of the method is facilitated, since an equilibrated adsorption of the Ag to the immunochemical device is expected when using a multiepitope chimera, and consequently the final cost of production should lessen (12, 25).
Evaluation of CP1 and CP2 as T. cruzi infection diagnostic tools.
Contrary to our expectations, the performance comparison of CP1 versus CP2, bearing RP1-RP2 and RP1-RP2-RP5, respectively, revealed that RP5 inclusion into the chimeric construction produced a discrimination efficiency decrease, due to the cutoff value rise from 0.162 for CP1 to 0.323 for CP2. Indeed, this is the same phenomenon we observed when the behavior of peptide assortments was analyzed during assessment of the performances of the RP1+RP2 mixture versus the RP1+RP2+RP5 mixture. The cutoff value increased from 0.163 to 0.480 when RP5 was added to the two-component mixture. Hence, both cases indicated that the presence of RP5 together with the other Ags favored unspecific Ag-Ab binding. This phenomenon is perhaps the consequence of interpeptide interactions leading to some epitope conformational arrangement, which allows further recognition not only by specific anti-T. cruzi Abs but also by nonspecific Abs commonly present in healthy individuals.
Even though the discrimination efficiency diminished from 7.059 to 5.591 when the sensitizing Ag was changed from CP1 to CP2, it should be pointed out that every low-reactivity serum sample tested produced higher OD values with CP2 than with CP1. A central drawback when diagnosing chagasic infection is the occurrence of inconclusive results due to OD values falling into the undetermined zone. For that reason, the capability to produce OD values far away from the cutoff, even for low-reactivity samples, is a highly appreciated Ag feature. Bearing this in mind, and as a means to evaluate this attribute, we calculated the discrimination limit value, +mOD/cutoff, i.e., the minimum OD produced by the least reactive Chagas' disease-positive serum sample divided by the cutoff value for CP1, CP2, and TPH (Table 2). CP2 rendered the highest discrimination limit value (4.571) compared to CP1 (2.874) and to TPH (3.454). This is a remarkable feature, since recombinant proteins expose less epitope variety than that shown by TPH, for which a higher signal should be expected. A hypothesis to explain this is that CP2 antigenicity is higher than that of other parasite proteins also present in TPH, which compete for the microplate binding sites. Thus, the use of TPH may lead to smaller amounts of highly antigenic peptides attaching compared to the use of CP2. Indeed, this is an outstanding attribute related to CP2's potential to reduce the number of undetermined results.
To preliminarily assess the immunochemical behavior of the best Ag obtained, CP2, we performed ELISAs with a complementary serum panel, which included 15 serum samples from patients suffering from leishmaniasis (Fig. 3), a disease known to cause false-positive results for T. cruzi infection diagnosis. When evaluating these serum samples against TPH, 60% of them gave false-positive results (data not shown). However, the outcome obtained with CP2 showed that the specificity rose to 93% for these leishmaniasic-conflictive serum samples, with an overall specificity of 99.4% for the 164 negative serum samples assessed. Moreover, most of the negative serum samples assayed with CP2 rendered OD values that were clearly below the cutoff line, even those serum samples that previously fell into the undetermined zone with the other Ags.
Summarizing, the whole set of results obtained in the present work allows the following conclusions to be drawn. (i) The multiepitope fused Ags obtained, CP1 and CP2, display better ELISA performances than mixtures containing the same antigenic peptides, each one as a single entity. (ii) The pilot assessment of CP2 showed that the new chimeric protein that gathers RP1, RP2, and RP5 in the same molecule was antigenic enough to produce a suitable sensitivity to diagnose T. cruzi infection for the samples studied. Moreover, CP2 displayed discrimination efficiency similar to that of TPH, with the added value of a higher capacity to solve out undetermined results for the serum panel tested.
Acknowledgments
We are grateful to Patricio Diosque (Instituto de Patología Experimental, Universidad Nacional de Salta) for providing CL Brener strain epimastigote DNA, Maria Edileuza Brito for providing samples from patients with clinical manifestations of cutaneous leishmaniasis, and Marcelo Gabriel Roma (Instituto de Fisiología Experimental, IFISE, CONICET) for his suggestions and comments.
This work was funded by CONICET (PIP no. 5303), ANPCyT (PICTR2002-00057), and UNL (CAI+D 12/B608).
Footnotes
Published ahead of print on 1 April 2009.
REFERENCES
- 1.Aguillon, J. C., R. Harris, M. C. Molina, A. Colombo, C. Cortes, T. Hermosilla, P. Carreno, A. Orn, and A. Ferreira. 1997. Recognition of an immunogenetically selected Trypanosoma cruzi antigen by seropositive chagasic human sera. Acta Trop. 63159-166. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre, S., A. M. Silver, M. E. F. Brito, M. E. Ribone, C. M. Lagier, and I. S. Marcipar. 2006. Design, construction, and evaluation of a specific chimeric antigen to diagnose chagasic infection. J. Clin. Microbiol. 441043-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida, E., M. A. Krieger, M. R. Carvalho, W. Oelemann, and S. Goldenberg. 1990. Use of recombinant antigens for the diagnosis of Chagas disease and blood bank screening. Mem. Inst. Oswaldo Cruz 85513-517. [DOI] [PubMed] [Google Scholar]
- 4.Anandarao, R., S. Swaminathan, S. Fernando, A. M. Jana, and N. Khanna. 2005. A custom-designed recombinant multiepitope protein as a dengue diagnostic reagent. Protein Expr. Purif. 41136-147. [DOI] [PubMed] [Google Scholar]
- 5.Aubert, D., G. T. Maine, J. C. Hunt, L. Howard, M. Sheu, S. Brojanac, L. E. Chovan, S. F. Nowlan, and J. M. Pinon. 2000. Recombinant antigens to detect Toxoplasma gondii-specific immunoglobulin G and immunoglobulin M in human sera by enzyme immunoassay. J. Clin. Microbiol. 381144-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Camargo, M. E. 1964. Growth and differentiation in Trypanosoma cruzi. I. Origin of metacyclic trypanosomes in liquid media. Rev. Inst. Med. Trop. Sao Paulo 1293-100. [PubMed] [Google Scholar]
- 8.Carvalho, M. R., M. A. Krieger, E. Almeida, W. Oelemann, M. A. Shikanai-Yassuda, A. Ferreira, J. B. Pereira, A. Saez-Alquezar, P. E. Dorlhiac-Llacer, D. F. Chamone, and S. Goldenberg. 1993. Chagas' disease diagnosis: evaluation of several tests in blood bank screening. Transfusion 33830-834. [DOI] [PubMed] [Google Scholar]
- 9.Cotrim, P. C., G. S. Paranhos, R. A. Mortara, J. Wanderley, A. Rassi, M. E. Camargo, and J. F. da Silveira. 1990. Expression in Escherichia coli of a dominant immunogen of Trypanosoma cruzi recognized by human chagasic sera. J. Clin. Microbiol. 28519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cura, E., S. Wendel, F. P. Pinheiro, and M. Weinserbacher. 1996. Manual de procedimientos de control de calidad para laboratorios de serologìas de los bancos de sangre. Pan American Health Organization, Washington, DC.
- 11.DaRocha, W. D., D. C. Bartholomeu, C. D. Macedo, M. F. Horta, E. Cunha-Neto, J. E. Donelson, and S. M. Teixeira. 2002. Characterization of cDNA clones encoding ribonucleoprotein antigens expressed in Trypanosoma cruzi amastigotes. Parasitol. Res. 88292-300. [DOI] [PubMed] [Google Scholar]
- 12.da Silveira, J. F., E. S. Umezawa, and A. O. Luquetti. 2001. Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol. 17286-291. [DOI] [PubMed] [Google Scholar]
- 13.Dipti, C. A., S. K. Jain, and K. Navin. 2006. A novel recombinant multiepitope protein as a hepatitis C diagnostic intermediate of high sensitivity and specificity. Protein Expr. Purif. 47319-328. [DOI] [PubMed] [Google Scholar]
- 14.El Sayed, N. M., P. J. Myler, D. C. Bartholomeu, D. Nilsson, G. Aggarwal, A. N. Tran, E. Ghedin, E. A. Worthey, A. L. Delcher, G. Blandin, S. J. Westenberger, E. Caler, G. C. Cerqueira, C. Branche, B. Haas, A. Anupama, E. Arner, L. Aslund, P. Attipoe, E. Bontempi, F. Bringaud, P. Burton, E. Cadag, D. A. Campbell, M. Carrington, J. Crabtree, H. Darban, J. F. da Silveira, P. de Jong, K. Edwards, P. T. Englund, G. Fazelina, T. Feldblyum, M. Ferella, A. C. Frasch, K. Gull, D. Horn, L. Hou, Y. Huang, E. Kindlund, M. Klingbeil, S. Kluge, H. Koo, D. Lacerda, M. J. Levin, H. Lorenzi, T. Louie, C. R. Machado, R. McCulloch, A. McKenna, Y. Mizuno, J. C. Mottram, S. Nelson, S. Ochaya, K. Osoegawa, G. Pai, M. Parsons, M. Pentony, U. Pettersson, M. Pop, J. L. Ramirez, J. Rinta, L. Robertson, S. L. Salzberg, D. O. Sanchez, A. Seyler, R. Sharma, J. Shetty, A. J. Simpson, E. Sisk, M. T. Tammi, R. Tarleton, S. Teixeira, S. Van Aken, C. Vogt, P. N. Ward, B. Wickstead, J. Wortman, O. White, C. M. Fraser, K. D. Stuart, and B. Andersson. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309409-415. [DOI] [PubMed] [Google Scholar]
- 15.Frasch, A. C., and M. B. Reyes. 1990. Diagnosis of Chagas disease using recombinant DNA technology. Parasitol. Today 6137-139. [DOI] [PubMed] [Google Scholar]
- 16.Gomes, Y. M., V. R. Pereira, M. Nakazawa, D. S. Rosa, M. D. Barros, A. G. Ferreira, E. D. Silva, S. F. Ogatta, M. A. Krieger, and S. Goldenberg. 2001. Serodiagnosis of chronic Chagas infection by using EIE-Recombinant-Chagas-Biomanguinhos kit. Mem. Inst. Oswaldo Cruz 96497-501. [DOI] [PubMed] [Google Scholar]
- 17.Goto, Y., D. Carter, and S. G. Reed. 2008. Immunological dominance of Trypanosoma cruzi tandem repeat proteins. Infect. Immun. 763967-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruber, A., and B. Zingales. 1993. Trypanosoma cruzi: characterization of two recombinant antigens with potential application in the diagnosis of Chagas' disease. Exp. Parasitol. 761-12. [DOI] [PubMed] [Google Scholar]
- 19.Guhl, F., C. Jaramillo, J. C. Carranza, and G. A. Vallejo. 2002. Molecular characterization and diagnosis of Trypanosoma cruzi and T. rangeli. Arch. Med. Res. 33362-370. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez, M., L. Pozo, I. Gomez, and A. Melchor. 2000. Chimeric synthetic peptide as antigen for immunodiagnosis of HIV-1 infection. Biochem. Biophys. Res. Commun. 272259-262. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez, M., M. E. Selles, P. L. Pozo, I. Gomez, and A. Melchor. 2000. Antigenicity of chimeric synthetic peptides based on HTLV-1 antigens and the impact of epitope orientation. Biochem. Biophys. Res. Commun. 2761085-1088. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez, M. M., P. P. Castellanos, B. Y. Marquez, P. L. Pozo, N. J. Diaz, and L. J. Gonzalez Lopez. 2001. Chimeric synthetic peptides containing two immunodominant epitopes from the envelope gp46 and the transmembrane gp21 glycoproteins of HTLV-I virus. Biochem. Biophys. Res. Commun. 2891-6. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez, M. M., P. P. Castellanos, B. Y. Marquez, P. L. Pozo, N. J. Diaz, and L. J. Gonzalez Lopez. 2001. Chimeric synthetic peptides from the envelope (gp46) and the transmembrane (gp21) glycoproteins for the detection of antibodies to human T-cell leukemia virus type II. Biochem. Biophys. Res. Commun. 2897-12. [DOI] [PubMed] [Google Scholar]
- 24.Hoft, D. F., K. S. Kim, K. Otsu, D. R. Moser, W. J. Yost, J. H. Blumin, J. E. Donelson, and L. V. Kirchhoff. 1989. Trypanosoma cruzi expresses diverse repetitive protein antigens. Infect. Immun. 571959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houghton, R. L., D. R. Benson, L. Reynolds, P. McNeill, P. Sleath, M. Lodes, Y. A. Skeiky, R. Badaro, A. U. Krettli, and S. G. Reed. 2000. Multiepitope synthetic peptide and recombinant protein for the detection of antibodies to Trypanosoma cruzi in patients with treated or untreated Chagas' disease. J. Infect. Dis. 181325-330. [DOI] [PubMed] [Google Scholar]
- 26.Houghton, R. L., D. R. Benson, L. D. Reynolds, P. D. McNeill, P. R. Sleath, M. J. Lodes, Y. A. Skeiky, D. A. Leiby, R. Badaro, and S. G. Reed. 1999. A multi-epitope synthetic peptide and recombinant protein for the detection of antibodies to Trypanosoma cruzi in radioimmunoprecipitation-confirmed and consensus-positive sera. J. Infect. Dis. 1791226-1234. [DOI] [PubMed] [Google Scholar]
- 27.Ibáñez, C. F., J. L. Affranchino, R. A. Macina, M. B. Reys, U. Aslund, U. Petterson, and A. A. C. Frasch. 1988. Multiple Trypanosoma cruzi antigens containing tandemly repeated amino acid sequence motifs. Mol. Biochem. Parasitol. 3027-34. [DOI] [PubMed] [Google Scholar]
- 28.Kemeny, D. J., and S. J. Challacombe. 1988. ELISA and other solid phase immunoassays: theoretical and practical aspects. John Wiley & Sons, New York, NY.
- 29.Kenny, G. E., and C. L. Dunsmoor. 1983. Principles, problems, and strategies in the use of antigenic mixtures for the enzyme-linked immunosorbent assay. J. Clin. Microbiol. 17655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerner, N., P. Liegeard, M. J. Levin, and M. Hontebeyrie-Joskowicz. 1991. Trypanosoma cruzi: antibodies to a MAP-like protein in chronic Chagas' disease cross-react with mammalian cytoskeleton. Exp. Parasitol. 73451-459. [DOI] [PubMed] [Google Scholar]
- 31.Krieger, M. A., E. Almeida, W. Oelemann, J. J. Lafaille, J. B. Pereira, H. Krieger, M. R. Carvalho, and S. Goldenberg. 1992. Use of recombinant antigens for the accurate immunodiagnosis of Chagas' disease. Am. J. Trop. Med. Hyg. 46427-434. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lafaille, J. J., J. Linss, M. A. Krieger, T. Souto-Padron, W. de Souza, and S. Goldenberg. 1989. Structure and expression of two Trypanosoma cruzi genes encoding antigenic proteins bearing repetitive epitopes. Mol. Biochem. Parasitol. 35127-136. [DOI] [PubMed] [Google Scholar]
- 34.Levin, M. J., J. F. da Silveira, A. C. Frasch, M. E. Camargo, S. Lafon, W. M. Degrave, and R. Rangel-Aldao. 1991. Recombinant Trypanosoma cruzi antigens and Chagas' disease diagnosis: analysis of a workshop. FEMS Microbiol. Immunol. 411-19. [DOI] [PubMed] [Google Scholar]
- 35.Levin, M. J., E. Mesri, R. Benarous, G. Levitus, A. Schijman, P. Levy-Yeyati, P. A. Chiale, A. M. Ruiz, A. Kahn, M. B. Rosenbaum, et al. 1989. Identification of major Trypanosoma cruzi antigenic determinants in chronic Chagas' heart disease. Am. J. Trop. Med. Hyg. 41530-538. [DOI] [PubMed] [Google Scholar]
- 36.Levitus, G., M. Hontebeyrie-Joskowicz, M. H. Van Regenmortel, and M. J. Levin. 1991. Humoral autoimmune response to ribosomal P proteins in chronic Chagas heart disease. Clin. Exp. Immunol. 85413-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorca, M., A. Gonzalez, C. Veloso, V. Reyes, and U. Vergara. 1992. Immunodetection of antibodies in sera from symptomatic and asymptomatic Chilean Chagas' disease patients with Trypanosoma cruzi recombinant antigens. Am. J. Trop. Med. Hyg. 4644-49. [DOI] [PubMed] [Google Scholar]
- 38.Lorca, M., C. Veloso, P. Munoz, M. I. Bahamonde, and A. Garcia. 1995. Diagnostic value of detecting specific IgA and IgM with recombinant Trypanosoma cruzi antigens in congenital Chagas' disease. Am. J. Trop. Med. Hyg. 52512-515. [DOI] [PubMed] [Google Scholar]
- 39.Marcipar, I. S., M. L. Olivares, L. Robles, A. Dekanty, A. Marcipar, and A. M. Silber. 2004. The diagnostic performance of recombinant Trypanosoma cruzi ribosomal P2β protein is influenced by its expression system. Protein Expr. Purif. 341-7. [DOI] [PubMed] [Google Scholar]
- 40.Marcipar, I. S., C. Roodveldt, G. Corradi, M. L. Cabeza, M. E. Brito, L. M. Winter, A. J. Marcipar, and A. M. Silber. 2005. Use of full-length recombinant calflagin and its C fragment for improvement of diagnosis of Trypanosoma cruzi infection. J. Clin. Microbiol. 435498-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paranhos-Bacalla, G. S., M. R. Santos, P. C. Cotrim, A. Rassi, M. Jolivet, M. E. Camargo, and J. F. da Silveira. 1994. Detection of antibodies in sera from Chagas' disease patients using a Trypanosoma cruzi immunodominant recombinant antigen. Parasite Immunol. 16165-169. [DOI] [PubMed] [Google Scholar]
- 42.Pastini, A. C., S. R. Iglesias, V. C. Carricarte, M. E. Guerin, D. O. Sanchez, and A. C. Frasch. 1994. Immunoassay with recombinant Trypanosoma cruzi antigens potentially useful for screening donated blood and diagnosing Chagas disease. Clin. Chem. 401893-1897. [PubMed] [Google Scholar]
- 43.Peralta, J. M., M. G. Teixeira, W. G. Shreffler, J. B. Pereira, J. M. Burns, Jr., P. R. Sleath, and S. G. Reed. 1994. Serodiagnosis of Chagas' disease by enzyme-linked immunosorbent assay using two synthetic peptides as antigens. J. Clin. Microbiol. 32971-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saez-Alquezar, A., E. C. Sabino, N. Salles, D. F. Chamone, F. Hulstaert, H. Pottel, E. Stoops, and M. Zrein. 2000. Serological confirmation of Chagas' disease by a recombinant and peptide antigen line immunoassay: INNO-LIA Chagas. J. Clin. Microbiol. 38851-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salles, N. A., E. C. Sabino, M. G. Cliquet, J. Eluf-Neto, A. Mayer, C. Almeida-Neto, M. C. Mendonca, P. Dorliach-Llacer, D. F. Chamone, and A. Saez-Alquezar. 1996. Risk of exposure to Chagas' disease among seroreactive Brazilian blood donors. Transfusion 36969-973. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor, New York, NY.
- 47.Shah, K., C. Davis, J. Wilson, and B. Parekh. 1996. Chimeric synthetic peptides as antigens for detection of antibodies to HIV-1 and HIV-2. East Afr. Med. J. 7363-66. [PubMed] [Google Scholar]
- 48.Soto, M., J. M. Requena, L. Quijada, and C. Alonso. 1996. Specific serodiagnosis of human leishmaniasis with recombinant Leishmania P2 acidic ribosomal proteins. Clin. Diagn. Lab Immunol. 3387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tijsen, P. 1985. Practice and theory of enzyme immunoassays. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 50.Umezawa, E. S., S. F. Bastos, M. E. Camargo, L. M. Yamauchi, M. R. Santos, A. Gonzalez, B. Zingales, M. J. Levin, O. Sousa, R. Rangel-Aldao, and J. F. da Silveira. 1999. Evaluation of recombinant antigens for serodiagnosis of Chagas' disease in South and Central America. J. Clin. Microbiol. 371554-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Umezawa, E. S., S. F. Bastos, J. R. Coura, M. J. Levin, A. Gonzalez, R. Rangel-Aldao, B. Zingales, A. O. Luquetti, and J. F. da Silveira. 2003. An improved serodiagnostic test for Chagas' disease employing a mixture of Trypanosoma cruzi recombinant antigens. Transfusion 4391-97. [DOI] [PubMed] [Google Scholar]
- 52.Umezawa, E. S., A. O. Luquetti, G. Levitus, C. Ponce, E. Ponce, D. Henriquez, S. Revollo, B. Espinoza, O. Sousa, B. Khan, and J. F. da Silveira. 2004. Serodiagnosis of chronic and acute Chagas' disease with Trypanosoma cruzi recombinant proteins: results of a collaborative study in six Latin American countries. J. Clin. Microbiol. 42449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vergara, U., M. Lorca, C. Veloso, A. Gonzalez, A. Engstrom, L. Aslund, U. Pettersson, and A. C. Frasch. 1991. Assay for detection of Trypanosoma cruzi antibodies in human sera based on reaction with synthetic peptides. J. Clin. Microbiol. 292034-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]