Abstract
The human cytomegalovirus (CMV) pp65 protein contains two bipartite nuclear localization signals (NLSs) at amino acids (aa) 415 to 438 and aa 537 to 561 near the carboxy terminus of CMV pp65 and a phosphate binding site related to kinase activity at lysine-436. A mutation of pp65 with K436N (CMV pp65mII) and further deletion of aa 537 to 561 resulted in a novel protein (pp65mIINLSKO, where NLSKO indicate NLS knockout) that is kinaseless and that has markedly reduced nuclear localization. The purpose of this study was to biologically characterize this protein and its immunogenicity compared to that of native pp65. Unlike the native CMV pp65, following either DNA- or recombinant adeno-associated virus-based transduction of CMV pp65mIINLSKO into cells in vitro, the first observation of pp65mIINLSKO expression was in the cytoplasm and pp65mIINLSKO was expressed at higher levels than the native protein. The CMV pp65mIINLSKO mRNA was more abundant earlier than CMV pp65 mRNA (at 4 h and 8 h, respectively), but the half-lives of the proteins were the same. This modification altered the antigenic processing of CMV pp65 in vitro, as measured by the improved efficiency of cytotoxic killing in a pp65mIINLSKO-transduced human HLA A*0201 target cell line. In HHDII mice expressing HLA A*0201, pp65mIINLSKO was as immunogenic as CMV pp65. By RNA microarray analysis, expression of the CMV pp65mIINLSKO had less of an effect on cell cycle pathways than the native CMV pp65 did and a greater effect on cell surface signaling pathways involving immune activity. It is concluded that the removal of the primary NLS motif from pp65 does not impair its immunogenicity and should be considered in the design of a vaccine.
A major immunodominant protein of human cytomegalovirus (CMV) is the tegument protein CMV pp65 (UL83) (15, 16, 23, 36). The biologic function of CMV pp65 is unclear, but as a nucleotropic protein which enters the nucleus immediately after infection (10, 28, 31, 33), CMV pp65 binds to polo-like kinase 1 (PLK-1), an enzyme important in mitosis (11), and it is likely that the protein has specific effects on cell cycle events (14, 22, 30). Despite this potential for cell toxicity, CMV pp65 has been proposed to be a critical antigen in any anti-CMV vaccine (17, 32). CMV pp65 has been shown to have protein kinase activity (8, 39), and a mutation at a critical phosphate binding site (CMV pp65mII) removed the kinase activity without altering the antigenicity (39). UL83 is considered an early-late gene, with synthesis beginning between 12 and 24 h after infection, during which time the protein product accumulates in the nucleus. However, at late times after infection, this CMV pp65 is exported back to the cytoplasm by means of the exportin system (31). CMV pp65 contains elements of the prototypic nuclear localization signal (NLS) in which arginine and lysine predominate within a bipartite motif in which short regions of basic amino acids are separated by 10 or more nonconserved amino acids (13, 24, 35). The nuclear localization signals of CMV pp65 consist of at least two such motifs located in the carboxy-terminal region of the polypeptide (33). One of these (termed the A-B motif by Schmolke et al. [33] but simplified to region A in this paper) is a classic bipartite signal located at amino acids (aa) 415 to 438, in which two arginine- and lysine-rich motifs are separated by 18 aa. When this region A is deleted, however, there is very little change in nuclear localization, indicating that there are other components to the NLS. A second NLS of CMV pp65 consists of a basic region of amino acids between aa 537 and 561; this region was termed the C-D motif by Schmolke et al. (33) and is termed region B in this paper. When this region was deleted by Schmolke et al. (33), there was a more dramatic reduction in the nuclear localization of CMV pp65, suggesting that this is the dominant NLS. However, the combined deletion of both the A and the B regions leads to the more complete inhibition of nuclear localization (33).
Although CMV pp65 has been the prototypic antigen for the demonstration of CMV-specific T-cell immunity (2, 17, 34), it is likely that other proteins of CMV will be necessary for the development of a vaccine that generates humoral and cellular protection. CMV pp65-specific T-cell responses have been used for the development of other immunotherapeutic approaches to the control of CMV infection (4, 19, 20). Because of the importance of CMV pp65 in a vaccine strategy, we have explored the effects of mutating elements of the protein in ways that preserve the class I-restricted cytotoxic T-cell epitopes while removing biologic signals that could have effects on normal cells. For example, we have demonstrated the immunogenicity of a kinaseless CMV pp65 in which the phosphate binding lysine-436 is mutated (8). By use of this approach, a phase I trial of an anti-CMV vaccine by using a mutation at this same active site has been completed (38). However, these mutations of CMVpp65 remain nucleotropic and, thus, potentially toxic to normal cellular processes.
This current study shows that the removal of the nucleotropic properties of CMV pp65 resulted in earlier RNA and protein synthesis posttransfection and abundant accumulation in the cytoplasm, without alteration of the immunogenicity of the protein.
MATERIALS AND METHODS
Plasmid constructs.
Native CMV pp65 (pp65N) was derived from CMV strain Towne DNA, as described previously (25), and inserted into plasmid BSpp65 18.1. This served as a template in which K436N, a phosphate binding site, was mutated, creating a kinaseless CMV pp65 (pp65mII) (39). By using a DNA expression plasmid (pVAXintApp65mII), this mutation was shown to have no detectable effect on nuclear localization or immunogenicity (8, 39). NLS motif B (NLS-B) was then removed from pVAXintApp65mII by a bidirectional reverse PCR with the following primers: 5′-TTGCGCAGCGGGCTGCCATACG (positions 1601 to 1622) and 5′-GACCCACGTCCACTCAGACACGCGAC (positions 1741 to 1764). The PCR cycles were 94°C for 1 min, followed by 18 cycles of 94°C for 30 s, 55°C for 1 min, and 68°C for 10 min. The parental DNA template, pVAXintApp65mII, was then digested with DpnI. The PCR product was recircularized by ligation to obtain pVAXintApp65mIINLSKO (KO represents knockout), from which NLS-B was deleted (Fig. 1). By using Escherichia coli DH5α competent cells, transformed clones were isolated and were confirmed to contain the NLSKO deletion by sequencing. The expression was verified by transfection of HEK293 cells and immunostaining with anti-CMV pp65 monoclonal antibody as described below. The control plasmid contained the intron A sequence of the CMV immediate-early gene, as described previously (8).
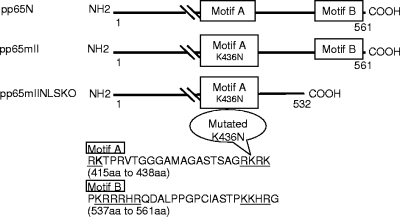
FIG. 1.
Schematic of the CMV pp65 expression vectors. The native CMV pp65 (pp65N) from the Towne strain (26) is shown with NLS-A and NLS-B. CMV pp65mII contains a mutation (K436N) in motif A (39), and CMV pp65mIINLSKO contains the same K436N mutation as pp65mII, but motif B is removed to obtain a truncated CMV pp65 (1 to 532 aa).
To prepare the shuttle plasmid for recombinant adeno-associated virus (rAAV) assembly, plasmid pCMVAAV2 (9) was cut with EcoRI and XbaI to accept the insert obtained by amplifying pVAXintApp65mIINLSKO with the following primers: EcoRI CMV pp65 (5′-TACGAATTCTACGCGCAGGCAGCATGGAG-3′) as the forward primer and CMV pp65 XbaI (5′-ACTTCTAGACCAAAAGTCGCGTGTCTGAGT-3′ as the reverse primer. The underlining indicates the restriction sites. The resulting plasmid, pCMVAAV-pp65mIINLSKO, does not contain intron A; and the integrity of the inverted terminal repeat was verified by BglII, MscI, and SmaI digestion, as described previously (9).
Encapsidation and purification of rAAV vector.
To assemble rAAV containing pp65mIINLSKO, the Helper-free system (Stratagene, San Diego, CA) was utilized with the following three plasmids: (i) pCMVAAV-pp65mIINLSKO flanked by the AAV inverted terminal repeat sequences; (ii) pAAV-RC containing the rep and cap genes from AAV type 2 (AAV2); and (iii) pHelper containing the E2A, E4, and VA RNA of adenovirus to provide the helper virus function (9). HEK293 cells were transfected with the three plasmids and collected after 72 h. The cell lysate was subjected to four rounds of freeze-thawing, sonicated six times, and then treated with Benzonase nuclease at 37°C for 1 h. The viral lysate was then purified with a ViraKit purification kit (Virapur, LLC, San Diego, CA), and the viral titer was measured both by the enumeration of the CMV pp65-positive plaques in transduced HT1080 cells stained with a CMV pp65-specific monoclonal antibody (Vector Laboratories Inc., Burlingame, CA) and by real-time PCR.
Cell culture and transfection.
The MRC-5 and HT1080 cells were grown in Dulbecco's modified Eagle's medium with a low glucose concentration supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). HEK293 cells were maintained in DMEM with a high glucose concentration with 10% FBS and 2 mM l-glutamine. HeLa cells were grown in RPMI with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were plated overnight and were transiently transfected by use of a calcium phosphate coprecipitation technique (Cell-Phect transfection kit; Pharmacia Biotech, Piscataway, NJ), according to the manufacturer's protocol. For rAAV transduction, MRC-5 or HT1080 cells were treated with permissive medium containing 40 mM hydroxyurea and 1 mM sodium butyrate for 5 to 6 h and were then transduced with rAAV for 16 to 48 h at a multiplicity of infection (MOI) of 10. At this MOI, the transduction efficiency was 80 to 100%. Three transduction experiments were performed for each condition.
Indirect immunofluorescence and immunoperoxidase staining.
The cells were plated on coverslips and transfected for 48 h, after which the coverslips were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde on ice for 20 min, and then permeabilized with 0.2% Triton X-100 in 1× PBS for 10 min at room temperature. After a thorough rinsing with PBS containing 0.05% Triton X-100 and 1% bovine serum albumin, the cells were incubated with primary mouse anti-CMV pp65 monoclonal antibody (Vector Laboratories Inc.) at a 1:200 dilution in PBS containing 0.5% Tween 20 (PBST) for 1 h at 37°C, washed, and then incubated with fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (IgG) specific for Fab (Sigma, St. Louis, MO) at 37°C for 30 min at a 1:160 dilution in PBST. During the last wash, 4′,6-diamidino-2-phenylindole (Sigma) in PBS (1:6,000) was added to stain the nuclei. Finally, the excess fluid was drained, the coverslip was mounted onto the slide, and photomicrographs were taken with an Olympus IX81 PA camera with Image Pro Plus 5.1 film (Media Cybernetics Inc.).
Immunoperoxidase staining of MRC-5 cells was performed as described above, except that the fixative consisted of 95% ethanol and 5% acetone and fixation was performed at room temperature for 15 min. After incubation with anti-CMV pp65 monoclonal antibody and then biotinylated anti-mouse IgG, the color was developed with avidin-peroxidase and the substrate aminoethylcarbazole (ABC kit; Vector Laboratories Inc.).
Total RNA extraction and reverse transcription-PCR (RT-PCR).
Total RNA from mock- or rAAV-transduced cells was extracted by using an RNeasy minikit (Qiagen, Valencia, CA), according to the manufacturer's instructions. One microgram total RNA was digested with DNase at 37°C for 1 h, and half of the RNA was converted to cDNA by using primer oligo(dT)12-18 (Invitrogen) and Superscript II reverse transcriptase (Invitrogen). The remaining RNA was used as a negative control without reverse transcriptase. One-tenth of the cDNA was subjected to PCR amplification with HotStart Taq DNA polymerase (Invitrogen). The PCR conditions were as follows: denaturation at 95°C for 2 min, followed by 40 cycles of 57°C for 30 s, 72°C for 30 s, and 95°C for 30 s. The primers used for CMV pp65 amplification were L1 (5′-AAAGAGCCCGACGTCTACTACACGT-3′) and L2 (5′-CCACGTACACCTTGACGTACTGGTC-3′), and those used for β-actin were 5′-GAAGTCCAGGGCGACGTAGCACAG-3′ and 5′-GCCCCCCTGAACCCCAAGGCCAACCG-3′ (40). The PCR products were resolved by electrophoresis on a 1.5% agarose gel containing ethidium bromide, and the bands were visualized under UV light.
To achieve better sensitivity and for quantitation, the same cDNA was also amplified by real-time quantitative PCR with a 7900 HT sequence detection system instrument (Applied Biosystems, Foster City, CA). The primers used were pp65F (5′-CAGATCTTCCTGGAGGTGCAA-3′) and pp65R (5′-CAGCCACGGGATCGTACTG-3′), and the probe was 5′-6-carboxyfluorescein-TACGCGAGACCGTGGAACTGCG-6-carboxytetramethylrhodamine-3′.
One microliter of each cDNA, 10 pmol primer, and 10 pmol probe mixed with the TaqMan universal PCR master mixture (Applied Biosystems) were amplified in a 30-μl reaction mixture. The results were analyzed with the software SDS2.3, provided with the ABI Prism 7900 HT sequence detection system instrument (Applied Biosystems).
Western blotting.
The rAAV- or mock-transduced cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.2, 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 1 mM EDTA containing 1 mM phenylmethylsulfonyl fluoride and 20 mM iodoacetamide plus protease inhibitors [1 μg/ml]), as described below. The protein concentrations were determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, IL). Equal amounts of protein were resolved by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a Hybond-ECL nitrocellulose membrane (Amersham, Piscataway, NJ). The membrane was blocked in 5% nonfat milk in PBST for 1 h at 37°C and incubated with primary antibody overnight at 4°C. After four washes with PBST, the blot was incubated with goat-anti mouse antibody conjugated to horseradish peroxidase (Sigma) at room temperature for 1 h in PBST and then detected with an enhanced chemiluminescence (ECL) Western blotting analysis system (Amersham). Briefly, the blot was submitted to the ECL detection reagent for 1 min and was then exposed to Hyperfilm, which was developed in a Kodak M35AX-OMAT processor. Epson Perfect 4490 Photo was used to scan the individual bands by densitometry.
Pulse-chase analysis.
MRC-5 cells were infected with rAAV pp65N, rAAV pp65mII, or rAAV pp65mIINLSKO at an MOI of 10 for 24 h and then rinsed twice and incubated for 1 h with methionine- and cysteine-free DMEM (Invitrogen) supplemented with 10% dialyzed fetal calf serum (Gibco-BRL) to which penicillin, streptomycin, glutamine, [l-35S]methionine, and [l-35S]cysteine Redivue Pro-mix in vitro cell labeling mix (0.1 mCi 35S/ml; GE Healthcare, Buckinghamshire, United Kingdom) were added. The cells were then washed and the medium was replaced with complete DMEM containing 5 mM cysteine and 5 mM methionine. At various time points, the cells were washed with ice-cold PBS and were then lysed in RIPA buffer. After a brief centrifugation at 14,000 × g for 10 min at 4°C, the cell lysate was pretreated with mouse IgG (Sigma) for 10 min, followed by incubation with 50 μl protein A-Sepharose fast-flow bead slurry (50%; GE Healthcare) at 4°C for 30 min. The supernatant was collected and immunoprecipitated by incubation with monoclonal anti-CMV pp65 antibody (Vector Laboratories Inc.) and then with 100 μl protein A beads for 3 h. After three washes with buffer A (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% Nonidet P-40, 0.1% SDS), the pellets were analyzed on a 10% SDS-polyacrylamide gel, dried, exposed to a PhosphorImager screen overnight, and analyzed with a Typhoon 9410 scanner (Amersham) and ImageQuant software.
In vitro cytotoxicity assay.
Antigen presentation by HLA A2-expressing MRC-5 cells was monitored by the standard 4-h 51Cr release assay, as described previously (8). MRC-5 cells were transduced with rAAV and were used 48 h later as target cells in triplicate wells. Cells of human CD8+ cytotoxic T-lymphocyte (CTL) clone 3.3F4, specific for the HLA A*02-restricted CMV pp65495 epitope, were used as effector T lymphocytes (5) to lyse the MRC-5 cells expressing the CMV epitope.
Mouse immunization.
Eight- to 10 week-old HHDII mice transgenic for the HLA A*0201 molecule (27) were immunized by priming the mice with DNA. followed by a booster with rAAV (9). In short, 50 μg of pVAXpp65mIINLSKO, pVAXpp65mII, or control pVAXintA, in addition to 50 μg of pVAX granulocyte-macrophage colony-stimulating factor (pVAXGM-CSF), were used for priming. This was followed, 4 weeks later, with a boost of 1 × 101 to 1 × 103 infectious unit (IU) of rAAV pp65mIINLSKO, rAAV pp65mII, or rAAV LacZ, as required by the experiment. The splenocytes were collected 20 to 30 days after the last injection and were processed either for ex vivo CTL detection or for in vitro stimulation (IVS) by using autologous blasts loaded with the CMV pp65495 epitope.
CTL detection in splenocytes collected from immunized HHDII mice.
The standard method of CTL detection in splenocytes collected from immunized HHDII mice consisted of a 4-h chromium release assay after one IVS at day 6 and a second IVS at day 12 postcollection, as described previously (8).
In addition, an enzyme-linked immunospot (ELISPOT) assay was performed at day 5 after stimulation by using in each well 5 × 105 cells that had been incubated overnight on a mixed cellulose ester membrane 96-well plate that had previously been coated with anti-gamma interferon (anti-IFN-γ; BD Pharmingen, Franklin Lakes, NJ). Spots were detected by incubation with biotinylated anti-IFN-γ (BD Pharmingen) overnight and were revealed with streptavidin-alkaline phosphatase (Vector Laboratories Inc.) and 5-bromo-4 chloro-3-indolylphosphate-nitroblue tetrazolium (Vector Laboratories Inc.).
In addition, a third method of detection of CTL activity was used: the intracellular cytokine assay by flow cytometry. For this method, the splenocytes were analyzed at day 5 after IVS, purified to remove dead cells by using a Ficoll density gradient, and incubated overnight in growth medium. The next day, the splenocytes were stimulated for 6 h with CMV pp65495 peptide. The cells were then permeabilized, stained with fluorescent labeled mouse anti-CD8 and anti-IFN-γ, and analyzed on a BD FACSCanto flow cytometer.
Microarray analysis.
The MRC-5 cells were transduced with rAAV IntA, rAAV pp65N, or rAAV pp65NLSKO for 48 h; and the total RNA from each transduction, done in triplicate, was purified by use of an RNeasy kit (Qiagen), according to the manufacturer's instructions. One microgram of each sample was processed by the GeneChip whole-transcript sense target labeling assay (Affymetrix, Inc., Santa Clara, CA). After reduction of the rRNA (Invitrogen) to remove most of the 18S and 28S rRNAs, the GeneChip whole-transcript cDNA synthesis kit, whole-transcript cDNA amplification kit, and whole-transcript terminal labeling kit (Affymetrix) were used for target preparation. Ten micrograms of cRNA (antisense RNA) was added to the second-cycle cDNA reaction, followed by fragmentation and the terminus-labeling process. Hybridization cocktails containing 2.5 μg of the fragmented, end-labeled cDNA were applied to the GeneChip Human Gene (version 1.0) ST arrays. The Genechip Human Gene (version 1.0) ST array was chosen because it offers whole-transcript coverage. The array contains 28,869 genes and approximately 26 probes spread across the full length of the gene. Hybridization was performed for 16 h. The arrays were washed and stained with the GeneChip Fluidics Station 450 by using the FS450_0007 script. The arrays were scanned by using the Affymetrix GCS 3000 7G and GeneChip operating software (version 1.4) to produce CEL intensity files. Raw intensity measurements of all probe sets were corrected for the background, normalized, and converted into expression measurements by using the Affymetrix expression console (version 1.1). The correlation between all replicates under each condition was above 0.94, which indicated that the experiment had good reproducibility. The Partek Genomic Suite (version 6.3) was then used to identify differentially expressed genes between each pair of sample groups. Genes with significantly differential expressions were selected by use of a cutoff of a P value of <0.05 and a 1.7-fold change in the level of expression. These genes were clustered with Pearson dissimilarity and average linkage in the Partek Genomic Suite. The genes in each cluster were analyzed by using David Bioinformatics Resources software (http://david.abcc.ncifcrf.gov/) to provide gene enrichment analysis, functional annotation clustering, and details of the gene families found within the gene clusters.
Microarray data accession number.
The data discussed in this report have been deposited in NCBI's Gene Expression Omnibus (GEO) database (6), are accessible through GEO series accession number GSE14347, and may be viewed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14347.
RESULTS
Removal of NLS from CMV pp65 protein.
The expression of the wild-type (WT) CMV pp65 protein in a transfected cell is restricted to the nucleus at early times and is detectable in the cytoplasm at late times (for a review, see reference 31). In an attempt to minimize the nuclear localization of the CMV pp65 protein and obtain primarily cytoplasmic expression, the NLS motifs were modified or removed from the CMV pp65 gene sequence. Motif A (location, aa 415 to 438) had already been mutated at the K436N site to remove the putative kinase domain II and was not further changed in the construct (Fig. 1, pp65mII) (39). The motif B (location, aa 537 to 561) was removed from the carboxy end of CMV pp65, resulting in a shorter form of the CMV pp65 (1 to 532 aa), called pp65mIINLSKO (see Fig. 1).
Cytoplasmic expression of pp65mIINLSKO and quantification by Western blotting.
The truncated pp65mIINLSKO DNA was inserted into the expression vector pVAX and into rAAV and tested for protein expression. Expression in transfected HeLa cells is shown in Fig. 2, for which the expression vector pVAXpp65mIINLSKO (Fig. 2B) and control vector pVAXpp65mII (Fig. 2A) were used. As expected, the alteration of the NLS motifs resulted in the cytoplasmic localization of the CMV pp65 protein, whereas CMV pp65mII localized to the nucleus. However, there were still stained nuclear cells with the deleted NLS motif at 48 h posttransfection (40% nucleated cells versus 60% cytoplasmic cells). The same was true for CMV pp65 expressed in rAAV-transduced MRC-5 cells (Fig. 2E and F). Although the stain was predominantly cytoplasmic with the removal of the NLS motif, there was still some nuclear localized CMV pp65 protein. The transduction efficiency was 80 to 100% at an MOI of 10.
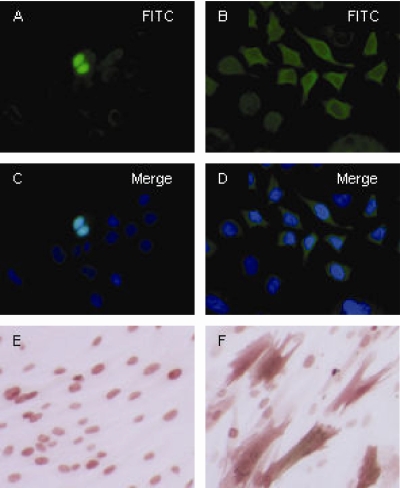
FIG. 2.
Cellular localization by immunostaining. HeLa cells were transfected with pVAXpp65mII (A and C) and showed a nuclear localization profile, and HeLa cells were transfected with pVAXpp65mIINLSKO (B and D) and primarily showed a cytoplasmic localization profile. MRC-5 cells were transduced with rAAV pp65mII (E) and showed a nuclear localization profile of CMV pp65, and MRC-5 cells were transduced with rAAV pp65mIINLSKO (F) and primarily showed a cytoplasmic localization profile.
Western blot analysis showed that the amount of CMV pp65 was increased in cells expressing pp65mIINLSKO, and to quantify this, serial dilutions of transduced MRC-5 cell extracts were separated by PAGE, as shown in Fig. 3. MRC-5 cells were transduced with rAAV pp65mII, rAAV pp65N, and rAAV pp65mIINLSKO for 48 h and lysed; and twofold dilutions of the lysate were analyzed by Western blotting. As shown in Fig. 3, pp65mIINLSKO yielded a CMV pp65-specific band that was present at an eightfold greater amount than either the pp65N or the pp65mII band (compare 1/8 and 1/64 serial dilutions, respectively). Of note, pp65N and pp65mII yielded the same amount of CMV pp65. The actin proteins were detected at up to 1/32 serial dilutions, which meant that less actin was present in the cellular lysate of the pp65mIINLSKO-transduced cells.
FIG. 3.
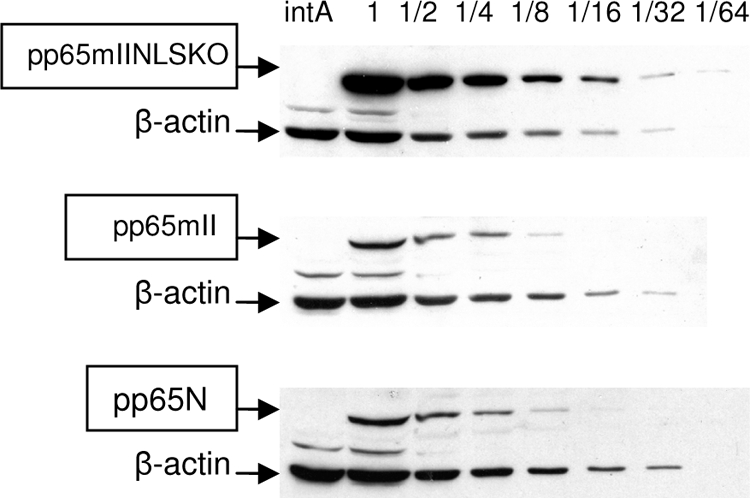
Quantification of pp65 protein by Western blotting. The expression of serial dilution of CMV pp65 in MRC-5 cells transduced at an MOI of 10 with rAAV pp65N, rAAV pp65mII, and rAAV pp65mIINLSKO is shown by Western blotting. The β-actin band served as a control for the amount of cellular protein.
Time required for CMV pp65mII and CMV pp65mIINLSKO expression.
The kinetics of the CMV pp65-specific mRNA and protein appearance could explain the relative abundance of pp65mIINLSKO in transduced cells. To check this, MRC-5 cells were transduced with rAAV pp65mII and rAAV pp65mIINLSKO and collected at 2, 4, 8, 16, 24, and 48 h postinfection. The input rAAV DNA was confirmed by quantitative PCR for all transductions, and no sample contained rAAV in an amount that varied by more than 10% of the amount of input rAAV, as measured by determination of the number of genome copies (gc) in each cell (data not shown). The mRNA was purified and amplified by RT-PCR for both CMV pp65- and actin-specific sequences, as described in Materials and Methods. The mRNA in the cells transduced with pp65mIINLSKO first appeared at 4 h postinfection and was present at 100 gc/μl, but only 10 gc/μl pp65mII and pp65N RNAs was present. As shown in Fig. 4A and B, the levels of pp65mII and pp65N reached 100 gc/μl 8 h later. Following the same pattern, the appearance of CMV pp65, as detected by Western blotting, first occurred at 16 h for pp65mIINLSKO and 24 h for pp65mII (Fig. 4C and D). This disparity in the time to the appearance of RNA and protein and the relative increase in the amount of CMV pp65, was observed in A293 cells and MRC-5 cells, but only the results from MRC-5 cell transduction are shown.
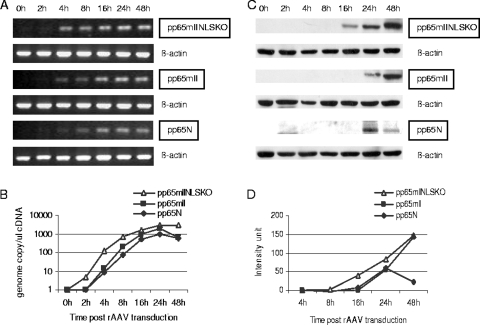
FIG. 4.
Time to CMV pp65mII and CMV pp65mIINLSKO expression. (A) mRNA purified from MRC-5 cells transduced with rAAV pp65mIINLSKO, rAAV pp65mII, or rAAV pp65N was collected at various times posttransduction and was analyzed by RT-PCR with CMV pp65-specific primers. β-Actin mRNA was used as a control in the RT-PCR. The product was run on a 1% agarose gel for visualization. (B) Quantitative PCR was used to show the number of gc/μl obtained after cDNA amplification at the indicated times posttransduction. (C) The level of expression of CMV pp65 by MRC-5 cells transduced with rAAV pp65mIINLSKO, rAAV pp65mII, and rAAV pp65N is shown by Western blotting. (D) Band intensity measured with Bio-Rad Quantity 1 software and graphed.
Stability of pp65mIINLSKO, pp65mII, and pp65N proteins.
To determine if the abundance of the pp65mIINLSKO protein could be explained by an increased protein stability in the cell, pulse-chase experiments were performed. MRC-5 cells transduced for 24 h were pulsed with [35S]cysteine and [35S]methionine for 1 h; and then the radiolabeled compound was chased with complete medium and the cells were collected at 0, 1, 4, 6, 8, and 24 h. As shown in Fig. 5, the pp65N, pp65mII, and pp65mIINLSKO proteins exhibited no difference in stability, even though more pp65 protein was present at the start of the chase with NLSKO (Fig. 5A). Counts, obtained with ImageQuant software, were normalized to 100 at time zero and are shown in Fig. 5B. All three pp65 proteins were extremely stable in transduced MRC-5 cells, and 50% degradation was not reached by 24 h.
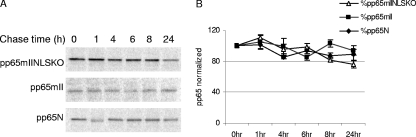
FIG. 5.
Stability of CMV pp65N protein compared to stabilities of pp65mII and pp65mIINLSKO determined by pulse-chase analysis. MRC-5 cells were transduced for 24 h with rAAV pp65N, rAAV pp65mII, and rAAV pp65mIINLSKO at an MOI of 10. The cells were pulsed for an 1 h with [35S]cysteine and [35S]methionine and chased with complete medium, and then the cells were collected at the indicated times for immunoprecipitation. (A) Radiolabeled proteins separated by PAGE analysis; (B) counts representing the mean and standard errors of the means of three experiments, with the values being normalized to a value of 100 at time zero.
Comparison of pp65mIINLSKO with pp65N by RNA microarray analysis.
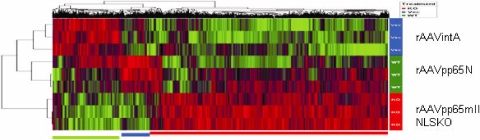
Finally, the question of whether pp65mIINLSKO had specific effects on the gene expression of transduced cells compared with the effects of native CMV pp65 was raised. Total RNAs were collected from each MRC-5 cell transduced for 48 h with rAAV IntA, rAAV pp65N, or rAAV pp65mIINLSKO and processed individually, in triplicate. A total of nine Affymetrix slides (GeneChip Human Gene, version 1.0, ST array) were hybridized with the processed product. Statistical analysis of the genes differentially expressed between each of the samples identified 989 genes, and the clustering diagram of these genes is shown in Fig. 6. The vector-only array was used as an rAAV reference for the other two constructs. Three distinct clusters were defined according to the downregulation of cellular genes by both pp65N (WT) and pp65mIINLSKO (KO) (176 probe sets representing 108 annotated genes in the David Bioinformatics Resources software; see the green cluster in Fig. 6), upregulation by pp65N but downregulation by pp65mIINLSKO (110 probe sets representing annotated 80 genes in the David Bioinformatics Resources software; see the blue cluster in Fig. 6), and upregulation of cellular genes by both constructs (703 probe sets representing annotated 389 genes in the David Bioinformatics Resources software; see the red cluster in Fig. 6). Table 1 shows the gene families affected in each cluster, as determined by the use of the David Bioinformatics Resources software. The enrichment score is a measure of the difference in P values defining differences within the cluster of gene families induced during transduction by native CMV pp65 and the NLS mutant. The green cluster (see the green bar in Fig. 6) indicated that 108 genes were downregulated, and within this group, 62 genes were involved in cellular metabolism and 34 genes were involved with nuclear proteins. In addition, the ZNF zinc finger family of proteins, the retinoblastoma binding protein 8 (RBBP8), and the E2f transcription factor were among the genes downregulated by both CMV pp65 and the NLSKO mutant. A blue cluster of genes that were upregulated by pp65N and downregulated by pp65NLSKO involved 28 genes of the mitotic cell cycle; 8 genes involved with chromosome segregation; 5 genes involved in the cell cycle check point; and others arbitrarily assigned to functions such as intracellular localization (17 genes), transport (11 genes), and ATP binding (15 genes). Here, the highest enrichment score was seen (13.65), confirming a role of CMV pp65 in cell cycle regulation, nuclear localization, and transport (14, 22, 30). Among the genes involved in this cluster were PLK-1, KIFxx (kinesin family), CDCA2 (cell division cycle associated 2), CCNB2 (cyclin b2), TOP2A (topoisomerase [DNA] II), and ABCA13/5 in the ATP-binding cassette transporter family.
FIG. 6.
Effects of pp65N and pp65mIINLSKO on cellular mRNA regulation by microarray analysis. Affymetrix slides (GeneChip Human Gene, version 1.0, ST) were hybridized with the mRNA of MRC-5 cells transduced with rAAV IntA, rAAV pp65N, and rAAV pp65mIInLSKO. The 989 probe sets were identified as being differentially expressed, and the hierarchical clustering of these genes is shown as a heat map with the three clusters labeled with colored bars: blue (pp65N and pp65 KO downregulated), green (pp65N upregulated and pp65 KO downregulated), and red (pp65N and pp65 KO upregulated). David Bioinformatics Resources software was used to determine the families of genes shown in Table 1. Data from the microarray are available under GEO accession number GSE14347.
TABLE 1.
Description of cluster families in pp65 WT versus NLSKOa
| Green cluster
|
Blue cluster
|
Red cluster
|
||||||
|---|---|---|---|---|---|---|---|---|
| Gene family affected (n = 108 genesb; ↓WT and KO↓) | No. of genes | Enrichment scorec | Gene family affected (n = 80 genes; ↑WT and KO↓) | No. of genes | Enrichment score | Gene family affected (n = 389 genes; ↑WT and KO↑) | No. of genes | Enrichment score |
| Cellular metabolism | 62 | 3.34 | Mitotic cell cycle | 28 | 13.65 | Glycoprotein | 143 | 12.32 |
| Nucleus | 34 | 1.65 | Cell cycle | 28 | 13.65 | Signal | 102 | 12.32 |
| Zinc finger C2H2 | 7 | 1.65 | Chromosome segregation | 8 | 5.84 | Secreted | 58 | 5.61 |
| Regulation of transcription | 17 | 1.65 | Cell cycle checkpoint | 5 | 4.87 | Signal transduction | 76 | 4.43 |
| Apoptosis | 8 | 0.63 | Microtubule cytoskeleton | 7 | 3.43 | Plasma membrane | 97 | 4.38 |
| Protein kinase | 5 | 0.77 | Localization | 17 | 3.43 | Cell differentiation | 52 | 4.72 |
| Cell cycle | 4 | 0.38 | Transport | 11 | 3.43 | Complement pathway | 5 | 2.47 |
| ATP binding | 15 | 3.57 | Humoral immune response | 5 | 2.47 | |||
| Chromosome | 11 | 2.12 | Immunoglobulin domain | 21 | 2.18 | |||
| Nuclear protein | 33 | 2.37 | Response to stimulus | 27 | 2.72 | |||
| Cellular process | 57 | 2.37 | Immune response | 26 | 2.47 | |||
| Immunoglobulin-like fold | 25 | 2.18 | ||||||
| Adaptive immune response | 9 | 2.47 | ||||||
| Innate immune response | 6 | 2.47 | ||||||
| Regulation of T-cell proliferation | 5 | 1.38 | ||||||
WT is pp65N; KO is pp65mIINLSKO. ↓, downregulation; ↑, upregulation.
Number of genes and gene families determined with David Bioinformatics Resources software.
The enrichment score is the geometric mean of each gene's P value in the cluster of the gene family.
The red cluster was the largest and contained 389 genes upregulated by both pp65N and pp65mIINLSKO. The enrichment score was 14.83. These genes comprised the glycoprotein families (143 genes); signaling pathways (102 genes); and also immune pathways, such as the complement pathway (5 genes), the humoral immune response (9 genes), the immunoglobulin domain (21 genes), and the response to stimulus (27 genes) (Fig. 6; Table 1). The immune response genes upregulated in this cluster included several which exhibited immunoglobulin-like structures, such as HLA-DQA2, CD1C, CD86, CD3e, CEACAM1, IGSF9B; other proteins involved in innate immunity; and the complement pathway proteins (C1QB, C2, C1QC, CR2, C4BPA).
Peptide presentation as measured by a 51Cr release assay in vitro.
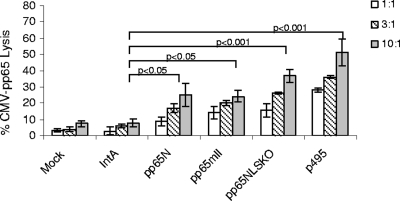
Since pp65mIINLSKO was more abundant in the cytoplasm, the next question was whether this immune response could be shown by increased antigen processing and presentation on the HLA of target cells. The human CD8+ CTL clone (clone 3.3F4), specific for pp65495 peptide presentation on the HLA A*0201 molecule (5), was used to determine the surface expression of the pp65495 peptide. MRC-5 cells expressing HLA A*0201 were transduced with rAAV pp65intA, rAAV pp65N, rAAV pp65mII, and rAAV pp65mIINLSKO. The cells obtained at effector cell/target cell ratios of 1:1, 3:1, and 10:1 are shown in Fig. 7. Statistical significance, as determined by analysis of variance, was reached for all groups by comparison of the results with those for the control vector, rAAV IntA. The level of the significant difference for positive control MRC-5 cells loaded with peptide pp65495 was the same as that as that for the MRC-5 cells transduced with pp65mIINLSKO (P < 0.001); the level of the significant difference for MRC-5 cells transduced with pp65N and pp65mII was lower (P < 0.05).
FIG. 7.
In vitro 51Cr release assay. A human CD8+ CTL clone specific to CMV pp65495 peptide (clone 3.3F4) was used as the effector cell. The target cells were MRC-5 cells transduced with rAAV IntA, rAAV pp65N, rAAV pp65mII, and rAAVpp65mIINLSKO or mock transduced for 48 h or were loaded with the pp65495 peptide as the control. The experiment was repeated three times, and the means ± standard errors are shown. The P values were calculated by one-way analysis of variance by the Bonferroni multiple-comparison test (with GraphPad Prism, version 5, software).
Prime-boost vaccination in HHDII mice.
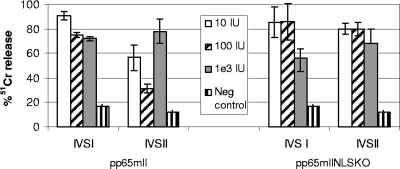
pp65mIINLSKO was further evaluated in vivo in HHDII mice transgenic for HLA A*0201. The vaccination consisted of a prime-boost strategy, as described for the pp65mII constructs (9). In this study, the mice were primed with DNA consisting of 50 μg pVAXpp65mIINLSKO or 50 μg pVAXpp65mII to which 50 μg pVAXGM-CSF was added as the adjuvant. Four weeks later, they were boosted with serial dilutions ranging from 1 × 101 to 1 × 103 IU rAAV expressing the same CMV pp65 mutant genes. Twenty days later, the splenocytes were tested by the chromium release assay after 6 days (IVS I) or 12 days (IVS II) of stimulation in culture with autologous blast cells loaded with the pp65495-specific peptide (Fig. 8). The response to pp65mIINLSKO reached 100% 51Cr release with a low vaccine dose of rAAV (10 to 100 IU) 6 days after IVS (Fig. 8, right panel, IVS I) and was down to 80% at day 12 after IVS (Fig. 8, right panel, IVSII). The CTL immune response was fairly similar whether pp65mIINLSKO or pp65mII was used and ranged from 30% to 100% when the means for two to four mice were used. The control mice immunized with the vector only showed less than 20% 51Cr release, and this was considered a negative result. We conclude that in HHDII mice expressing HLA A*0201, pp65mIINLSKO was as immunogenic as CMV pp65.
FIG. 8.
Prime-boost vaccination of HHDII mice with pp65mII and pp65mIINLSKO. HHDII mice transgenic for HLA A*0201 were inoculated subcutaneously with 50 μg pVAXpp65mII or pVAXpp65mIINLSKO and 50 μg pVAXGM-CSF. This was followed 4 weeks later by the intramuscular injection of a total of 1 × 101, 1 × 102, or 1 × 103 IU of rAAV pp65mII or rAAV pp65mIINLSKO in each thigh muscle. Controls received an injection of 1 × 105 IU (the highest experimental dose) of rAAV LacZ. The splenocytes were collected 20 days after the last injection, and a chromium release assay was performed after day 6 (IVS I) and day 12 (IVS II), as described in Materials and Methods. The means and standard errors for two to four mice per dilution are shown (effector cell/target cell ratio, 100:1). The experiments were repeated three times.
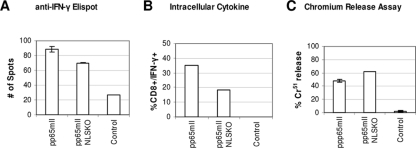
In addition to the 51Cr release assay, CTL responses were also detected by the ELISPOT assay for the IFN-γ response after in vitro stimulation and by an intracellular cytokine assay to measure the number of cells releasing IFN-γ in CD8+ cells by fluorescence-activated cell sorter analysis. This is shown in Fig. 9 for mice (two mice in each group) immunized with DNA followed by a booster of 10 IU rAAV pp65mII or rAAV pp65mIINLSKO. In summary, it was confirmed that the two forms of CMV pp65 have similar immunogenicities.
FIG. 9.
Immune response with a low dose of the rAAV pp65 vaccine. Mice were immunized with 50 μg of pVAXpp65mII, 50 μg of pVAXpp65mIINLSKO, or 50 μg of pVAXintA. They were boosted at 4 weeks with 10 IU rAAV pp65mII, 10 IU of rAAV pp65mIINLSKO, or 100 IU of rAAV LacZ. Splenocytes were collected at day 30 and stimulated with CMV pp65495 peptide for 5 days. (A) Anti-IFN-γ ELISPOT assay; (B) intracellular cytokine (IFN-γ) detection by fluorescence-activated cell sorter analysis; (C) chromium release assay. Each bar represents the mean for two samples and the standard error of the mean.
DISCUSSION
Since CMV pp65 is a likely component of an eventual anti-CMV vaccine, it might become necessary to produce a protein with intact immunologic domains but little remaining biologic function. Little is known about the biologic properties of CMV pp65, but once it localizes to the nucleus, it does bind to PLK-1 and thus could have effects on the cell cycle (11). We have previously created a kinaseless CMV pp65 with the K436N mutation (pp65mII) for use in a vaccine, and the resultant pp65mII appeared to have a nuclear localization and immunogenicity similar to those of native CMV pp65 (8). A similarly designed CMV DNA vaccine is currently in clinical trials (38). On a theoretical basis, a kinaseless CMV pp65 from which NLS is deleted would likely further improve the safety of an anti-CMV vaccine. Thus, this work was motivated by the need to determine if removal of the nuclear localizing properties of CMV pp65 influenced either its antigenicity or its immunogenicity.
Because the C-terminal portion of the protein is rich in cytotoxic T-cell epitope sequences (7, 15), we were able to alter only the K436N/NLS-B region in order to preserve these potential epitopes. Yet, removal of NLS-B from pp65mII had a dramatic effect on the cytoplasmic accumulation of protein and increased the antigenic presentation of the cytotoxic T-cell epitope. Target cells transduced with either a DNA expression vector or rAAV encoding pp65mIINLSKO were more efficiently lysed in vitro by CTLs than the unmodified CMV pp65. More importantly, the use of these expression vectors in a vaccine model showed that the ability of pp65mIINLSKO to induce cellular immunity was as good as that of the native protein.
An unexpected effect of the deletion of NLS-B was the effect on transcription and expression of CMV pp65. RNA specific for CMV pp65 appeared earlier in the deletion vectors than in control CMV pp65, and the protein was detected approximately 8 h before control CMV pp65 was detected. These effects were seen with multiple cell types. The reason for the enhancement of CMV pp65 expression after NLS-B deletion is not known, but on the basis of the concomitant enhancement of CMV pp65-specific RNA levels, it is possible that nuclear localization of the protein affects RNA transcription by a feedback loop, which has been shown for other systems (12, 21). This deserves further study with CMV pp65 to better understand the role of this abundant structural protein in CMV infection.
There is no explanation for why the level of expression of the CMV pp65 NLS mutant was greater than that of native CMV pp65. We explored the proteins for regions which could alter protein degradation, such as motifs rich in amino acids proline, glutamic acid, serine, and threonine (PEST), which are known to be associated with reduced protein half-lives (29). A PEST motif, KAESTVAPEEDTDEDSDNEI, exists at aa 457 to 477 and is unaltered in both the native and the mutant CMV pp65 forms. Pulse-chase experiments with the 35S-labeled protein confirmed that there were no differences in the stabilities of the CMV pp65 form with the NLS deletion and the control CMV pp65. There appear to be no differences in CMV pp65 degradation that would explain the variation in the heightened protein levels associated with the NLSKO mutant.
Since pp65N was sequestered in the nucleus and pp65mIINLSKO was mainly found in the cytoplasm, we investigated the potential effects of these two proteins on cellular RNA expression. An Affymetrix gene microarray was used to hybridize cDNA from cells transduced with either native CMV pp65 or the NLSKO mutant for 48 h.
A potentially important observation is that the genes upregulated by WT pp65 and downregulated by NLSKO included genes of the cell cycle clusters, including genes such as PLK-1, UBE2C (ubiquitin-conjugating enzyme e2C), CCNB2, and KIFxx. Some of these genes have already been associated either with viral infection events or with binding to CMV pp65 (11, 31). One cannot know at this time if this change would translate into a safer vaccine, but it seems likely that the downregulation of families of mRNA related to the cell cycle and mitotic events could be protective. This will require further investigation, but clearly, pp65mIINLSKO differs from native CMV pp65 in this regard and could support application to improved anti-CMV vaccine design.
The upregulation of cellular genes by both the pp65 WT and the NLSKO genes also deserves attention, especially because of its number: 389 human genes were identified by the mRNA microarray to be upregulated. This cluster is characterized by numerous families of genes in the signaling, secretion pathway, and glycoprotein gene families. The impact of pp65 on inflammatory gene expression has been shown by others (1, 3). By comparing by DNA array analysis the effect of a pp65-deficient mutant virus with that of a virus with WT pp65 on fibroblast cells, those investigators showed that there was a much stronger induction of many IFN responses and proinflammatory chemokine RNAs in the absence of pp65. It is possible that these alterations in gene expression explain the immunodominance of CMV pp65 (18, 37) but also suggest a much larger role of the nuclear localization signal of pp65 than was previously anticipated.
In summary, the removal of the NLSs from CMV pp65 can be accomplished without significant alteration of the immunogenicity of the protein. In the absence of the nuclear localization of CMV pp65, there is modification of cellular events which appear to influence the appearance mRNA and the amount of protein expressed. However, the ability of the protein to be processed and antigenically presented to the class I system is actually enhanced by these changes, and the immunogenicity of the mutant CMV pp65 in a vaccine model is comparable to that of WT CMV pp65. These data suggest that the NLSKO mutation of CMV pp65 be evaluated in future candidate CMV vaccination strategies.
Acknowledgments
This study was supported in part by U.S. Public Health Service grant AI58148 (to J.A.Z.) from the National Institutes of Health.
We are grateful to Don J. Diamond for providing the 3.3F4 cells and to the Functional Genome Core facility at the Beckman Research Institute of the City of Hope, in particular, Ning Ye for her excellent technical expertise in microarray technology.
Footnotes
Published ahead of print on 15 April 2009.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 7810995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berencsi, K., Z. Gyulai, E. Gonczol, S. Pincus, W. I. Cox, S. Michelson, L. Kari, C. Meric, M. Cadoz, J. Zahradnik, S. Starr, and S. Plotkin. 2001. A canarypox vector-expressing cytomegalovirus (CMV) phosphoprotein 65 induces long-lasting cytotoxic T cell responses in human CMV-seronegative subjects. J. Infect. Dis. 1831171-1179. [DOI] [PubMed] [Google Scholar]
- 3.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 10011439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobbold, M., N. Khan, B. Pourgheysari, S. Tauro, D. McDonald, H. Osman, M. Assenmacher, L. Billingham, C. Steward, C. Crawley, E. Olavarria, J. Goldman, R. Chakraverty, P. Mahendra, C. Craddock, and P. A. Moss. 2005. Adoptive transfer of cytomegalovirus-specific CTL to stem cell transplant patients after selection by HLA-peptide tetramers. J. Exp. Med. 202379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond, D. J., J. York, J. Y. Sun, C. L. Wright, and S. J. Forman. 1997. Development of a candidate HLA A*0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood 901751-1767. [PubMed] [Google Scholar]
- 6.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elkington, R., S. Walker, T. Crough, M. Menzies, J. Tellam, M. Bharadwaj, and R. Khanna. 2003. Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J. Virol. 775226-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallez-Hawkins, G., N. A. Lomeli, X. Li, Z. Q. Yao, C. La Rosa, D. J. Diamond, and J. A. Zaia. 2002. Kinase-deficient CMVpp65 triggers a CMVpp65 specific T-cell immune response in HLA-A*0201.Kb transgenic mice after DNA immunization. Scand. J. Immunol. 55592-598. [DOI] [PubMed] [Google Scholar]
- 9.Gallez-Hawkins, G., X. Li, A. E. Franck, L. Thao, S. F. Lacey, D. J. Diamond, and J. A. Zaia. 2004. DNA and low titer, helper-free, recombinant AAV prime-boost vaccination for cytomegalovirus induces an immune response to CMV-pp65 and CMV-IE1 in transgenic HLA A*0201 mice. Vaccine 23819-826. [DOI] [PubMed] [Google Scholar]
- 10.Gallina, A., E. Percivalle, L. Simoncini, M. G. Revello, G. Gerna, and G. Milanesi. 1996. Human cytomegalovirus pp65 lower matrix phosphoprotein harbours two transplantable nuclear localization signals. J. Gen. Virol. 77(Pt 6)1151-1157. [DOI] [PubMed] [Google Scholar]
- 11.Gallina, A., L. Simoncini, S. Garbelli, E. Percivalle, G. Pedrali-Noy, K. S. Lee, R. L. Erikson, B. Plachter, G. Gerna, and G. Milanesi. 1999. Polo-like kinase 1 as a target for human cytomegalovirus pp65 lower matrix protein. J. Virol. 731468-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu, W., A. L. Wells, F. Pan, and R. H. Singer. 2008. Feedback regulation between zipcode binding protein 1 and beta-catenin mRNAs in breast cancer cells. Mol. Cell. Biol. 284963-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39499-509. [DOI] [PubMed] [Google Scholar]
- 14.Kapasi, A. J., and D. H. Spector. 2008. Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 82394-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kern, F., T. Bunde, N. Faulhaber, F. Kiecker, E. Khatamzas, I. M. Rudawski, A. Pruss, J. W. Gratama, R. Volkmer-Engert, R. Ewert, P. Reinke, H. D. Volk, and L. J. Picker. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J. Infect. Dis. 1851709-1716. [DOI] [PubMed] [Google Scholar]
- 16.Khan, N., R. Bruton, G. S. Taylor, M. Cobbold, T. R. Jones, A. B. Rickinson, and P. A. Moss. 2005. Identification of cytomegalovirus-specific cytotoxic T lymphocytes in vitro is greatly enhanced by the use of recombinant virus lacking the US2 to US11 region or modified vaccinia virus Ankara expressing individual viral genes. J. Virol. 792869-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanna, R., and D. J. Diamond. 2006. Human cytomegalovirus vaccine: time to look for alternative options. Trends Mol. Med. 1226-33. [DOI] [PubMed] [Google Scholar]
- 18.Lacey, S. F., D. J. Diamond, and J. A. Zaia. 2004. Assessment of cellular immunity to human cytomegalovirus in recipients of allogeneic stem cell transplants. Biol. Blood Marrow Transplant. 10433-447. [DOI] [PubMed] [Google Scholar]
- 19.Leen, A. M., G. D. Myers, U. Sili, M. H. Huls, H. Weiss, K. S. Leung, G. Carrum, R. A. Krance, C. C. Chang, J. J. Molldrem, A. P. Gee, M. K. Brenner, H. E. Heslop, C. M. Rooney, and C. M. Bollard. 2006. Monoculture-derived T lymphocytes specific for multiple viruses expand and produce clinically relevant effects in immunocompromised individuals. Nat. Med. 121160-1166. [DOI] [PubMed] [Google Scholar]
- 20.Lucas, K. G., Q. Sun, R. L. Burton, A. Tilden, W. P. Vaughan, M. Carabasi, D. Salzman, and A. Ship. 2000. A phase I-II trial to examine the toxicity of CMV- and EBV-specific cytotoxic T lymphocytes when used for prophylaxis against EBV and CMV disease in recipients of CD34-selected/T cell-depleted stem cell transplants. Hum. Gene Ther. 111453-1463. [DOI] [PubMed] [Google Scholar]
- 21.Luo, C., J. J. Loros, and J. C. Dunlap. 1998. Nuclear localization is required for function of the essential clock protein FRQ. EMBO J. 171228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McElroy, A. K., R. S. Dwarakanath, and D. H. Spector. 2000. Dysregulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130. J. Virol. 744192-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaughlin-Taylor, E., H. Pande, S. J. Forman, B. Tanamachi, C. R. Li, J. A. Zaia, P. D. Greenberg, and S. R. Riddell. 1994. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J. Med. Virol. 43103-110. [DOI] [PubMed] [Google Scholar]
- 24.Nigg, E. A., P. A. Baeuerle, and R. Luhrmann. 1991. Nuclear import-export: in search of signals and mechanisms. Cell 6615-22. [DOI] [PubMed] [Google Scholar]
- 25.Pande, H., T. D. Lee, M. A. Churchill, and J. A. Zaia. 1990. Structural analysis of a 64-kDa major structural protein of human cytomegalovirus (Towne): identification of a phosphorylation site and comparison to pp65 of HCMV (AD169). Virology 1786-14. [DOI] [PubMed] [Google Scholar]
- 26.Pande, H., S. W. Baak, A. D. Riggs, B. R. Clark, J. E. Shively, and J. A. Zaia. 1984. Cloning and physical mapping of a gene fragment coding for a 64-kilodalton major late antigen of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 814965-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascolo, S., N. Bervas, J. M. Ure, A. G. Smith, F. A. Lemonnier, and B. Perarnau. 1997. HLA-A2.1-restricted education and cytolytic activity of CD8+ T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J. Exp. Med. 1852043-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prichard, M. N., W. J. Britt, S. L. Daily, C. B. Hartline, and E. R. Kern. 2005. Human cytomegalovirus UL97 kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J. Virol. 7915494-15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers, S., R. Wells, and M. Rechsteiner. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234364-368. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez, V., A. K. McElroy, and D. H. Spector. 2003. Mechanisms governing maintenance of Cdk1/cyclin B1 kinase activity in cells infected with human cytomegalovirus. J. Virol. 7713214-13224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez, V., J. A. Mahr, N. I. Orazio, and D. H. Spector. 2007. Nuclear export of the human cytomegalovirus tegument protein pp65 requires cyclin-dependent kinase activity and the Crm1 exporter. J. Virol. 8111730-11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schleiss, M. R., J. C. Lacayo, Y. Belkaid, A. McGregor, G. Stroup, J. Rayner, K. Alterson, J. D. Chulay, and J. F. Smith. 2007. Preconceptual administration of an alphavirus replicon UL83 (pp65 homolog) vaccine induces humoral and cellular immunity and improves pregnancy outcome in the guinea pig model of congenital cytomegalovirus infection. J. Infect. Dis. 195789-798. [DOI] [PubMed] [Google Scholar]
- 33.Schmolke, S., P. Drescher, G. Jahn, and B. Plachter. 1995. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J. Virol. 691071-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selinsky, C., C. Luke, M. Wloch, A. Geall, G. Hermanson, D. Kaslow, and T. Evans. 2005. A DNA-based vaccine for the prevention of human cytomegalovirus-associated diseases. Hum. Vaccin. 116-23. [DOI] [PubMed] [Google Scholar]
- 35.Stochaj, U., and P. Silver. 1992. Nucleocytoplasmic traffic of proteins. Eur. J. Cell Biol. 591-11. [PubMed] [Google Scholar]
- 36.Sylwester, A. W., B. L. Mitchell, J. B. Edgar, C. Taormina, C. Pelte, F. Ruchti, P. R. Sleath, K. H. Grabstein, N. A. Hosken, F. Kern, J. A. Nelson, and L. J. Picker. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter, E. A., P. D. Greenberg, M. J. Gilbert, R. J. Finch, K. S. Watanabe, E. D. Thomas, and S. R. Riddell. 1995. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 3331038-1044. [DOI] [PubMed] [Google Scholar]
- 38.Wloch, M. K., L. R. Smith, S. Boutsaboualoy, L. Reyes, C. Han, J. Kehler, H. D. Smith, L. Selk, R. Nakamura, J. M. Brown, T. Marbury, A. Wald, A. Rolland, D. Kaslow, T. Evans, and M. Boeckh. 2008. Safety and immunogenicity of a bivalent cytomegalovirus DNA vaccine in healthy adult subjects. J. Infect. Dis. 1971634-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao, Z. Q., G. Gallez-Hawkins, N. A. Lomeli, X. Li, K. M. Molinder, D. J. Diamond, and J. A. Zaia. 2001. Site-directed mutation in a conserved kinase domain of human cytomegalovirus-pp65 with preservation of cytotoxic T lymphocyte targeting. Vaccine 191628-1635. [DOI] [PubMed] [Google Scholar]
- 40.Zaia, J. A. 1990. Understanding human cytomegalovirus infection. UCLA Symp. Mol. Cell. Biol. New Ser. 137319-334. [Google Scholar]