Abstract
Mycobacterium leprae is the noncultivable pathogen of leprosy. Since the genome sequence of an isolate of M. leprae has become available, multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) has been explored as a tool for strain typing and identification of chains of transmission of leprosy. In order to discover VNTRs and develop methods transferable to clinical samples, MLVA was applied to a global collection of M. leprae isolates derived from leprosy patients and propagated in armadillo hosts. PCR amplification, agarose gel electrophoresis, and sequencing methods were applied to DNA extracts from these infected armadillo tissues (n = 21). We identified polymorphisms in 15 out of 25 short-tandem-repeat (STR) loci previously selected by in silico analyses of the M. leprae genome. We then developed multiplex PCR for amplification of these 15 loci in four separate PCRs suitable for fluorescent fragment length analysis and demonstrated STR profiles highly concordant with those from the sequencing methods. Subsequently, we extended this method to DNA extracts from human clinical specimens, such as skin biopsy specimens (n = 30). With these techniques, mapping of multiple loci and differentiation of genotypes have been possible using total DNA extracts from limited amounts of clinical samples at a reduced cost and with less time. These practical methods are therefore available and applicable to answer focused epidemiological questions and to allow monitoring of the transmission of M. leprae in different countries where leprosy is endemic.
The causative pathogen of leprosy is Mycobacterium leprae. A continued incidence, defying global campaigns to eliminate leprosy even after years of rigorous case finding and the availability of multidrug therapy regimens (28, 29, 30, 31), is attributed to subclinical human and environmental reservoirs of the pathogen (1, 8, 13). In recent years, molecular strain-typing methodologies have complemented conventional infectious disease epidemiology. With the publication in 2001 of the complete genome sequence of an isolate from Tamil Nadu, India, called the TN strain (4), selection of potential polymorphic genomic markers for strain typing was feasible. The first genetic markers that showed polymorphism were short tandem repeats (STRs) in the M. leprae genome. One was a 6-bp intragenic sequence in the rpoT gene, and the second, a trinucleotide (TTC) repeat element upstream of a pseudogene (17, 23). These sequences exhibit variable numbers of tandem repeats (VNTRs) when sequenced in different isolates. Based on these observations, we short-listed 44 loci (including the rpoT and TTC loci) by in silico analyses of the M. leprae genome and accomplished the screening of 11 STR loci, of which 9 were polymorphic when tested in a small panel of four human isolates derived from passage through armadillos (6). Five were minisatellites (6- to 50-bp repeat units), and four were microsatellites (1- to 5-bp repeat units). Since then, others have also shown that VNTR loci exist in M. leprae isolates (25, 33, 34). Three single-nucleotide polymorphisms have also been discovered by comparing sequences of a limited number of strains (20).
The goal of our work has been to discover and apply DNA variation among M. leprae isolates to identify sources and chains of transmission of leprosy in regions of endemicity. There are, however, physiological and practical issues relevant to strain typing of M. leprae in the clinical setting, such as the long incubation period and low transmissibility of leprosy and the requirement for clinical specimens such as slit skin smears and skin biopsy specimens from leprosy patients due to the inability of M. leprae to grow in culture. During the course of the last 4 years, field studies in which STR mapping was implemented have been reported. Matsuoka et al. (16) applied the microsatellite locus (TTC)21 to type M. leprae strains obtained from nasal swabs and slit skin smears from patients grouped by village, dwelling, or household in Indonesia, while Young et al. (33) combined (AT)15, (GTA)9, and (TTC)21 VNTR loci for the identification of short- and long-range M. leprae transmission chains in areas within and surrounding the city of Hyderabad, India. Monot et al. mapped five M. leprae STR loci in patients from Mali, Africa (19). The results from those studies demonstrated heterogeneity in prevalent haplotypes, indicating that genotype mapping with a small panel of one to five microsatellite VNTR loci was insufficient to discern strain relatedness. However, within an intrafamilial case, three markers were congruent (33). The authors of those studies concluded that in these areas of endemicity, multiple rather than single dominant isolates are found and that additional genomic markers are necessary for strain typing.
For these reasons, assays for the amplification and differentiation of multiple genomic loci are needed. When these requirements have been met, it becomes possible to undertake systematic strain-typing studies that include suitable sampling strategies and conventional epidemiology methods for monitoring transmission and detecting clusters of cases. In light of these laboratory, field, and clinical issues, we further explored multiple-locus VNTR analysis (MLVA) techniques. In this paper, we report the development and testing of multiplex-PCR methods for MLVA for reference armadillo-derived M. leprae isolates and clinical materials and address allelic properties of individual loci and the reproducibility and feasibility of the techniques. In a future paper, we will apply and extend these methods to the data from population-based studies in Cebu, the Philippines (R. M. Sakamuri, M. Kimura, W. Li, K. Madanahally, H.-C. Kim, H. Lee, M. Balagon, R. Gelber, W. C. Black, S.-N. Cho, P. J. Brennan, and V. Vissa, submitted for publication).
MATERIALS AND METHODS
Armadillo-derived human M. leprae isolates.
Clinical isolates of M. leprae, passaged in an armadillo, were obtained from the National Hansen's Disease Program (NHDP), Louisiana State University, Baton Rouge, LA, in the form of infected liver and spleen tissues, as reported in an earlier study (6). Frozen, infected-armadillo-passaged clinical isolates obtained during the 1970s and 1980s from the National Institute of Medical Research, Mill Hill, London, United Kingdom (courtesy of the late Richard Rees and Joseph Colston), and from the Florida Institute of Technology, Melbourne, FL (courtesy of Eleanor Storrs), were selected from the remaining inventory of tissues shipped to Colorado State University (CSU) following the discontinuation of the armadillo colonies at these institutes.
Clinical samples.
A set of 30 skin biopsy specimens from leprosy patients were selected from previous approved studies. These specimens were from patients attending the Leonard Wood Memorial Skin Clinic in Cebu, the Philippines, collected in collaboration with Yonsei University. These biopsy specimens were frozen immediately upon collection for storage and sectioning.
DNA purification.
The Qiagen DNeasy tissue kit (Qiagen, CA) was employed to obtain total DNA from small portions of infected armadillo tissues (∼25 to 100 mg). DNA was eluted in 100 to 200 μl elution buffer, and typically 1 to 2 μl of DNA was sufficient for one PCR. Genomic DNA from the Philippines sample collection was extracted from cryosections at Yonsei University using a previously described method (32). DNA extracts were provided to CSU. Reference M. leprae NHDP63 DNA was obtained from a large-scale preparation of bacteria from infected liver or spleen obtained from the NHDP as described previously (6).
VNTR mapping.
The methods for STR amplification by PCR using high-fidelity SuperMix (Invitrogen, CA) and a touch-down thermocycler program have been described previously (6). The copy numbers (alleles) for all microsatellite loci were determined by direct sequencing of PCR products. Minisatellite copy numbers were inferred by 3% agarose gel electrophoresis using the EZ Load 20-bp molecular ruler (Bio-Rad) for product length determination. Sequences were obtained for representative products at the Macromolecular Resource Facility at CSU and SeqWright (Fisher Biotechnology, Houston, TX).
M. leprae primers for multiplex PCR and fragment length analysis (FLA).
Fluorescent 5′-end-labeled forward primers were custom synthesized at Applied Biosystems, CA (Table 1), and the reverse, desalted primers were custom synthesized at Integrated DNA Technologies, Coralville, IA. These primers were reconstituted in 1× TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) at a 100 μM concentration. Aliquots of these master stocks were stored at −20°C. Separate working stocks of the forward- and reverse-primer sets were prepared according to the combinations listed in Table 1 and adjusted to a 200-μl final volume with TE buffer, such that each primer was at a concentration of 2 μM.
TABLE 1.
Multiplex STR primers in four different combinations
| Multiplex PCR combination no. | Locusa | Forward primerd | Tm (°C)e | Reverse primer | Tm (°C) | Amplicon sizeb (bp) |
|---|---|---|---|---|---|---|
| 1 | (AC)8b | VIC-GCCCACTTACCTCAACCAAC | 65.4 | CCTATAACGGCACTCAGTCCA | 61.4 | 390 |
| (GTA)9 | NED-AGCCTTAGTCGCGCAGATG | 62.6 | TCCGCTGTCCGTCCGCTGA | 68.8 | 307 | |
| (GGT)5 | 6FAM-GCAGCGGTGTAACAGCATAGC | 68.4 | TGTCTGCCTTGCGAAACGGTC | 65.5 | 242 | |
| (AT)17 | PET-TCTCCAACATGCTGCGACA | 62.1 | GTACAGCGGCCTGATCGAA | 60.0 | 181 | |
| 6-3ac (rpoT) | VIC-ATGCCGAACCGGACCTCGACGTTGA | 60.0 | TCGTCTTCGAGGTCGTCGAGA | 59.5 | 91 | |
| 2 | 21-3 | 6FAM-GAATCTGACCTTTCGGAAATG | 56.8 | CGATGCAGCTTCCTACGG | 60.7 | 312 |
| (AC)9 | NED-AGCGCCCGTTGTCGATAGA | 63.9 | GACTGGATGTCGGCACCCC | 65.5 | 236 | |
| (AT)15 | PET-CAATATGCGGGTTGGCGCTTCTG | 66.2 | CCGTCTGGCTCGATGGCTGGATTC | 68.6 | 168 | |
| (AC)8ac | VIC-GTGTTACGCGGAACCAGGCA | 65.5 | CCATCTGTTGGTACTACTGA | 53.5 | 124 | |
| 3 | 27-5 | 6FAM-ATTGAGCAGATGGCCGGTC | 62.8 | AGCAGTCGGCACGCCCTT | 67.8 | 327 |
| 6-7 | VIC-GCCATCGTTGTCGGTTCATC | 61.5 | CGGAGGAGGTGGGTACGGT | 66.1 | 268 | |
| (TA)18 | NED-CGTGCGTCGTGTGTAGGC | 63.9 | GACGTGGCAACATCGAAGTT | 61.0 | 230 | |
| (GAA)21 | PET-CTACAGGGGGCACTTAGCTC | 62.1 | GGACCTAAACCATCCCGTTTT | 60.4 | 201 | |
| 4 | 18-8 | PET-GCCCGTCTATCCGCATCAA | 62.5 | GCAAAGATCAGCACGCCAAT | 61.8 | 348 |
| 12-5 | VIC-CTGGTCCACTTGCGGTACGAC | 65.1 | GGAGAAGGAGGCCGAATACA | 61.4 | 289 | |
| 23-3 | 6FAM-CCGAAGCCCTGGACGAAG | 63.1 | GCCGTAAATCCGCTCCC | 56.0 | 243 | |
| (TA)10c | PET-TAGATTCAAACGACCATGCA | 60.0 | TGATAATCACGTGTTTCCGC | 60.0 | 185 |
Amplification and detection of STRs using multiplex PCR.
Multiplex PCR was performed using the multiplex-PCR kit (Qiagen). Each reaction mixture (20-μl final volume) was assembled in a PCR cabinet at room temperature and comprised 10 μl of 2× Qiagen multiplex-PCR master mix, 2 μl Q solution, 2 μl (each) of the forward- and reverse-primer working stocks, and 1 to 2 μl of template DNA; the volume was adjusted with PCR-grade water. The final concentration of each primer was thus 0.2 μM. Following an activation step at 95°C for 15 min, 40 cycles of PCR were run as follows: denaturation at 94°C for 30 s, primer annealing at 60°C for 90 s, and primer extension at 72°C for 90 s. The PCR was terminated with a final extension at 72°C for 10 min. For DNA sequencing, the multiplex-PCR sample was simply diluted 10-fold; 1 to 2 μl was combined with 10 pmol of the forward primer for the BigDye cycle sequencing reaction at the Macromolecular Resource Facility at CSU.
FLA.
After the multiplex PCR, 1 μl of the PCR product was diluted 30- to 60-fold, and 1 μl of the diluted PCR product was combined with 12 μl deionized formamide (Applied Biosystems) and 0.3 μl of the LIZ-500 DNA standard (Applied Biosystems). The sample was denatured at 94°C for 5 min and subjected to capillary electrophoresis on the Applied Biosystems genetic analyzer 3130 at the Macromolecular Resource Facility at CSU. The samples were injected into the capillary (50-cm length, POP-7 polymer) by applying an injection voltage of 1 kV for 22 s. The capillary electrophoresis was run at a voltage of 15 kV at 60°C for 45 min. Following the separation, the electropherograms were visualized and analyzed using GeneMapper version 3.7 software (Applied Biosystems) to determine the major allele for each VNTR locus in each multiplex-PCR combination.
VNTR data analyses.
The microsatellite tool kit (http://animalgenomics.ucd.ie/sdepark/ms-toolkit/) was used to calculate allele frequency and average allelic diversity.
RESULTS
Discovery of new VNTRs.
In our earlier study (6), 44 loci suitable for VNTR strain typing were listed. VNTRs were discovered at seven microsatellite loci containing 1-, 2-, and 3-bp repeat units in a panel of four M. leprae isolates. In the present study, we screened for polymorphisms within more microsatellite loci in the same set of isolates (Table 2, taxa 15 to 18). Reliable sequence results for the monomeric G/C-rich tracts, such as for the (C)20, (G)22, (C)18, and (G)8 loci, were not obtained even though we were able to obtain PCR products as reported by other groups (25, 34). Previously, we did not confirm VNTR polymorphism at (AC)9; however, sequencing of the products revealed two alleles with eight or nine copies (6). When we examined the dinucleotide (AC)8b, we found the seven-copy and not the eight-copy allele, as seen in the TN isolate and in all four isolates with microsatellite loci. Of the AT-rich loci, (AT)17, (TA)18, and (AT)15 have already been proven to be polymorphic (6, 34), so we did not screen any more AT dinucleotide STR loci at this stage. Four trinucleotide microsatellites [(AGT)5a, (AGT)5b, (ACT)5, and (GGT)5] were screened; only (GGT)5 was polymorphic, with two alleles, 4 and 5. All four isolates had three copies of the pentanucleotide (CACCG) STR, as in the TN strain. From these screens, (AC)9, (AC)8b, and (GGT)5 have therefore been included in the panel of microsatellite VNTR loci.
TABLE 2.
Origins and MLVA results for the armadillo-derived M. leprae isolates used in this studya
| Taxonb | Isolate or animal code (passage no.) | Other isolate(s) (passage no.) | Yr of origin | Country of origin of donor | Region | VNTR pattern at locusc:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (AT)17 | (TA)18 | (AC)8a | (AT)15 | (TA)10 | (AC)8b | (AC)9 | (TTC)21 | (GTA)9 | (GGT)5 | 6-7 | 12-5 | 18-8 | 21-3 | 23-3 | 27-5 | ||||||
| 1 | TN | 1980 | India | IN | 17 | 18 | 8 | 15 | 10 | 8 | 9 | 21 | 9 | 5 | 7 | 5 | 8 | 3 | 3 | 5 | |
| 2 | CD236 (0) | 1977 | India | IN | 16 | N | 8 | 15 | 10 | 8 | 9 | 21 | 9 | 5 | 7 | 5 | 8 | 3 | 3 | 5 | |
| 3 | 3045/334 (2) | 336 (2) | 1987 | India | IN | N | 11 | 8 | 13 | 8 | 7 | 10 | 15 | 10 | 5 | 5 | 4 | 7 | 1 | 2 | 5 |
| 4 | 3044/329 (2) | 333 (2), 325 (2), 264 (1), 267 (1) | 1988 | India | IN | N | 9 | 9 | 17 | 10 | 7 | 8 | 15 | 11 | 4 | 7 | 4 | 7 | 2 | 2 | 5 |
| 5 | I480 (1) | I478 (1) | 1985 | Philippines | OR | 16 | 19 | 8 | 15 | 10 | 8 | 9 | 21 | 9 | 5 | 7 | 5 | 8 | 3 | 3 | 5 |
| 6 | A606 (2) | NA | Philippines | OR | 11 | 17 | 11 | 12 | 13 | 6 | 7 | 13 | 9 | 5 | 5 | 4 | 8 | 3 | 2 | 5 | |
| 7 | W88 (1) | 1976 | Philippines | OR | 12 | 18 | 11 | 16 | 11 | 8 | 8 | 28 | 15 | 5 | 7 | 4 | 8 | 3 | 2 | 5 | |
| 8 | W112 (2) | NA | Philippines | OR | 11 | 18 | 9 | 16 | 11 | 8 | 8 | N | 14 | 5 | 7 | 4 | 8 | 3 | 2 | 5 | |
| 9 | I226 (1) | A308 (2) | NA | Thailand | OR | 11 | 16 | 10 | 12 | 13 | 6 | 7 | 13 | 9 | 5 | 5 | 4 | 8 | 3 | 2 | 5 |
| 10 | A344 (3) | 1980 | Thailand | OR | 11 | 21 | 10 | 14 | 10 | 8 | 7 | 14 | 9 | 4 | 8 | 4 | 8 | 2 | 2 | 5 | |
| 11 | 3039/321 (2) | 3017 (1) | 1987 | Ethiopia | AFR | 13 | 13 | 8 | 20 | 11 | 7 | 10 | 29 | 8 | 5 | 6 | 4 | 8 | 3 | 3 | 5 |
| 12 | I487 (1) | NA | Guyana | SAM | 12 | 17 | 10 | 12 | 13 | 6 | 7 | 13 | 9 | 5 | 5 | 4 | 8 | 3 | 2 | 5 | |
| 13 | 2936/41 (0) | 1987 | Malawi | AFR | 12 | 16 | 8 | 13 | N | 8 | 7 | 13 | 10 | 5 | 8 | 3 | 7 | 2 | 2 | 5 | |
| 14 | 3035/299 (2) | NA | Malawi | AFR | 12 | 16 | 8, 9 | 12, 13 | 8, 9 | 7, 8 | 7, 8 | 13, 16 | 8, 10 | 4, 5 | 6, 8 | 3, 4 | 6, 7 | 2 | 2 | 5 | |
| 15 | NHDP98 | 1990 | United States | NAM | 11 | 12 | 8 | 14 | 11 | 7 | 8 | 10 | 9 | 4 | 7 | 5 | 8 | 2 | 2 | 4 | |
| 16 | NHDP63 | NA | United States | NAM | 13 | 16 | 10 | 16 | 13 | 7 | 8 | 10 | 10 | 4 | 7 | 5 | 8 | 2 | 2 | 4 | |
| 17 | BR4923 | 1996 | Brazil | SAM | 15 | 18 | 7 | 20 | 8 | 7 | 8 | 12 | 12 | 4 | 6 | 4 | 3 | 2 | 2 | 5 | |
| 18 | Thai-53 | NA | Thailand | OR | 10 | 13 | 11 | 17 | 11 | 7 | 9 | 15 | 9 | 5 | 6 | 5 | 8 | 3 | 2 | 5 | |
| 19 | NHDP10 | 1996 | United States | NAM | 13 | 16 | 10 | 17 | 12 | 7 | 8 | 11 | 10 | 4 | 7 | 5 | 8 | 2 | 2 | 4 | |
| 20 | N1192 | NA | NA | NAM | 10 | 13 | 11 | 17 | 11 | 7 | 9 | 15 | 9 | 5 | 6 | 5 | 8 | 3 | 2 | 5 | |
| 21 | N1200 | NA | NA | NAM | 13 | 16 | 10 | 16 | 14 | 7 | 8 | 10 | 10 | 4 | 7 | 5 | 8 | 2 | 2 | 4 | |
| 22 | NHDP55 | 1996 | United States | NAM | 13 | 20 | 10 | 17 | 13 | 7 | 8 | 12 | 10 | 4 | 7 | 5 | 8 | 2 | 2 | 4 | |
N, not tested, PCR negative, or sequence negative; NA, not available; AFR, Africa; IN, India; NAM, North America; OR, Orient; SAM, South America.
Based on the reference sequenced strain (4) 2-14, obtained from Mill Hill and FIT, and 15-22 Armadillo-derived human leprosy specimens obtained from NHDP (25).
VNTR pattern indicates the number(s) of copies found at the specified VNTR locus.
Likewise, we screened the minisatellite 6-3b, 7-3, 10-4, 15-3, and 23-3 loci for VNTRs within the four standard isolates. Only the 23-3 locus was polymorphic, showing the two-copy allele instead of the three-copy allele seen in the TN strain (Table 2). Similar trends of polymorphisms in STR loci were detected in an independent study that included Asian isolates maintained by the system using footpad inoculation of nude mice (34).
VNTR mapping of an archived collection of M. leprae isolates from various parts of the world.
We expanded our reference panel with a collection of frozen armadillo tissues sent from the National Institute of Medical Research, Mill Hill, London, United Kingdom, and the Florida Institute of Technology, Melbourne, FL. The M. leprae inocula were originally obtained from leprosy patients from India, Ethiopia, Malawi, Guyana, the Philippines, and Thailand; such animal infections were performed during WHO studies to generate sufficient bacilli for vaccine production (22). M. leprae isolates representing the original armadillo infection (from an armadillo infected with a human biopsy suspension, i.e., passage 0) and isolates from subsequent armadillo-to-armadillo passages (first, second, or third) in these residual tissue banks were tested if available. We utilized these specimens to assess the purity and stability of VNTR patterns (Table 2). Tissue records indicated that in some instances, biopsy homogenates from multiple patients and/or biopsy specimens were combined for the first armadillo inoculation. A panel of 16 VNTR loci [10 microsatellites, including (TA)10, and 6 minisatellites] was amplified and analyzed. All loci with the exception of 6-3a were polymorphic, with new alleles being identified within this expanded collection.
With regard to the purity and stability of VNTR profiles of M. leprae in these specimens, except in two instances (compare I480 with A606 and 2936/41 with 3035/299 in Table 2), we found a characteristic VNTR pattern for 16 loci for each tissue, despite the possibility that multiple biopsy specimens were used in the first inoculation. In this VNTR mapping exercise, we were able to find two isolates (CD236 and I480) that closely matched the profile of the original sequenced TN isolate (4). The combined presence of the TN-like 23-3 locus and 21 copies of the TTC triplet is a signature not found in any of the other isolates. In this collection of 21 armadillo-derived M. leprae isolates and the in silico data from the TN genome, the number of haplotypes was 16, with a genetic diversity of 0.5463, which is a measure of diversity across the 15 VNTR loci (Table 2), and was calculated in the microsatellite tool kit that uses Nei's definition of gene diversity (21).
Selection of VNTR loci and primer combinations for multiplex PCR.
We then developed a multiplex-PCR approach for strain typing of clinical isolates. We utilized primer sequences described by us or others in various combinations to enable simultaneous amplification of three or four loci, such that the products were detectable in the 3% agarose gel, with minimum overlap (Fig. 1). The combinations listed in Table 1 proved to be satisfactory. At present, there are four combinations for amplification of 15 loci. Primer sets for loci 6-3a and (TA)10 have been subsequently added to combinations 1 and 4, respectively, allowing mapping at 17 STR loci.
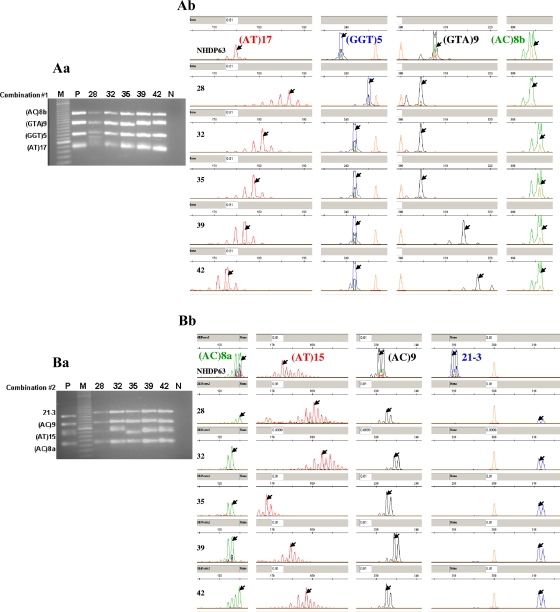
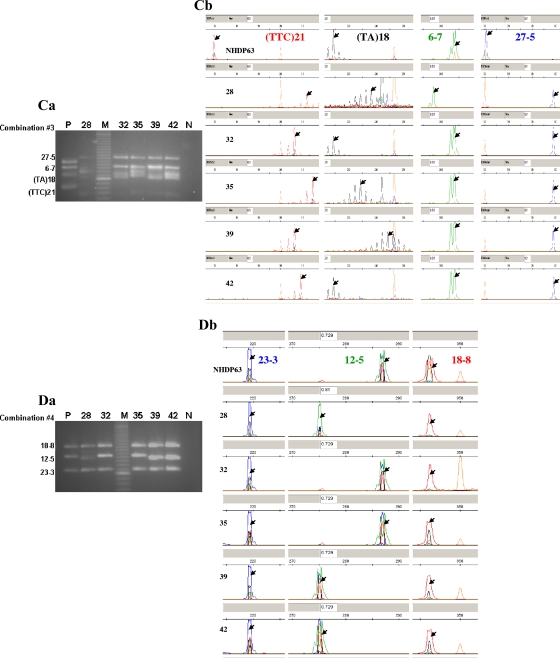
FIG. 1.
Gel electrophoresis and FLA of 15 VNTR loci from five Cebu, the Philippines, clinical isolates amplified using the combination 1, 2, 3, and 4 primer sets. (Aa, Ba, Ca, and Da) Gel electrophoresis of multiplex-PCR products (combinations 1 to 4). Lanes: M, 20-bp DNA marker (Bio-Rad); P, multiplex-PCR products from NHDP63 (positive control); 28, 32, 35, 39, and 42, multiplex-PCR products from clinical isolates; N, negative control. (Ab, Bb, Cb, and Db) Electropherograms obtained from FLA, where the orange peaks represent the LIZ-500 size standards and red, blue, black, and green peaks represent the VNTR products in each of the combinations. Arrows indicate the peaks selected as the allele for each locus.
Multiplex PCR and FLA for clinical samples.
All the combinations were optimized with NHDP63 DNA obtained from the tissue of an armadillo infected with the isolate from a leprosy patient and subsequently utilized as our reference DNA for VNTR mapping (6). In order to test the feasibility of multiplex PCR and FLA for clinical samples, we used DNA from 30 stored biopsy samples from leprosy patients (Fig. 1Aa, Ba, Ca, and Da). The VNTR alleles for NHDP63 and the 30 clinical isolates are shown in Table 3.
TABLE 3.
VNTR data from clinical samples using FLA from Cebu, the Philippines
| Strain or taxon | No. of copies found at the indicated locus with multiplex-PCR combination:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
||||||||||||
| (AC)8b | (GTA)9 | (GGT)5 | (AT)17 | 21-3 | (AC)9 | (AT)15 | (AC)8a | 27-5 | 6-7 | (TA)18 | (TTC)21 | 18-8 | 12-5 | 23-3 | |
| NHDP63 | 7 | 10 | 4 | 13 | 2 | 8 | 16 | 10 | 4 | 7 | 16 | 10 | 8 | 5 | 2 |
| 13 | 8 | 9 | 5 | 18 | 3 | 9 | 15 | 9 | 5 | 7 | 26 | 31 | 8 | 5 | 2 |
| 14 | 8 | 11 | 5 | 15 | 3 | 9 | 16 | 9 | 5 | 7 | 18 | 24 | 8 | 4 | 2 |
| 15 | 8a | 11a | 5a | 12a | 3 | 8 | 17 | 9 | 5 | 7a | 18a | 22a | 8a | 4a | 2a |
| 16 | 8 | 9 | 5 | 14 | 3 | 10 | 13 | 9 | 5 | 7 | 21 | 19 | 8 | 5 | 2 |
| 17 | 7 | 10 | 4 | 13 | 1a | 9a | 13a | 9a | 5a | 8 | 15 | 14 | 7 | 4 | 2 |
| 18 | 8a | 10a | 5a | 14a | 3 | 8 | 16 | 9 | 5 | 7 | 20 | 36 | 8 | 4a | 2a |
| 19 | 7 | 8a | 4 | 14 | 1a | 9a | 17a | 9a | 5a | 8a | 14a | 16a | 7a | 4a | 2a |
| 20 | 8a | 9a | 5a | 16a | 3a | 9a | 12a | 8a | 5a | 8a | 18a | 23a | 8a | 4a | 2a |
| 21 | 6 | 11 | 5 | 12 | 3a | 8a | 19a | 9a | 5a | 7 | 25 | 29 | 8 | 4a | 2a |
| 22 | 6a | 11a | 5a | 12a | 2a | 9a | 13a | 11a | 5a | 6a | 13a | 20a | 8a | 4a | 2a |
| 23 | 8 | 9 | 5 | 16 | 3 | 8 | 11 | 9 | 5 | 8 | 17 | 28 | 8 | 4 | 2 |
| 24 | 8 | 9 | 5 | 17 | 3 | 9 | 14 | 9 | 5 | 7 | 16 | 22a | 8 | 5 | 3 |
| 25 | 7 | 10 | 4 | 14 | 1 | 8 | 15 | 9 | 5 | 8 | 15 | 20 | 7 | 4 | 2 |
| 26 | 8 | 8a | 5 | 18 | 3 | 9 | 13 | 10 | 5 | 7 | 18 | 19a | 8 | 5 | 3 |
| 27 | 8 | 8 | 5 | 16 | 3 | 10 | 15 | 9 | 5a | 7a | 18a | 31a | 8 | 5 | 2 |
| 28c | 7 | 9 | 6 | 19 | 3 | 9 | 20 | 10 | 5 | 6 | 22 | 25 | 8 | 4 | 2 |
| 29 | 7 | 9 | 6 | 16 | 3b | 9b | 19b | 10b | 5b | 6b | 20b | 24b | 8 | 4 | 2 |
| 30 | 7 | 11 | 5 | 13 | 3b | 8b | 15b | 8b | 5b | 7b | 23b | 24b | 8 | 4 | 2 |
| 31 | NDd | 11 | ND | 17 | 3 | ND | 15 | 9 | 4 | ND | 23 | 28 | ND | 5 | 2 |
| 32c | 8 | 9 | 5 | 16 | 3 | 10 | 21 | 9 | 5 | 7 | 16 | 23 | 8 | 5 | 2 |
| 33 | 7b | 9b | 4b | 13b | 1b | 9b | 14b | 9b | 5b | 8b | 13b | 16b | 7b | 4b | 2b |
| 34 | 8 | 9 | 5 | 15 | 3b | 9b | 15b | 9b | 5b | 7b | 21b | 27b | 8 | 5 | 2 |
| 35c | 8 | 9 | 5 | 15 | 3 | 9 | 14 | 9 | 5 | 7 | 21 | 26 | 8 | 5 | 2 |
| 36 | 8 | 10 | 5 | 15 | 3b | 9b | 21b | 9b | 5b | 6b | 20b | 26b | 8 | 5 | 2 |
| 37 | 8 | 9 | 5 | 15 | 3b | 9b | 15b | 10b | 5b | 7b | 18b | 24b | 8 | 5 | 2 |
| 38c | 8 | 9 | ND | 16 | 3 | 9 | 15 | 9 | 5 | 7 | 17 | 27 | 8 | 5 | 2 |
| 39c | 8 | 12 | 5 | 14 | 3 | 10 | 17 | 9 | 5 | 7 | 26 | 23 | 8 | 4 | 2 |
| 40 | ND | ND | ND | 15 | 1b | 9b | 14b | 9b | 4b | 4b | 15b | 25b | 8 | 5b | 2b |
| 41 | 8 | 10 | 5 | 18 | 3b | 9b | 14b | 9b | 5b | 8b | 21b | 31b | 8 | 5 | 2 |
| 42c | 8 | 13 | 5 | 12 | 3 | 9 | 19 | 10 | 5 | 7 | 16 | 24 | 8 | 4 | 2 |
Also sequenced.
Two FLA runs using the same PCR sample were carried out.
Three PCRs and three FLA runs (two PCRs with a Qiagen multiplex-PCR kit and one with an ABI AmpliTaq Gold PCR master kit) were performed.
ND, not detected.
The addition of Q solution, a cosolvent that can alleviate amplification problems associated with secondary structure and/or high GC content, was found to be useful for some loci and was therefore routinely included in the PCR mixture. In addition to being subjected to conventional agarose gel electrophoresis, the products of the multiplex PCRs were subjected to FLA (Fig.1Ab, Bb, Cb, and Db).
We also retested several of the armadillo-derived M. leprae isolates (Table 2) by this multiplex-PCR-FLA method and obtained concordant alleles. Interestingly, the FLA profile from the tissue infected with strain 3035/299 from Malawi, which was previously thought to be different from 2936/41 (0-passage specimen) at multiple loci, was found to carry two alleles at these loci, one the same as that found in 2936/41, along with secondary alleles. The results were reproduced when a different scraping from the same frozen tissue was used for DNA extraction and typing.
Stutter phenomena and 3′ base tailing and allele determination.
Stutter products, defined as products formed by slipped-strand DNA synthesis at repeat regions, were commonly observed for the microsatellite loci. We observed both minus and plus stutters ranging from −3 to +3 repeats with reference and clinical template DNAs (Table 4). The addition of an extra base at the 3′ end of PCR products (nontemplate 3′ extension) was also observed. The extent and pattern of stutters and 3′ tailing tended to be characteristic for each locus. Despite the stutter and 3′ base tailing, we were able to select the peaks, as indicated by the arrows in the electropherograms of Fig.1 Ab, Bb, Cb, and Db for the reportable alleles of the majority of the loci. A nonspecific product peak at 242 bp (NED label, black peak) in combination 3 can be seen with human-derived specimens.
TABLE 4.
Amplicon sizes, 3′ tailing effect, and stutter products detected by FLA for NHDP63e
| Multiplex-PCR combination | Locus | Amplicon sized | 3′ tailing effect | Presence or absence of stutter products:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| −3 | −2 | −1 | +1 | +2 | +3 | ||||
| 1 | (AC)8b | 384.15 ± 0.1589 | +b | − | − | + | + | − | − |
| (GTA)9 | 307.86 ± 0.4416 | +b | − | − | + | + | − | − | |
| (GGT)5 | 238.53 ± 0.3047 | +b | − | − | − | − | − | − | |
| (AT)17 | 174.83 ± 0.0961 | − | + | + | + | + | + | + | |
| 2 | 21-3 | 289.75 ± 0.3965 | +b | − | − | + | − | − | − |
| (AC)9 | 231.58 ± 0.2792 | +a | − | − | + | − | − | − | |
| (AT)15 | 173.54 ± 0.2251 | +b | + | + | + | + | + | + | |
| (AC)8a | 125.17 ± 0.1555 | +b | − | + | + | + | + | − | |
| 3 | 27-5 | 300.61 ± 0.1253 | +b | − | − | + | − | − | − |
| 6-7 | 263.95 ± 0.2714 | +a | − | − | + | − | − | − | |
| (TA)18 | 224.51 ± 0.1618 | − | + | + | + | + | + | + | |
| (GAA)21 | 169.80 ± 0.1354 | − | + | + | + | − | − | ||
| 4 | 18-8 | 346.20 ± 0.2149 | − | +c | +c | +c | − | − | − |
| 12-5 | 286.96 ± 0.2742 | +b | − | − | − | − | − | − | |
| 23-3 | 219.40 ± 0.2408 | +b | − | − | − | − | − | − | |
3′-end tailed and blunt PCR products are detected in approximately equal proportions.
3′ tailing proceeds to near completion; the blunt PCR product is limited in abundance.
Position −1, −2, and −3 stutter products are weak. Instead, peaks at the two-copy and three-copy allele positions can be seen, which would be the −5 and −6 stutter products for NHDP63.
Average amplicon size ± standard deviation from 10 independent multiplex-PCR and FLA experiments. FLA was performed on the ABI Genetic analyzer 3130.
+ and − signs indicate the presence and absence, respectively, of 3′ tailing or stutter products.
Concordance between VNTR alleles determined by fragment length analysis and DNA sequencing.
In order to verify the FLA-based allele calls, a subset of samples was submitted to multiplex PCR using unlabeled primers and DNA sequencing of amplicons. For each combination, we randomly tested at least three DNA isolates. The alleles determined by sequencing of the amplicons were in concordance with those estimated by the FLA (Table 3).
Reproducibility of multiplex PCR and FLA.
In order to test for the run-to-run reproducibility of the multiplex PCR and FLA, multiplex PCR was performed several times using NHDP63 DNA and the products from each PCR were submitted to FLA on separate days. Overall, the peak sizes and patterns remained stable between independent PCRs and FLA experiments. The average amplicon sizes and standard deviations for each locus determined from 10 such experiments are shown in Table 4. The multiplex-PCR and FLA reproducibility was also verified for clinical isolates. Samples 28, 32, 35, 39, and 42 were subjected to multiplex PCR and FLA at least three times, twice with Qiagen multiplex enzyme and once with a high-fidelity enzyme (AmpliTaq Gold; Applied Biosystems). Concordant FLA peaks were observed from all three runs, which also demonstrated that the variation due to the DNA polymerase is minimal. Furthermore, when stored multiplex-PCR products were rerun for FLA, there were no significant changes in the profiles, indicating that products can be stored (up to a year at −20°C) and analyzed at a subsequent convenient time and location.
DISCUSSION
The goal of our studies is to generate useful molecular epidemiological tools for monitoring the residual leprosy burden in countries of endemicity to complement local surveillance and control programs. Prolonged contact with a leprosy patient is a known risk factor, but only 10 to 30% of the new cases can be linked to an index case (2, 26). The number of new cases among children is not insignificant (a sign of recent transmission). Isolates resistant to one or more antileprosy drugs are being detected in different countries of endemicity, and the relapse rates among treated cases are ∼13% (5, 7, 9, 14). In addition, the concept of zoonotic transmission has not been excluded (18). Thus, finding a source(s) and chains of transmission by strain typing is a practical application. Special considerations for the molecular epidemiology of leprosy include the long doubling time of M. leprae, followed by an incubation period of 2 to 8 years before symptoms may be perceived or diagnosed and the lack of laboratory cultivation methods for M. leprae.
Furthermore, outbreaks are not common, though intrafamilial clusters exist and pockets of hyperendemicity have been reported (11, 12).
In this context, we pursued our strategy to identify polymorphic markers for M. leprae for strain-typing applications (6). Of the common methods for infectious pathogens, we and others have explored STR analyses for leprosy transmission, because nucleotide substitutions, inversions, recombinations, and transpositions had been found to be rare (3, 27). The preferred method for measuring evolutionary distance between two samples takes into account the allele sizes (number of repeating units) at each VNTR locus. During the course of our experiments, three novel single-nucleotide polymorphisms which led to a model for the origin and global spread of leprosy were discovered (20).
In light of field studies indicating that one to three microsatellites were insufficient for the detection of strain diversity and the transmission of M. leprae, we exploited the availability of a laboratory collection of 21 human clinical specimens passaged in armadillos for the purpose of identifying new genomic loci. Independently, a study performed with a different sample set composed of 27 mouse-derived human isolates, primarily from Japan but also including one isolate each from the Republic of Korea and Indonesia and three isolates from Thailand, showed that the list of polymorphic loci are equivalent to that from our study. Locus 10-4 is comprised of four 10-base repeats, with a consensus repeat sequence: ttATTAATAA (6). The lowercase “tt” in the minisatellite 10-4 represents nonconserved bases within the tandem repeat unit sequence. The repeat units 1, 2, and 4 have TT, while repeat unit 3 has CG. Polymorphisms were not found either in the copy number or in the nonconserved nucleotides present at the first and/or second position in any of the repeats, even though locus 10-4 is situated in a noncoding region. The 7-3 locus is contained within a putative functional open reading frame in which VNTRs would cause frameshifting, perhaps the reason that the 7-3 locus was not polymorphic. The 23-3 locus was peculiar because the TN pattern with three copies was rare. We discovered a 5-base deletion in the third repeat in two isolates from Ethiopia. We have termed this allele 23-3′. Zhang et al., showed that 32 of the 44 loci in our first list are polymorphic; of these, they selected 9 loci (all microsatellites) for determining strain identity in isolates collected from families with multiple cases (34).
To improve throughput, we have developed a multiplex-PCR approach suitable for rapid amplification of 15 VNTR loci from M. leprae-containing specimens like slit skin smears and skin biopsy specimens. The benefits are reduced PCR reagent consumption, including of the DNA template, and ease of setup. Of these 15 VNTR loci, 6 are minisatellites and 9 are microsatellites. Two more loci [6-3a and (TA)10] have since been included in the PCR. Microsatellites tend to exhibit higher allelic diversity than minisatellites, even in localized collections, and when combined with minisatellites are useful in differentiating specimens from different countries/geographical regions (Table 5). For the 21 armadillo samples plus that of strain TN, a gene diversity of 0.5463 was observed for 16 loci. A similar trend was seen for the 30 clinical isolates from the Philippines (gene diversity of 0.54 for 15 loci). For the same loci, the gene diversity of the 27 isolates (mainly of Japanese origin) reported by Zhang et al. (34) is 0.6. These diversity indices are comparable to those in other bacterial VNTR-based typing systems. With Bacillus anthracis, an eight-VNTR-locus system was used for strain typing. An average diversity of 0.52 for 89 isolates was shown (10). With Salmonella enterica serotype Enteritidis, Malorny et al. demonstrated a typing system based on nine VNTR loci (with individual allelic diversity ranging from 0.07 to 0.65) which allowed further discrimination of isolates within single phage types (15). The average diversity was 0.42 for this collection of 240 isolates. In the context of the strain typing of Mycobacterium tuberculosis, different genomic markers and combinations have been utilized. The locus diversity varied from 0.1 to 0.8 when 24 mycobacterial interspersed repetitive unit VNTR loci were applied to a set of 90 isolates from a standardized worldwide collection. These loci have been recommended for high-resolution phylogenetic studies (24). This data set represents an average genetic diversity of 0.58 for the 24 loci.
TABLE 5.
Allele diversity detected for M. leprae STR loci in leprosy patients from the Philippines
| Diversity at indicated STR locus
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Allele | Frequency | Allele | Frequency | Allele | Frequency | |||
| (AC)8a | (AC)8b | (AC)9 | ||||||
| 8 | 6.90 | 6 | 7.14 | 8 | 20.69 | |||
| 9 | 72.41 | 7 | 25.00 | 9 | 65.52 | |||
| 10 | 17.24 | 8 | 67.86 | 10 | 13.79 | |||
| 11 | 3.45 | |||||||
| (AT)15 | (AT)17 | (TA)18 | ||||||
| 11 | 3.45 | 12 | 13.79 | 13 | 3.45 | |||
| 12 | 3.45 | 13 | 13.79 | 14 | 6.90 | |||
| 13 | 13.79 | 14 | 17.24 | 15 | 6.90 | |||
| 14 | 3.45 | 15 | 20.69 | 16 | 6.90 | |||
| 15 | 24.14 | 16 | 20.69 | 17 | 13.79 | |||
| 16 | 20.69 | 17 | 3.45 | 18 | 17.24 | |||
| 17 | 6.90 | 18 | 10.34 | 19 | 3.45 | |||
| 18 | 3.45 | 20 | 3.45 | |||||
| 19 | 6.90 | 21 | 13.79 | |||||
| 20 | 6.90 | 22 | 10.34 | |||||
| 22 | 6.90 | 25 | 3.45 | |||||
| 26 | 10.34 | |||||||
| (GGT)5 | (GTA)9 | (GAA)21 | ||||||
| 4 | 15.38 | 8 | 10.71 | 14 | 3.45 | |||
| 5 | 80.77 | 9 | 46.43 | 16 | 6.90 | |||
| 6 | 3.85 | 10 | 17.86 | 19 | 6.90 | |||
| 11 | 17.86 | 20 | 6.90 | |||||
| 12 | 3.57 | 22 | 13.79 | |||||
| 13 | 3.57 | 23 | 13.79 | |||||
| 24 | 13.79 | |||||||
| 25 | 6.90 | |||||||
| 26 | 6.90 | |||||||
| 28 | 3.45 | |||||||
| 29 | 3.45 | |||||||
| 30 | 3.45 | |||||||
| 31 | 6.90 | |||||||
| 36 | 3.45 | |||||||
| 6-7 | 12-5 | 18-8 | ||||||
| 4 | 3.45 | 4 | 55.17 | 7 | 14.29 | |||
| 6 | 13.79 | 5 | 44.83 | 8 | 85.71 | |||
| 7 | 58.62 | |||||||
| 8 | 24.14 | |||||||
| 21-3 | 23-3 | 27-5 | ||||||
| 1 | 17.24 | 2 | 93.10 | 4 | 3.45 | |||
| 2 | 3.45 | 3 | 6.90 | 5 | 96.55 | |||
| 3 | 75.86 | |||||||
| 4 | 3.45 | |||||||
In addition, we developed the FLA method for mapping VNTR alleles, which is more rapid than the conventional sequencing method. To validate and standardize the technique, reference strain NHDP63 was used (Table 4) and included every time clinical samples were analyzed. Although the minisatellite banding profiles can easily be interpreted by visual inspection after electrophoresis on conventional agarose gels, it was convenient to include these in the multiplex-PCR combinations for FLA. Rapid mapping of 15 VNTRs in a set of 30 clinical specimens obtained from one clinic has been demonstrated using the multiplex-PCR-FLA technique. The amplicon sizes were found to be reproducible from run to run for the reference stain and the clinical isolates. However, the sizes (in bp) do not correspond to the calculated size of the DNA fragments (Table 4). This could be due to the nature of the gel matrix, differences during migration between labeled PCR fragments, capillary length, the ladder used for calibration, the sequence composition, and the models of Applied Biosystems genetic analyzers. So we recommend that a reference strain (such as NHDP63 in our experiments) be used in all of the runs for consistency in allele definitions for a given instrument within a laboratory. The allelic diversity for M. leprae in the Philippine sample collection is shown in Table 5.
For the minisatellite 18-8, minus stutter peaks were observed. These appear at copy positions 2 and 3. Even though some of the microsatellites are prone to the stutter phenomenon (in vitro and in vivo) and 3′ tailing effect, electropherograms can be interpreted to assign alleles. The removal of the 3′ tail artifacts by a high-fidelity enzyme minimized the complexity of the peaks, as was seen for the (AT)15 products (not shown). Stutter products are also often observed in conventional sequence traces, wherein the stutter product sequences merge, making it difficult to read the sequence at the end of the repeat region. However, in FLA, the main product and the stutter products are clearly discernible. This suggests that the FLA technique is more convenient for detecting such products and reporting the predominant alleles than conventional sequencing. Strain typing allows us also to compare and confirm the identities of laboratory reference strains. For example, the VNTR pattern of the Thai-53 isolate in our study is quite different from that described by Zhang et al. (34) but is probably the same as that discussed by Monot et al. (20). The original strain was isolated in Japan from a Thai patient (25, 34).
VNTR stability throughout two passages in armadillo was noted in our studies, except with two M. leprae suspensions. With the information available to us for the archived tissues, we surmise that 2936/41 and 3035/299 originated from one common original inoculation in 1983. Tissue or animal 2936/41 represents passage 0 (patient to armadillo), while 3035/299 is that of passage 2 (armadillo to armadillo) (date not known). Unfortunately, the intermediate “passage 1 tissue” was not in our collection when we started this study. Therefore, we are unable to test or demonstrate when or how an additional VNTR profile emerged in the passage 2 isolate. We can propose only that the alternate alleles in 3035/299 represent a secondary strain rather than that they arise from VNTR instability sometime during incubation or passage because 11 of 16 loci carried two alleles. Endogenous infection of the host armadillos is another possibility. Similarly, regarding samples I480, I478, and A606, we inferred that they were derived from the same human inoculum. The passage 0 animal was also common for all three. I480 and I478 are from “passage 1” infections but in two different animals. These tissue specimens gave matching M. leprae VNTR patterns. A606, which is a passage 2 tissue, did not originate directly from either I480 or I478 but from a different animal. The tissues corresponding to passage 0 and the precursor to A606 were not present in our sample bank for this study, so we cannot speculate on the origins of the different M. leprae genotypes seen in A606 (alleles at 11 of 16 loci were altered) compared to I480 and I478. For those armadillos infected with approximately three patient biopsy homogenates, it appears that one isolate was dominant in the first two passages. Truman et al. demonstrated that microsatellites were highly stable after multiple passages in armadillo and/or nude mice, albeit allele shifts by one or more repeat units were not infrequent (25). Matsuoka et al. demonstrated that the TTC (GAA) locus was stable after 11 passages in nude mice (16).
This study was undertaken mainly for developing methods for VNTR-based strain typing. There was limited epidemiological or clinical information associated with the 30 coded clinical frozen samples (Table 3). However, it was possible to identify four isolates (17, 19, 25, and 33) which have alleles 4 and 1 for the loci (GGT)5 and 21-3, respectively. These isolates cluster and separate when analyzed by the neighbor-joining method (data not shown). For the same loci, the alleles are 6 and 3 for the pair of isolates (28 and 29) which also cluster but into a different branch. There was no association with genotype and village or city. In a larger study which included these 30 samples and 170 more that were collected from patients in the same province since 2003, a well-defined population structure with distinct VNTR signatures was identified. Several pairs of isolates with matching genotypes associated with multicase families or communities could be noted (Sakamuri et al., submitted).
Our future work is focused on improving the sensitivity of, detecting mixed VNTR patterns in, automating allele calling in, and eliminating nonspecific peaks (usually seen in samples with low bacterial numbers) in multiplex PCR and FLA. We have initiated the generation of databases that include all available polymorphism data along with relevant clinical information for each sample so as to enable molecular geneticists and clinical epidemiologists objective means for tracing leprosy and for implementing preventive measures. These approaches will be explored in a future paper (Sakamuri et al., submitted).
Acknowledgments
This work was supported by NIH NIAID grants AI-59644, AI-063457, and AI-47197 and contract NOI AI-25469.
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Blake, L. A., B. C. West, C. H. Lary, and J. R. Todd, IV. 1987. Environmental nonhuman sources of leprosy. Rev. Infect. Dis. 9562-577. [DOI] [PubMed] [Google Scholar]
- 2.Chen, S., L. Zhang, D. Liu, and B. Liu. 2003. Should household contact examination in a low endemic situation of leprosy continue? Int. J. Lepr. Other Mycobact. Dis. 7195-100. [DOI] [PubMed] [Google Scholar]
- 3.Cole, S. T., P. Supply, and N. Honoré. 2001. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr. Rev. 72449-461. [PubMed] [Google Scholar]
- 4.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honoré, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 4091007-1011. [DOI] [PubMed] [Google Scholar]
- 5.dela Cruz, E., R. V. Cellona, M. V. Balagon, L. G. Villahermosa, T. T. Fajardo, Jr., R. M. Abalos, E. V. Tan, and Walsh, G. P. 1996. Primary dapsone resistance in Cebu, The Philippines; cause for concern. Int. J. Lepr. Other Mycobact. Dis. 64253-256. [PubMed] [Google Scholar]
- 6.Groathouse, N. A., B. Rivoire, H. Kim, H. Lee, S. N. Cho, P. J. Brennan, and V. D. Vissa. 2004. Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae. J. Clin. Microbiol. 421666-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Federation of Anti-Leprosy Associations. 2003. Annual report. The situation of activities supported by ILEP in 2002. International Federation of Anti-Leprosy Associations, London, United Kingdom.
- 8.Izumi, S. 1999. Subclinical infection by Mycobacterium leprae. Int. J. Lepr. Other Mycobact. Dis. 67S67-S71. [PubMed] [Google Scholar]
- 9.Jain, S., R. G. Reddy, S. N. Osmani, D. N. Lockwood, and S. Suneetha. 2002. Childhood leprosy in an urban clinic, Hyderabad, India: clinical presentation and the role of household contacts. Lepr. Rev. 73248-253. [PubMed] [Google Scholar]
- 10.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 1822928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerr-Pontes, L. R., A. C. Montenegro, M. L. Barreto, G. L. Werneck, and H. Feldmeier. 2004. Inequality and leprosy in Northeast Brazil: an ecological study. Int. J. Epidemiol. 33262-269. [DOI] [PubMed] [Google Scholar]
- 12.Kumar, A., A. Girdhar, and B. K. Girdhar. 2003. Epidemiology of leprosy in urban Agra. Lepr. Rev. 7431-34. [PubMed] [Google Scholar]
- 13.Levy, L. 1976. Studies of the mouse foot pad technique for cultivation of Mycobacterium leprae. 3. Doubling time during logarithmic multiplication. Lepr. Rev. 47103-106. [DOI] [PubMed] [Google Scholar]
- 14.Maeda, S., M. Matsuoka, N. Nakata, M. Kai, Y. Maeda, K. Hashimoto, H. Kimura, K. Kobayashi, and Y. Kashiwabara. 2001. Multidrug resistant Mycobacterium leprae from patients with leprosy. Antimicrob. Agents Chemother. 453635-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malorny, B., E. Junker, and R. Helmuth. 2008. Multi-locus variable-number tandem repeat analysis for outbreak studies of Salmonella enterica serotype Enteritidis. BMC Microbiol. 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuoka, M., L. Zhang, T. Budiawan, K. Saeki, and S. Izumi. 2004. Genotyping of Mycobacterium leprae on the basis of the polymorphism of TTC repeats for analysis of leprosy transmission. J. Clin. Microbiol. 42741-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuoka, M., S. Maeda, M. Kai, N. Nakata, G.-T. Chae, T. P. Gillis, K. Kobayashi, S. Izumi, and Y. Kashiwabara. 2000. Mycobacterium leprae typing by genomic diversity and global distribution of genotypes. Int. J. Lepr. 68121-128. [PubMed] [Google Scholar]
- 18.Meyers, W. M., B. J. Gormus, and G. P. Walsh. 1992. Nonhuman sources of leprosy. Int. J. Lepr. Other Mycobact. Dis. 60477-480. [PubMed] [Google Scholar]
- 19.Monot, M., N. Honoré, N. Balière, B. Ji, S. Sow, P. J. Brennan, and S. T. Cole. 2008. Are variable-number tandem repeats appropriate for genotyping Mycobacterium leprae? J. Clin. Microbiol. 462291-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monot, M., N. Honoré, T. Garnier, R. Araoz, J. Y. Coppée, C. Lacroix, S. Sow, J. S. Spencer, R. W. Truman, D. L. Williams, R. Gelber, M. Virmond, B. Flageul, S. N. Cho, B. Ji, A. Paniz-Mondolfi, J. Convit, S. Young, P. E. Fine, V. Rasolofo, P. J. Brennan, and S. T. Cole. 2005. On the origin of leprosy. Science 3081040-1042. [DOI] [PubMed] [Google Scholar]
- 21.Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, NY.
- 22.Rees, R. J. 1983. Progress in the preparation of an antileprosy vaccine from armadillo-derived Mycobacterium leprae. Int. J. Lepr. Other Mycobact. Dis. 51515-518. [PubMed] [Google Scholar]
- 23.Shin, Y. C., H. Lee, H. Lee, G. P. Walsh, J. D. Kim, and S. N. Cho. 2000. Variable numbers of TTC repeats in Mycobacterium leprae DNA from leprosy patients and use in strain differentiation. J. Clin. Microbiol. 384535-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rüsch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 444498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truman, R., A. B. Fontes, A. B. De Miranda, P. Suffys, and T. Gillis. 2004. Genotypic variation and stability of four variable-number tandem repeats and their suitability for discriminating strains of Mycobacterium leprae. J. Clin. Microbiol. 422558-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Beers, S. M., M. Hatta, and P. R. Klatser. 1999. Patient contact is the major determinant in incident leprosy: implications for future control. Int. J. Lepr. Other Mycobact. Dis. 67119-128. [PubMed] [Google Scholar]
- 27.Williams, D. L., T. P. Gillis, and F. Portaels. 1990. Geographically distinct isolates of Mycobacterium leprae exhibit no genotypic diversity by restriction fragment-length polymorphism analysis. Mol. Microbiol. 41653-1659. [DOI] [PubMed] [Google Scholar]
- 28.World Health Assembly, World Health Organization. 1991. Elimination of leprosy: resolution of the 44th World Health Assembly. Resolution WHA 44.9. World Health Organization, Geneva, Switzerland.
- 29.World Health Organization. 2008. Global leprosy situation, beginning of 2008. Wkly. Epidemiol. Rec. 83(33)293-300. [PubMed] [Google Scholar]
- 30.World Health Organization. 1982. Chemotherapy of leprosy for control programme. WHO Tech. Rep. Ser. 675. [PubMed]
- 31.World Health Organization. 2003. The final push strategy to eliminate leprosy as a public health problem. Questions and answers, 2nd ed. World Health Organization, Geneva, Switzerland.
- 32.Yoon, K. H., S. N. Cho, M. K. Lee, R. M. Abalos, R. V. Cellona, T. T. Fajardo, Jr., L. S. Guido, E. C. Dela Cruz, G. P. Walsh, and J. D. Kim. 1993. Evaluation of polymerase chain reaction amplification of Mycobacterium leprae-specific repetitive sequence in biopsy specimens from leprosy patients. J. Clin. Microbiol. 31895-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young, S. K., G. M. Taylor, S. Jain, L. M. Suneetha, S. Suneetha, D. N. Lockwood, and D. B. Young. 2004. Microsatellite mapping of Mycobacterium leprae populations in infected humans. J. Clin. Microbiol. 424931-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, L., T. Budiawan, and M. Matsuoka. 2005. Diversity of potential short tandem repeats in Mycobacterium leprae and application for molecular typing. J. Clin. Microbiol. 435221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]