Abstract
Quantification of hepatitis C virus (HCV) RNA is essential for the everyday management of chronic hepatitis C therapy. “Real-time” PCR techniques are potentially more sensitive than classical PCR techniques, are not prone to carryover contamination, and have a consistently wider dynamic range of quantification. Thus, they are rapidly replacing other technologies for routine quantification of HCV RNA. We extensively evaluated the intrinsic characteristics and clinical performance of the m2000sp-m2000rt Abbott real-time PCR platform for HCV RNA quantification. The study shows that the m2000sp-m2000rt platform is sensitive, specific, and precise; that the results are reproducible; and that the platform has a broad dynamic range of quantification. When comparing HCV RNA levels measured in the same individuals with the m2000sp-m2000rt platform and the third-generation branched-DNA assay, a trend toward a modest overestimation of HCV RNA levels was observed in the m2000sp-m2000rt platform in all genotypes except genotype 5. The differences, however, were unlikely to have any impact in clinical practice. In conclusion, our study shows that the Abbott m2000 real-time PCR system for HCV RNA quantification is sensitive, specific, and precise; that the results are reproducible; and that the platform's broad dynamic range of quantification is well suited to HCV RNA monitoring in the clinical setting.
Monitoring of hepatitis C virus (HCV) RNA levels is essential for the management of chronic hepatitis C therapy with the combination of pegylated alpha interferon and ribavirin. Indeed, the rapid (undetectable HCV RNA at week 4 of therapy) and early (more than 2 log10 drop or undetectable HCV RNA at week 12 of therapy) virologic response is a strong predictor of the likelihood of sustained viral eradication and is used to tailor the treatment duration and to improve cure rates (2, 6, 7, 10, 14, 16, 23, 26, 27). HCV RNA quantification will continue to be of major importance in the management of therapy with the new HCV drugs in development to assess the virologic response and to detect virologic breakthroughs related to viral resistance early enough to alter therapy and rescue antiviral efficacy (20).
Ideally, HCV RNA quantification assays should be sensitive, specific, accurate, and precise, and the results should be reproducible. They should have a broad range of linear quantification that fully covers the HCV RNA levels observed in clinical practice in both untreated and treated patients, and quantification should be independent of the HCV genotype. Real-time PCR techniques currently are replacing classical PCR methods and the branched-DNA (bDNA) technology, which did not fulfill all of these criteria. Real-time PCR methods are sensitive, with lower limits of detection/quantification on the order of 10 to 15 HCV RNA international units (IU)/ml, and are not prone to carryover contamination. They benefit from a broad dynamic range of quantification of 7 to 8 log10 units that covers most of the HCV RNA levels encountered in clinical practice.
In this study, we investigated the intrinsic and clinical performances of the recently developed Abbott m2000 real-time PCR system for HCV RNA quantification, which uses the automated extractor m2000sp and the m2000rt device for automated real-time PCR amplification and detection of PCR products (m2000sp-m2000rt; Abbott Diagnostic, Chicago, IL).
MATERIALS AND METHODS
Standards.
A standard panel (OptiQuant HCV RNA; AcroMetrix, Benicia, CA) containing different concentrations of HCV RNA from a single source, i.e., an HCV-infected individual with HCV genotype 1b (15), was used to study the analytical performance of the assay. The seven panel members were NAP-000, NAP-HCV5E1, NAP-HCV5E2, NAP-HCV5E3, NAP-HCV5E4, NAP-HCV5E5, and NAP-HBV5E6, which contain no HCV RNA, 5 × 101 IU/ml (1.7 log10 IU/ml), 5 × 102 IU/ml (2.7 log10 IU/ml), 5 × 103 IU/ml (3.7 log10 IU/ml), 5 × 104 IU/ml (4.7 log10 IU/ml), 5 × 105 IU/ml (5.7 log10 IU/ml), and 5 × 106 IU/ml (6.7 log10 IU/ml), respectively.
Clinical specimens.
Serum samples were obtained from patients followed in the Department of Hepatology and Gastroenterology of Henri Mondor Hospital, Créteil, France. Group A comprised 202 HCV-seronegative individuals, i.e., subjects without total anti-HCV antibodies in a third-generation enzyme immunoassay (Vitros ECi; Ortho-Clinical Diagnostics, Raritan, NJ). Group B comprised 141 patients with chronic HCV infection, characterized by the presence of both total anti-HCV antibodies and HCV RNA. The HCV genotype was determined in all cases by means of direct sequence analysis of a portion of the nonstructural 5B (NS5B) gene that encodes the RNA-dependent RNA polymerase, followed by phylogenetic analysis, as recently described (3). Group B included 55 patients infected with HCV genotype 1, 21 with genotype 2, 29 with genotype 3, 24 with genotype 4, 9 with genotype 5, and 3 with genotype 6. Serum was separated from whole blood by centrifugation, placed into sterile tubes, and frozen at −70°C until it was used in the study. Group C included 10 patients infected with HCV genotype 1, 7 with genotype 2, 8 with genotype 3, 9 with genotype 4, and 1 with genotype 5, whose sera had been tested for HCV RNA with the Cobas Ampliprep/Cobas TaqMan (CAP/CTM) real-time PCR assay (Roche Molecular Systems, Pleasanton, CA).
Assessment of m2000sp-m2000rt platform performance. (i) Analytical sensitivity.
In order to evaluate the analytical sensitivity of the m2000sp-m2000rt assay relative to the manufacturer's stated lower limit of detection (12 IU/ml), serial 1/2 dilutions of the NAP-HCV5E4 standard down to 6.25 IU/ml were tested 20 times.
(ii) Specificity.
The specificity of the m2000sp-m2000rt assay was assessed by testing the 202 HCV-seronegative clinical specimens from group A.
(iii) Linearity, accuracy, and influence of the HCV genotype.
The linearity of quantification in the m2000sp-m2000rt platform was assessed by testing the seven samples of the standard panel OptiQuant HCV RNA, which contain up to 5 × 106 IU/ml (6.7 log10 IU/ml). Each panel member dilution was tested six times in the same experiment with the m2000sp-m2000rt platform and three times in the same experiment with the third-generation bDNA-based assay, the Versant HCV RNA 3.0 assay. The average measured values were compared with the expected HCV RNA levels. In addition, serial one-fifth dilutions down to signal extinction were tested in 10 genotype 1, 10 genotype 2, 9 genotype 3, 10 genotype 4, 4 genotype 5, and 3 genotype 6 samples from group B. The dilutions were made with the Nucleic Acid Test dilution matrix (AcroMetrix), a defibrinated, delipidized normal human plasma.
We also compared the results of the m2000sp-m2000rt platform with those obtained for the same samples with the third-generation bDNA-based assay in the 141 samples from group B. The bDNA assay can be confidently used as a comparator, since it has been shown to be precise and accurate and to equally quantify HCV genotypes 1 to 6, due to the use of a set of 6 capture and 17 extender oligonucleotide probes spanning the full-length 5′-noncoding region and the 5′ third of the core coding region for hybridization of the HCV genome (1, 8, 9, 12, 13, 17-19, 24).
Finally, HCV RNA levels obtained with the m2000sp-m2000rt platform were compared with those obtained in the same samples with the CAP/CTM assay and the third-generation bDNA-based assay in the 35 HCV-positive samples from group C.
(iv) Precision and reproducibility.
In order to assess precision (or intra-assay reproducibility), each member of the OptiQuant HCV RNA panel was tested six times. In order to assess interassay reproducibility, the low positive control and the high positive control provided in the kits were tested 38 times in corresponding runs on different days.
HCV RNA quantification. (i) m2000sp-m2000rt.
HCV RNA was extracted from 500 μl of serum in the automated extractor m2000sp, according to the manufacturer's instructions. The m2000rt device was then used for automated real-time PCR amplification and detection of PCR products according to the manufacturer's instructions. HCV RNA levels were expressed in IU/ml.
(ii) bDNA.
In the Versant HCV RNA 3.0 Assay, HCV RNA was recovered from 50 μl of serum and quantified in the semiautomated System 340 bDNA analyzer (Siemens Medical Solutions Diagnostics, Tarrytown, NY), according to the manufacturer's instructions. HCV RNA levels were expressed in IU/ml.
(iii) CAP/CTM.
HCV RNA was extracted from 850 μl of serum in the automated Cobas AmpliPrep extractor, according to the manufacturer's instructions. The Cobas TaqMan 48 Analyzer was used for automated real-time PCR amplification and detection of PCR products. The generated data were analyzed with Amplilink software. HCV RNA levels were expressed in IU/ml.
Statistical analysis.
Descriptive statistics are shown as the mean ± standard deviation (SD) or as medians and interquartile ranges as appropriate. Comparisons between groups were made using the Kruskall-Wallis test or the Mann-Whitney test. The relationship between quantitative variables was studied by means of regression analysis. P values of <0.05 were considered significant.
RESULTS
Intrinsic performance of the m2000sp-m2000rt assay. (i) Analytical sensitivity.
Serial 1/2 dilutions of a standard containing 50 IU/ml HCV RNA down to 6.25 IU/ml were tested 20 times in different experiments with the m2000sp-m2000rt platform. HCV RNA was detected in 20/20, 20/20, 13/20, and 13/20 replicate tests of samples containing 50 IU/ml (1.7 log10 IU/ml), 25 IU/ml (1.4 log10 IU/ml), 12.5 IU/ml (1.1 log10 IU/ml), and 6.25 IU/ml (0.8 log10 IU/ml), respectively. This result was in keeping with the manufacturer's stated lower limit of detection of the assay (12 IU/ml).
(ii) Specificity.
The specificity of the m2000sp-m2000rt platform was assessed by testing 202 samples from anti-HCV-seronegative patients (group A). No sample tested positive above the lower limit of detection of 12 IU/ml (specificity, 100%; 95% confidence interval, 99 to 100%).
(iii) Precision and reproducibility.
Precision (intra-assay reproducibility) was assessed by testing the seven members of the OptiQuant HCV RNA standard that contained 0, 1.7, 2.7, 3.7, 4.7, 5.7, and 6.7 log10 IU/ml, respectively, six times in the same experiment. As shown in Table 1, the coefficients of variation varied from 0.76% to 7.68%. Interassay variability was assessed by testing both the high positive control and the low positive control, extracted like clinical samples, 38 times in different experiments. The coefficients of variation were 1.98% and 3.91%, respectively (Table 1).
TABLE 1.
Intra-assay (precision) and interassay reproducibility of m2000sp-m2000rt HCV real-time PCR assay results
| Reproducibilitya | Standard | Control | Target HCV RNA (log10 IU/ml) | No. of determinations | Mean (SD) measured HCV RNA (log10 IU/ml) | Coefficient of variation (%) |
|---|---|---|---|---|---|---|
| Intra-assay | NAP-000 | 0.00 | 6 | Target not detected | ||
| NAP-HCV5E1 | 1.70 | 6 | 1.39 (0.11) | 7.68 | ||
| NAP-HCV5E2 | 2.70 | 6 | 2.31 (0.06) | 2.74 | ||
| NAP-HCV5E3 | 3.70 | 6 | 3.30 (0.03) | 0.76 | ||
| NAP-HCV5E4 | 4.70 | 6 | 4.27 (0.05) | 1.28 | ||
| NAP-HCV5E5 | 5.70 | 6 | 5.13 (0.28) | 5.47 | ||
| NAP-HCV5E6 | 6.70 | 6 | 6.47 (0.14) | 2.14 | ||
| Interassay | LPC | 2.63 | 38 | 2.64 (0.10) | 3.91 | |
| HPC | 5.87 | 38 | 5.98 (0.12) | 1.98 |
For intra-assay reproducibility, the seven members of the standard panel (NAP-000 to NAP-HCV5E6) containing 0 to 5 × 106 IU/ml, i.e., 6.70 log10 IU/ml, respectively, were tested six times in the same experiment. For interassay reproducibility, the assay low positive control (LPC) and high positive control (HPC) were tested 38 times in different experiments.
Accuracy, linear quantification, and influence of the HCV genotype. (i) Linear quantification of standard panel dilutions.
The OptiQuant HCV RNA genotype 1b standard panel, containing 5 × 101 IU/ml (1.7 log10 IU/ml) to 5 × 106 IU/ml (6.7 log10 IU/ml), was used to assess the linearity of HCV RNA quantification in the m2000sp-m2000rt assay. The panel was tested six times and three times in the same experiments with the m2000sp-m2000rt platform and bDNA, respectively. As shown in Fig. 1A, a significant relationship was found between the average measured and the expected HCV RNA levels (r = 0.9980; P < 0.0001). The difference between the average measured and the expected HCV RNA levels varied from 0.23 to 0.57 log10 IU/ml. A significant relationship was also found between the average measured and the expected HCV RNA levels in the bDNA assay (r = 0.9997; P < 0.0001) (Fig. 1B). The difference between the average measured and the expected HCV RNA levels varied from 0.21 to 0.33 log10 IU/ml. The HCV RNA levels of three standards, NAP-000, NAP-HCV5E1, and NAP-HCV5E2, was below the limit of detection of the bDNA assay (615 IU/ml, i.e., 2.79 log10 IU/ml).
FIG. 1.
HCV RNA quantification of a commercial standard panel containing 5 × 101 (1.7 log10) to 5 × 106 (6.7 log10) HCV RNA IU/ml (OptiQuant HCV RNA; AcroMetrix, Benicia, CA) by the m2000sp-m2000rt platform (top) and bDNA (bottom). The average measured results are shown as a function of the expected results (solid lines). The dotted lines are the equality lines.
(ii) Quantification of HCV RNA in clinical samples containing HCV genotypes 1 to 6.
One hundred and forty-one samples from patients with chronic hepatitis C infected with HCV genotypes 1 to 6 (group B) (see Materials and Methods) were tested with both the m2000sp-m2000rt platform and the third-generation bDNA assay. All of these samples fell within the dynamic ranges of quantification of both assays. As shown in Fig. 2, there was a significant relationship between the HCV RNA levels obtained in the same samples with the m2000sp-m2000rt platform and bDNA for genotypes 1 to 5 (only three genotype 6 samples were tested). The regression lines were slightly above the expected equality line, as a result of higher values obtained with the m2000sp-m2000rt platform than with bDNA for the same sample in the majority of cases (131 out of 141 clinical specimens tested) (Fig. 2).
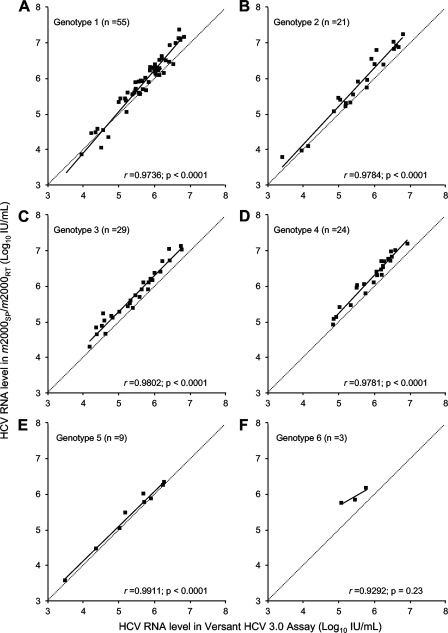
FIG. 2.
Correlation between HCV RNA levels measured by the m2000sp-m2000rt platform and bDNA in the same samples from group B, consisting of 141 clinical samples containing HCV genotype 1 (A), 2 (B), 3 (C), 4 (D), 5 (E), and 6 (F). The dotted lines are the equality lines.
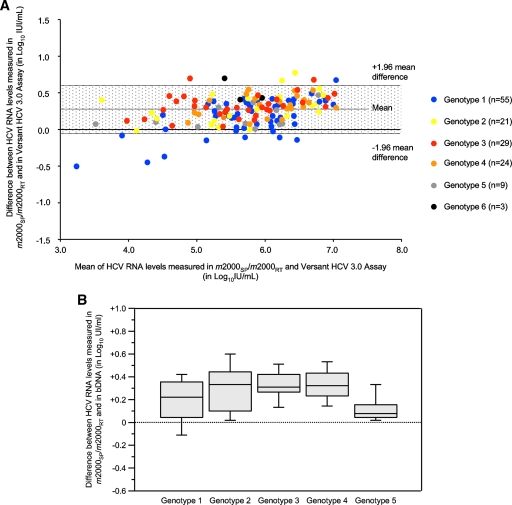
Figure 3A shows a Bland-Altman analysis of the HCV RNA levels measured in the 141 samples from group B by both the m2000sp-m2000rt platform and bDNA. The figure plots the difference between the two measured values (the m2000sp-m2000rt platform minus bDNA) as a function of the mean of both measurements. A moderate overestimation of HCV RNA levels by the m2000sp-m2000rt platform compared to bDNA was observed in 131 of the 141 samples (92.9%) containing all HCV genotypes (median m2000sp-m2000rt minus bDNA difference, 0.30 log10 IU/ml). Six samples (one genotype 1, two genotype 2, two genotype 4, and one genotype 6) had an m2000sp-m2000rt minus bDNA difference below −1.96 times the mean difference, whereas eight samples (all of genotype 1) had an m2000sp-m2000rt minus bDNA difference above +1.96 times the mean difference. The individual differences between the m2000sp-m2000rt platform and bDNA, however, were always below 1.0 log10 IU/ml in these samples. Box plots of individual differences between the two methods are shown for each genotype in Fig. 3B. They confirm the global, very moderate overestimation of HCV RNA levels in the m2000sp-m2000rt platform compared to bDNA independent of the HCV genotype. The median differences were 0.22 log10 IU/ml for genotype 1, 0.33 log10 IU/ml for genotype 2, 0.31 log10 IU/ml for genotype 3, 0.32 log10 IU/ml for genotype 4, and −0.08 log10 IU/ml for genotype 5 (P = 0.013). HCV genotype 6 is not shown in Fig. 3B because only three samples were tested.
FIG. 3.
(A) Bland-Altman plot analysis of HCV RNA levels measured by both the m2000sp-m2000rt platform and bDNA in the 141 samples from group B. The difference between HCV RNA levels measured by the m2000sp-m2000rt platform and bDNA is represented as a function of the mean of the two values. Different genotypes are represented by different colors. The gray area corresponds to the mean difference ± 1.96 SD. (B) Distribution of the differences between HCV RNA levels measured in the m2000sp-m2000rt platform and bDNA in the same samples according to the HCV genotype (1 to 5). The results are presented as box plots, where the horizontal line represents the median value, the gray boxes the 50th percentiles, and the upper and lower bars the 95th percentiles.
(iii) Linear quantification of serial dilutions of HCV-infected sera.
Serial one-fifth dilutions down to signal extinction were tested in 10 genotype 1, 10 genotype 2, 9 genotype 3, 10 genotype 4, 4 genotype 5, and 3 genotype 6 samples from group B. Serial dilution quantification was linear in all cases, whatever the HCV genotype, with Pearson's coefficient ranging from 0.9940 to 0.9996 for HCV genotype 1, 0.9930 to 0.9996 for HCV genotype 2, 0.9971 to 0.9996 for HCV genotype 3, 0.9812 to 0.9997 for HCV genotype 4, 0.9938 to 0.9989 for HCV genotype 5, and 0.9975 to 0.9983 for HCV genotype 6. The expected difference between two successive one-fifth dilutions is 0.70 log10 IU/ml. The mean ± SD differences between the undiluted sample and the first one-fifth dilution, and between each dilution and the subsequent one, were 0.81 ± 0.12, 0.76 ± 0.12, 0.74 ± 0.08, 0.77 ± 0.13, 0.76 ± 0.11, and 0.75 ± 0.11, respectively (not significantly different).
Comparison with CAP/CTM.
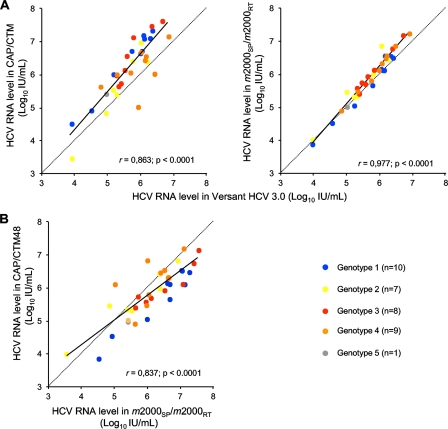
Sera from 35 patients with chronic HCV infection with different genotypes (group C) were tested with the m2000sp-m2000rt assay, the CAP/CTM assay, and the third-generation bDNA-based assay. Figure 4A shows the better correlation between the m2000sp-m2000rt platform and bDNA than between CAP/CTM and bDNA. The greatest differences between CAP/CTM and bDNA were seen for HCV genotype 2 and 4 samples, as reported previously (4). Figure 4B shows the relationship between HCV RNA levels measured in the same samples by the m2000sp-m2000rt platform and CAP/CTM.
FIG. 4.
Correlation between HCV RNA levels measured by CAP/CTM and bDNA (left) and by the m2000sp-m2000rt platform and bDNA (right) (A), and in CAP/CTM and the m2000sp-m2000rt platform (B) in the same samples from group C, consisting of 35 clinical samples containing HCV genotypes 1 to 5. The dotted lines are the equality lines.
DISCUSSION
Our assessment of the intrinsic performance of the m2000sp-m2000rt platform for HCV RNA quantification shows the excellent specificity, precision, and reproducibility of the technique, in keeping with previous reports (11, 22). The manufacturer's stated lower limit of detection of the assay is 12 IU/ml, as confirmed in the present study.
In clinical samples from patients with chronic hepatitis C infected with the six HCV genotypes, we observed a significant relationship between the HCV RNA levels measured in the same samples with the m2000sp-m2000rt platform and the third-generation bDNA assay. The use of the third-generation bDNA assay as the comparator was justified by the fact that the assay is accurate and reproducible, quantifies HCV RNA independently of the HCV genotype, and has been well calibrated to the World Health Organization (WHO) international HCV RNA standard, as confirmed in this study (1, 8, 9, 12, 13, 24, 25). The correlation was found for all HCV genotypes (it was not significant for genotype 6, but only three samples could be tested), and the median individual differences between the HCV RNA levels measured in the two assays were below ±0.5 log10 IU/ml (i.e., a threefold difference) in all but 10 clinical samples.
The HCV RNA quantification panel used in this study was calibrated with the WHO international HCV RNA standard by means of the second-generation bDNA assay (15, 21). A minor underestimation of HCV RNA levels was observed with both the m2000sp-m2000rt platform and the third-generation bDNA assay relative to the expected HCV RNA values, in spite of a linear relationship. When comparing HCV RNA levels measured in the same individuals by both techniques, we observed a trend toward a modest overestimation of HCV RNA levels in the m2000sp-m2000rt platform compared to the third-generation bDNA in all genotypes except genotype 5. The differences, however, were unlikely to have any impact on the clinical use of the m2000sp-m2000rt platform. Importantly, no sample was substantially underquantified in the m2000sp-m2000rt real-time PCR assay relative to bDNA. In contrast, we have shown that another real-time PCR platform for HCV RNA quantification, CAP/CTM, underestimates HCV RNA levels in approximately 30% of HCV genotype 4 and 15% of HCV genotype 2 infections as a probable result of mispairing of the PCR primers and/or TaqMan probe (4). We recently reported two patients infected with HCV genotype 4 with undetectable HCV RNA in CAP/CTM and high viral levels in the m2000sp-m2000rt platform and bDNA (5). When testing the same patients' samples with the m2000sp-m2000rt platform, CAP/CTM, and bDNA in the present study, we observed better accuracy of the m2000sp-m2000rt platform than CAP/CTM for HCV RNA quantification, especially for HCV genotype 2 and 4 samples.
We tested serial dilutions of clinical samples of the different genotypes. Quantification was linear in all cases, whatever the HCV genotype. This is unlike the CAP/CTM assay, in which we found that HCV RNA levels were overestimated in undiluted clinical samples, but not when the same samples were tested after dilution, a phenomenon observed with all six HCV genotypes (4). These findings explain the greater overestimation of HCV RNA levels relative to bDNA in CAP/CTM than with the m2000sp-m2000rt platform. The molecular basis for the HCV RNA overestimation by CAP/CTM in undiluted samples is unknown. We hypothesized a biochemical interaction with a blood component during one of the reaction steps, an interaction that vanishes when the concentration of this putative component is reduced in diluted samples (4). The results of the present study suggest that this putative interaction does not occur during the m2000sp-m2000rt reaction.
In conclusion, our study shows that the Abbott m2000 real-time PCR system for HCV RNA quantification (the m2000sp-m2000rt platform) is sensitive, specific, and precise; that the results are reproducible; and that its broad dynamic range of quantification is well suited to HCV RNA monitoring in the clinical setting. This is particularly important in the current clinical context. Indeed, novel therapeutic strategies that include HCV protease and polymerase inhibitors will require sensitive and accurate assays with a broad dynamic range of quantification to monitor the virological responses and to tailor treatment in order to prevent resistance.
Acknowledgments
The m2000sp-m2000rt kits used in this study were kindly provided by Abbott Molecular Diagnostics (Rungis, France).
This work is part of the activity of the VIRGIL European Network of Excellence on Antiviral Drug Resistance supported by a grant (LSHM-CT-2004-503359) from the Priority 1 Life Sciences, Genomics and Biotechnology for Health program in the Sixth Framework Programme of the European Union.
We are grateful to Françoise Darthuy and Guillaume Dameron for helpful technical assistance.
Footnotes
Published ahead of print on 15 April 2009.
REFERENCES
- 1.Beld, M., R. Sentjens, S. Rebers, C. Weegink, J. Weel, C. Sol, and R. Boom. 2002. Performance of the New Bayer Versant HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche Cobas Amplicor HCV Monitor, version 2.0, assay. J. Clin. Microbiol. 40788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, T., M. von Wagner, S. Nasser, C. Sarrazin, T. Heintges, T. Gerlach, P. Buggisch, T. Goeser, J. Rasenack, G. R. Pape, W. E. Schmidt, B. Kallinowski, H. Klinker, U. Spengler, P. Martus, U. Alshuth, and S. Zeuzem. 2006. Extended treatment duration for hepatitis C virus type 1: comparing 48 versus 72 weeks of peginterferon-alfa-2a plus ribavirin. Gastroenterology 1301086-1097. [DOI] [PubMed] [Google Scholar]
- 3.Bronowicki, J. P., D. Ouzan, T. Asselah, H. Desmorat, J. P. Zarski, J. Foucher, M. Bourlière, C. Renou, A. Tran, P. Melin, C. Hézode, M. Chevallier, M. Bouvier-Alias, S. Chevaliez, F. Montestruc, I. Lonjon-Domanec, and J. M. Pawlotsky. 2006. Effect of ribavirin in genotype 1 patients with hepatitis C responding to pegylated interferon alpha-2a plus ribavirin combination. Gastroenterology 1311339-1341. [DOI] [PubMed] [Google Scholar]
- 4.Chevaliez, S., M. Bouvier-Alias, R. Brillet, and J. M. Pawlotsky. 2007. Overestimation and underestimation of hepatitis C virus RNA levels in a widely used real-time polymerase chain reaction-based method. Hepatology 4622-31. [DOI] [PubMed] [Google Scholar]
- 5.Chevaliez, S., M. Bouvier-Alias, L. Castéra, and J. M. Pawlotsky. 2009. The Cobas Ampliprep-Cobas Taqman real-time polymerase chain reaction assay fails to detect hepatitis C virus RNA in highly viremic genotype 4 clinical samples. Hepatology 491397-1398. [DOI] [PubMed] [Google Scholar]
- 6.Dalgard, O., K. Bjoro, K. B. Hellum, B. Myrvang, S. Ritland, K. Skaug, N. Raknerud, and H. Bell. 2004. Treatment with pegylated interferon and ribavarin in HCV infection with genotype 2 or 3 for 14 weeks: a pilot study. Hepatology 401260-1265. [DOI] [PubMed] [Google Scholar]
- 7.Davis, G. L., J. B. Wong, J. G. McHutchison, M. P. Manns, J. Harvey, and J. Albrecht. 2003. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology 38645-652. [DOI] [PubMed] [Google Scholar]
- 8.Elbeik, T., N. Markowitz, P. Nassos, U. Kumar, S. Beringer, B. Haller, and V. Ng. 2004. Simultaneous runs of the Bayer Versant HIV-1 version 3.0 and HCV bDNA version 3.0 quantitative assays on the system 340 platform provide reliable quantitation and improved work flow. J. Clin. Microbiol. 423120-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbeik, T., J. Surtihadi, M. Destree, J. Gorlin, M. Holodniy, S. A. Jortani, K. Kuramoto, V. Ng, R. Valdes, Jr., A. Valsamakis, and N. A. Terrault. 2004. Multicenter evaluation of the performance characteristics of the Bayer Versant HCV RNA 3.0 assay (bDNA). J. Clin. Microbiol. 42563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferenci, P., M. W. Fried, M. L. Shiffman, C. I. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, M. Chaneac, and K. R. Reddy. 2005. Predicting sustained virological responses in chronic hepatitis C patients treated with peginterferon alfa-2a (40 KD)/ribavirin. J. Hepatol. 43425-433. [DOI] [PubMed] [Google Scholar]
- 11.Fytili, P., C. Tiemann, C. Wang, S. Schulz, S. Schaffer, M. P. Manns, and H. Wedemeyer. 2007. Frequency of very low HCV viremia detected by a highly sensitive HCV-RNA assay. J. Clin. Virol. 39308-311. [DOI] [PubMed] [Google Scholar]
- 12.Germer, J. J., P. J. Heimgartner, D. M. Ilstrup, W. S. Harmsen, G. D. Jenkins, and R. Patel. 2002. Comparative evaluation of the Versant HCV RNA 3.0, Quantiplex HCV RNA 2.0, and Cobas Amplicor HCV Monitor version 2.0 assays for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendricks, D. A., M. Friesenhahn, L. Tanimoto, B. Goergen, D. Dodge, and L. Comanor. 2003. Multicenter evaluation of the Versant HCV RNA qualitative assay for detection of hepatitis C virus RNA. J. Clin. Microbiol. 41651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, D. M., T. R. Morgan, P. Marcellin, P. J. Pockros, K. R. Reddy, S. J. Hadziyannis, P. Ferenci, A. M. Ackrill, and B. Willems. 2006. Early identification of HCV genotype 1 patients responding to 24 weeks peginterferon alpha-2a (40 kd)/ribavirin therapy. Hepatology 43954-960. [DOI] [PubMed] [Google Scholar]
- 15.Jorgensen, P. A., and P. D. Neuwald. 2001. Standardized hepatitis C virus RNA panels for nucleic acid testing assays. J. Clin. Virol. 2035-40. [DOI] [PubMed] [Google Scholar]
- 16.Mangia, A., R. Santoro, N. Minerva, G. L. Ricci, V. Carretta, M. Persico, F. Vinelli, G. Scotto, D. Bacca, M. Annese, M. Romano, F. Zechini, F. Sogari, F. Spirito, and A. Andriulli. 2005. Peginterferon alfa-2b and ribavirin for 12 vs. 24 weeks in HCV genotype 2 or 3. N. Engl. J. Med. 3522609-2617. [DOI] [PubMed] [Google Scholar]
- 17.Morishima, C., M. Chung, K. W. Ng, D. J. Brambilla, and D. R. Gretch. 2004. Strengths and limitations of commercial tests for hepatitis C virus RNA quantification. J. Clin. Microbiol. 42421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pawlotsky, J. M. 2002. Molecular diagnosis of viral hepatitis. Gastroenterology 1221554-1568. [DOI] [PubMed] [Google Scholar]
- 19.Pawlotsky, J. M. 2002. Use and interpretation of virological tests for hepatitis C. Hepatology 36S65-S73. [DOI] [PubMed] [Google Scholar]
- 20.Pawlotsky, J. M., S. Chevaliez, and J. G. McHutchison. 2007. The hepatitis C virus life cycle as a target for new antiviral therapies. Gastroenterology 1321979-1998. [DOI] [PubMed] [Google Scholar]
- 21.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76149-158. [DOI] [PubMed] [Google Scholar]
- 22.Schutten, M., E. Fries, C. Burghoorn-Maas, and H. G. Niesters. 2007. Evaluation of the analytical performance of the new Abbott RealTime RT-PCRs for the quantitative detection of HCV and HIV-1 RNA. J. Clin. Virol. 4099-104. [DOI] [PubMed] [Google Scholar]
- 23.Shiffman, M. L., F. Suter, B. R. Bacon, D. Nelson, H. Harley, R. Sola, S. D. Shafran, K. Barange, A. Lin, A. Soman, and S. Zeuzem. 2007. Peginterferon alpha-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N. Engl. J. Med. 357124-134. [DOI] [PubMed] [Google Scholar]
- 24.Trimoulet, P., P. Halfon, E. Pohier, H. Khiri, G. Chene, and H. Fleury. 2002. Evaluation of the Versant HCV RNA 3.0 assay for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 402031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsongalis, G. J. 2006. Branched DNA technology in molecular diagnostics. Am. J. Clin. Pathol. 126448-453. [DOI] [PubMed] [Google Scholar]
- 26.von Wagner, M., M. Huber, T. Berg, H. Hinrichsen, J. Rasenack, T. Heintges, A. Bergk, C. Bernsmeier, D. Haussinger, E. Herrmann, and S. Zeuzem. 2005. Peginterferon-alpha-2a (40KD) and ribavirin for 16 or 24 weeks in patients with genotype 2 or 3 chronic hepatitis C. Gastroenterology 129522-527. [DOI] [PubMed] [Google Scholar]
- 27.Zeuzem, S., M. Buti, P. Ferenci, J. Sperl, Y. Horsmans, J. Cianciara, E. Ibranyi, O. Weiland, S. Noviello, C. Brass, and J. Albrecht. 2006. Efficacy of 24 weeks treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C infected with genotype 1 and low pretreatment viremia. J. Hepatol. 4497-103. [DOI] [PubMed] [Google Scholar]