Abstract
Human rhinoviruses (HRV), and to a lesser extent human enteroviruses (HEV), are important respiratory pathogens. Like other RNA viruses, these picornaviruses have an intrinsic propensity to variability. This results in a large number of different serotypes as well as the incessant discovery of new genotypes. This large and growing diversity not only complicates the design of real-time PCR assays but also renders immunofluorescence unfeasible for broad HRV and HEV detection or quantification in cells. In this study, we used the 5′ untranslated region, the most conserved part of the genome, as a target for the development of both a real-time PCR assay (Panenterhino/Ge/08) and a peptide nucleic acid-based hybridization oligoprobe (Panenterhino/Ge/08 PNA probe) designed to detect all HRV and HEV species members according to publicly available sequences. The reverse transcription-PCR assay has been validated, using not only plasmid and viral stocks but also quantified RNA transcripts and around 1,000 clinical specimens. These new generic detection PCR assays overcame the variability of circulating strains and lowered the risk of missing emerging and divergent HRV and HEV. An additional real-time PCR assay (Entero/Ge/08) was also designed specifically to provide sensitive and targeted detection of HEV in cerebrospinal fluid. In addition to the generic probe, we developed specific probes for the detection of HRV-A and HRV-B in cells. This investigation provides a comprehensive toolbox for accurate molecular identification of the different HEV and HRV circulating in humans.
Human rhinoviruses (HRV) are the most frequent etiologic agents of respiratory tract infections, while human enteroviruses (HEV) replicate predominantly in the gastrointestinal tract. A large proportion of HEV can also disseminate, causing viremia and potentially invading the central nervous system (13, 20). It is also well established that some enteroviruses can exhibit respiratory tropism, acid lability, and a preference for ICAM-I (the receptor of most rhinoviruses), thus attenuating the frontier between these two viral species (4, 15, 18, 19). HRV and HEV are now classified as members of the same Enterovirus genus.
Like many RNA viruses, rhino- and enteroviruses have an intrinsic propensity to variability, as demonstrated by the very large number of different serotypes identified (http://www.picornaviridae.com/enterovirus/enterovirus.htm). This number is constantly increasing with the continuous discovery of new genotypes or even new species (1, 6, 7, 9-11, 14, 16, 17, 22, 23, 24). As a result, the use of specific diagnostic tools designed to discriminate between viral species may result in underdetection of divergent variants or new species, as exemplified by the delayed discovery of the HRV-C species. This is in parallel with the insensitivity in cell culture for both HRV and HEV isolation.
The extreme diversity of these viruses not only complicates the design of real-time PCR assays but also renders immunofluorescence almost unfeasible for broad HRV and HEV detection in cells. This latter point is of importance, since the identification of full virions remains an invaluable tool in clinical virology. In addition, the cytopathic effects associated with HRV are often spurious.
In this study, the increasing number of HRV and HEV sequences available in GenBank (http://www.ncbi.nlm.nih.gov/Genbank/index.html) was taken into account to define the most conserved regions within the genus. Once defined, these sequences were used as targets for the development of both generic real-time PCR assays and peptide nucleic acid (PNA)-based hybridization probes designed to detect all HRV and HEV species members according to publicly available sequences. By overcoming the variability of circulating strains and by targeting the internal ribosomal entry site, a functionally important and thus highly conserved region of the HRV and HEV genomes, these new detection methods will lower the risk of missing divergent HRV and HEV.
Each real-time PCR assay developed here has been designed to be adapted to the different clinical conditions and biological specimens for which these viruses need to be screened. Importantly, these assays have been adapted to the new variants recently identified. We also developed a real-time PCR assay specifically designed to provide a sensitive and targeted detection of HEV, as well as PNA-based oligoprobe assays aiming to specifically detect HRV-A and -B in cells. This investigation aims to provide a comprehensive toolbox for an accurate and sensitive identification of the different HEV and HRV.
MATERIALS AND METHODS
Virus strains.
The virus strains used to test real-time PCR and PNA probe specificities were purchased at ATCC-LGC Promochem Sarl (Molsheim, France) for the rhinovirus reference strains or were obtained from the collection of the University of Geneva Hospitals virology laboratory for the enterovirus and human parechovirus (HPeV) strains.
Virus stocks of HRV-14 and EV-71 were grown in 75-cm2 flasks (HeLa Ohio [HeLa-OH] cells and Vero cells, respectively). The virus supernatants were clarified, aliquoted, and frozen at −80°C. Both virus stocks were serially diluted by 10-fold and titrated on their respective cell lines to determine their 50% cell culture infectious doses (CCID50) (21). One hundred ninety microliters of each dilution was used for RNA extraction in duplicate. CCID50 values included in each reverse transcription (RT) reaction are shown (see Fig. 2D to F). One-eighth of the RT product was used in each PCR.
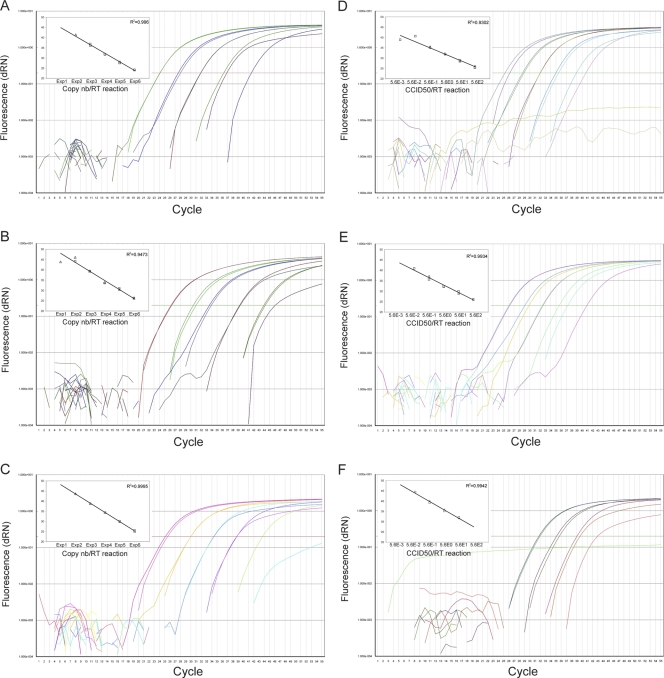
FIG. 2.
Sensitivities of the Panenterhino (A and D), Panenterhino/Ge/08 (B and E), and Entero/Ge/08 (C and F) real-time PCRs tested on serial dilutions of HRV-14 (A, B, D, and E) and EV-71 (C, F) using in vitro transcripts (A to C) and quantified virus stocks (D to F). The insets show the standard curves for each of the standard dilution series. Extractions were performed in duplicate (□, first round of extractions; ▵, second round of extractions). The viral copy number (nb) (A to C) and the CCID50 value (D to F) input in the RT reaction are indicated on the x axis.
Nucleotide sequence alignment and primer and probe design.
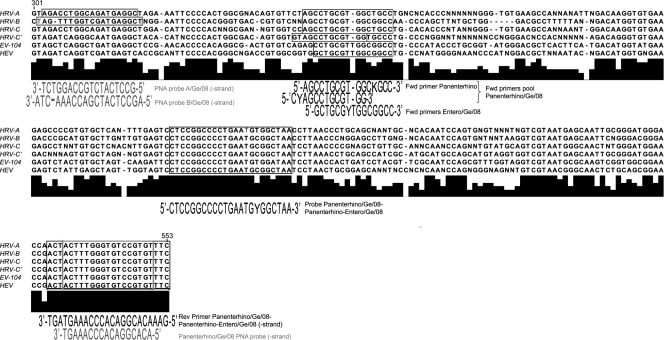
Available 5′ nontranslated region (NTR) sequences for all HRV-A, HRV-B, and 86 HEV reference strains (http://www.picornaviridae.com/sequences/sequences.htm), as well as sequences from clinical strains of EV-104 (a HEV-C genotype presenting a divergent 5′ NTR; GenBank accession no. EU840733) (24), HRV-C, and HRV-C with divergent 5′ NTR sequences (GenBank no. EU126764, EU126772, EU126775, EU126777, EU126781, EU126783-4, EU126786, EU126788, EU840728, EU840729, and EU840974; named here HRV-C′) (11, 24), were downloaded from GenBank (August 2008). The 5′ NTR sequences from each of these six virus groups were aligned independently with the Vector NTI Advance 10 AlignX module (Invitrogen, Basel, Switzerland). The residue fraction for consensus calculation was set to 0.6 for each alignment, and residues presenting a fraction inferior to this value were replaced by degenerate nucleotides. The resulting six consensus sequences were aligned together and used for real-time PCR primers and probes as well as for PNA probe design (Table 1). Real-time PCR primers were ordered from Microsynth AG (Balgach, Switzerland), and probes were ordered from Applied Biosystems (Rotkreuz, Switzerland). PNA probes sequences were designed with the help of the Applied Biosystems online design tool (http://www.appliedbiosystems.com/support/pnadesigner.cfm) and ordered from Panagene Inc. (Daejeon, Korea).
TABLE 1.
Primers, probes, and PNA probe sequences
| Type of assay | Target(s) | Sequence (5′ to 3′) (nt positions relative to HRV-2)a
|
||
|---|---|---|---|---|
| Forward primer(s) | Probe | Reverse primer | ||
| PNA probe | ||||
| A/Ge/08 | HRV-A | [Fam]-OO-GCCTCATCTGCCAGGTCT (nt 320 to 303)d | ||
| B/Ge/08 | HRV-B | [Fam]-OO-AGCCTCATCGACCAAACTA (nt 321 to 302)d | ||
| Panenterhino/Ge/08 | HRV/HEV | [Fam]-OO-ACACGGACACCCAAAGT (nt 550 to 534)d | ||
| Real-time PCR | ||||
| Panenterhino | HRV/HEV | AGCCTGCGTGGCKGCC (nt 350 to 365) | CTCCGGCCCCTGAATGYGGCTAA (nt 436 to 458)b | GAAACACGGACACCCAAAGTAGT (nt 553 to 531)d |
| Panenterhino/Ge/08 | HRV/HEVc | AGCCTGCGTGGCKGCC (nt 350 to 365) | CTCCGGCCCCTGAATGYGGCTAA (nt 436 to 458)b | GAAACACGGACACCCAAAGTAGT (nt 553 to 531)d |
| CYlnaAGCClnaTGCGTGG (nt 348 to 360)e | ||||
| Entero/Ge/08 | HEV | GCTGCGYTGGCGGCC (nt 352 to 365) | CTCCGGCCCCTGAATGYGGCTAA (nt 436 to 458)b | GAAACACGGACACCCAAAGTAGT (nt 553 to 531)d |
nt, nucleotide; [Fam], 6-carboxyfluorescein tag.
The common real-time PCR probe was labeled at the 5′ end with 6-carboxyfluorescein and at the 3′ end with the 6-carboxytetramethylrhodamine fluorescent quencher.
Improved sensitivity for HRV-C′.
Negative-strand sequence.
lna, locked nucleic acid.
Plasmids.
Positive controls for the different real-time PCR assays were constructed as follows: HRV-2 and HRV-14 viral stocks were amplified with forward primer P10 (5′-GTACWCTRKTAYTMYGGTAMYYTTGTACGCC-3′) and reverse primer P1.12 (5′-AGTGCATCTGGTAATTTCCA-3′); CL-Lba 170085 (belonging to HRV-C species; GenBank no. EU840952) was amplified with the same forward primer (P10) and with reverse primer P23 (5′-GAAACACGGACACCCAAAGTAGT-3′); CL-Fnp5 (belonging to HRV-C subgroup viruses with divergent 5′ NTRs; GenBank no. EU840728) was amplified with forward primer P11 (5′-GCACTTCTGTTTCCCC-3′) and reverse primer P23; CL-04993 (EV-71 clinical strain; GenBank no. EU414333) was amplified with forward primer Ent_5′NTR (5′-TTAAAACAGCTGTGGGTTGTTCCC-3′) and reverse primer Ent_P1.3 (5′-AAGTAGTGAAACTGTGCATTCTGGCC-3′); and CL-1231094 (EV-104 clinical strains; GenBank no. EU840733) was amplified with forward primer P10 and reverse primer P1.152 (5′-GCAAATGAATTTTTGGCGGG-3′). The different PCR products were cloned with TOPO TA technology in the pCR2.1-TOPO and pCR-Blunt II-TOPO (for CL-1231094) vectors (Invitrogen).
Production of in vitro RNA transcripts for RT-PCR standard curves.
EV-71 (kindly provided by M. Arita, National Institute of Infectious Diseases, Tokyo, Japan) and HRV-14 infectious clones (2) were linearized at a unique MluI restriction site downstream of the 3′ viral poly(A). RNA transcripts were synthesized in vitro with the T7 RNA polymerase from their respective linear templates using the MEGAscript high yield transcription T7 kit (Ambion, Rotkreuz, Switzerland) for 3 h at 37°C and purified with the RNeasy minikit (Qiagen, Hombrechtikon, Switzerland). Transcribed RNA was quantified and checked by 0.1% sodium dodecyl sulfate-1% agarose gel analysis. RNA transcript concentrations were then adjusted and serially diluted by 10-fold in order to use 106 to 10−1 viral RNA copies per RT reaction. One-fourth of the RT product was used in each PCR.
RNA extraction, RT, and real-time PCR.
Virus isolates as well as clinical specimens were extracted by the HCV Amplicor specimen preparation kit (Roche, Rotkreuz, Switzerland), TRIzol kit (Invitrogen), or QIAamp viral RNA minikit (Qiagen), according to the manufacturers' instructions. Extracted RNA, as well as in vitro transcribed RNA, was used as a template for the synthesis of cDNA with random hexamers (Roche) at 42°C using the reverse transcriptase SuperScript II (Invitrogen), according to the manufacturers' instructions. The cDNA was then amplified using a TaqMan 7500 (Applied Biosystems) thermocycler under the following cycling conditions: 50°C for 2 min, 95°C for 10 min, 55 cycles of 95°C for 15 sec, and 60°C for 1 min. Results were analyzed using the SDS version 1.4 program (Applied Biosystems).
Hybridization in situ using PNA probes.
HRV infections were done with HeLa-OH cells, whereas EV-69 was grown on A549 or Vero cells, EV-70 and CV-A21 on A549 cells only, and EV-71 on Vero cells only. In situ hybridization of paraffin-embedded cells was performed with the PNA ISH detection kit protocol (DakoCytomation, Baar, Switzerland), according to the manufacturer's instructions. The same protocol was adapted for in situ hybridization on growing cells, as follows. PNA probes were first diluted in water to 10 μM, and then the working dilutions (10 nM for A/Ge/08 and B/Ge/08 PNA probes and 1 nM for the Panenterhino/Ge/08 PNA probe) were made in the hybridization mix (30% formamide, 10% dextran sulfate, 0.1% sodium pyrophosphate, 0.2% polyvinylpyrrolidone, 0.2% Ficoll, 5 mM EDTA [pH 8.8], 50 mM Tris-HCl [pH 7.5], 10 mM NaCl). Cells were washed twice with phosphate-buffered saline (PBS) lacking Ca2+ and Mg2+ (PBS−) and fixed with 4% paraformaldehyde for 10 min at room temperature at 16 h postinfection for titrations and A/Ge/08 and B/Ge/08 PNA probe specificity tests and 4 days postinfection for the Panenterhino/Ge/08 PNA probe specificity test. After being briefly washed with PBS−, cells were rehydrated in the presence of 1 ml of PBS− for 10 min at room temperature. Cells were then permeabilized with proteinase K provided by the kit (diluted 1:100 in Tris-buffered saline) for 30 min at room temperature in a humidity chamber. Cells were rinsed with PBS−, briefly fixed again for 5 min with 4% paraformaldehyde at room temperature, and washed for 5 min in PBS−. After two immersions in RNase-free water for 3 min, the remaining protocol followed the kit procedure. Hybridization was made at 55°C for 1.5 h for the three probes, and washes were performed at 60°C.
Immunofluorescence.
Cells were washed twice with PBS− and fixed for 1.5 h in methanol-acetone (50:50 dilution) at −20°C. They were then air dried for a few minutes at room temperature before incubation for 45 min at 37°C in a humidity chamber with the following primary antibodies: a rabbit anti-HRV-14 serum, diluted 1/1,000 (VR-284AS/Rb; ATCC) for the detection of HRV-14; a mouse anti-EV-71 monoclonal antibody, diluted 1/50 (MAB979; Chemicon-Millipore, Zug, Switzerland) for the detection of EV-71; and the undiluted panenterovirus blend (Chemicon-Millipore) for the detection of other enteroviruses. After the cells were intensively washed with PBS, Alexa Fluor 488 goat anti-rabbit antibody (Invitrogen) and anti-mouse immunoglobulin G-fluorescein isothiocyanate conjugate (Chemicon-Millipore) were added, and the cells were incubated for 45 min at 37°C in the dark. After a final rinsing with PBS, coverslips were mounted in Fluoprep embedding medium (bioMérieux, Marcy l'Etoile, France).
RESULTS
Panenterhino, Panenterhino/Ge/08, and Entero/Ge/08 real-time PCR assays.
Based on a careful search of all HRV and HEV sequences either available in public libraries or generated by our ongoing investigations, we first identified potential highly conserved sequence targets within the 5′ NTR that could be used to design different sets of primers and probes (Fig. 1). The so-called Panenterhino real-time PCR was first developed in order to target all members of the Enterovirus genus (HRV and HEV) that could be potentially detected in respiratory specimens. Panenterhino/Ge/08 is an evolution of this assay and aims not only to keep the large spectrum of the Panenterhino assay but also to target a subgroup of HRV-C viruses presenting a divergent 5′ NTR (11, 24) and named here HRV-C′. Entero/Ge/08 was specifically designed to provide sensitive and targeted detection of HEV. Two previously validated real-time PCR assays, RHP1 and RHP2, detecting HRV-A and HRV-B, as well as an enterovirus real-time PCR (HEV assay) (3), were used in the present study as comparative assays for the validation procedure. Primers and probe sequences developed in this study are detailed in Table 1.
FIG. 1.
Alignment of 5′ NTR consensus sequences obtained for HRV and HEV members. The primers, probes, and PNA probes developed in this study are boxed, and the corresponding sequences are depicted below the boxes. The different assays used are described in Table 1.
The three new assays developed, Panenterhino, Panenterhino/Ge/08, and Entero/Ge/08 real-time PCR, were first tested for optimal primers and probe concentrations (data not shown). Their analytical sensitivity was then determined on serially diluted concentrations by 10-fold of HRV-14-, HRV-2-, and EV-71-derived plasmids. All assays showed the conservation of a wide linearity, ranging from 1 to 106 plasmid copies/reaction, with the lowest limit of detection being 1 to 10 copies of input (data not shown). In vitro transcription of full-length HRV-14 and EV-71 infectious clones allowed us to define the analytical sensitivity of the whole RT-PCR procedure in terms of virus copy number detected from the RT to the final amplicon detection step. As shown in Fig. 2, the detection limit is 100 copies per RT reaction (equivalent to 25 copies of input per PCR, assuming a 100% RT efficacy) (see Materials and Methods) for Panenterhino and Entero/Ge/08 and 10 copies per RT reaction (equivalent to 2.5 copies per PCR) for Panenterhino/Ge/08 (Fig. 2A to C). Finally, analytical sensitivity was compared to cell culture infectivity. Serial dilutions of titrated virus stocks were extracted in duplicate, reverse transcribed, and PCR amplified. The limit of detection of an HRV-14 viral stock is 5.6 ×10−3 CCID50 per RT reaction (equivalent to 7 × 10−4 CCID50 per PCR) (see Materials and Methods) for the Panenterhino assay (Fig. 2D) and 5.6 × 10−2 CCID50 (equivalent to 7 × 10−3 CCID50 per PCR) for the Panenterhino/Ge/08 (Fig. 2E). The limit of detection of the EV-71 stock for the Entero/Ge/08 assay is 5.6 × 10−2 CCID50 per RT reaction (equivalent to 7 × 10−3 CCID50 per PCR) (Fig. 2F). In summary, the real-time PCR assays presented here are around 100-fold more sensitive overall than cell culture.
The specificities and sensitivities of the new assays were then compared with previous real-time PCR assays on plasmids containing 5′ NTRs originating from HRV-2, HRV-14, EV-71, EV-104 (a newly described member of the HEV-C species [24]), and the new HRV-C, as well as HRV-C′. The results presented in Table 2 show that both Panenterhino and Panenterhino/Ge/08 assays are able to detect all HEV- and HRV-derived plasmids. However, the addition of a second forward primer, adapted from a recently published HRV-specific real-time PCR (12), enables the Panenterhino/Ge/08 assay to detect the HRV-C′ in a sensitive manner. These analytical validations revealed that the previously developed assays targeting specifically HRV-A and HRV-B do not detect the divergent HRV-C and HRV-C′ groups. Similarly, the previously used HEV assay does not detect EV-104 strains.
TABLE 2.
Specificities and sensitivities of different real-time PCR assays on rhinovirus and enterovirus plasmid-derived constructs
| Targeta | Results from indicated assaysb
|
|||||
|---|---|---|---|---|---|---|
| RHP1 | RHP2 | Panenterhino | Panenterhino/Ge/08 | HEV | Entero/Ge/08 | |
| HRV-2 | 34.7 (±0.2) | Neg | 34.2 (±0.2) | 34.5 (±0.2) | Neg | Neg |
| HRV-14 | Neg | 42.7 (±2.1) | 34.3 (±0.5) | 35.3 (±1.3) | Neg | 36.3 (±2.3) |
| HRV-C | Neg | Neg | 34.9 (±0.1) | 34.7 (±0.1) | Neg | Neg |
| HRV-C′ | Neg | Neg | 46.5 (±6.5) | 37.4 (±0.7) | Neg | Neg |
| EV-71 | Neg | Neg | 35.7 (±0.2) | 35.6 (±0.4) | 33.9 (±0.3) | 31.9 (±0.2) |
| EV-104 | Neg | Neg | 34.7 (±0.2) | 34.6 (±0.2) | Neg | 35.2 (±0.1) |
HRV- and HEV-derived plasmids were used at a uniform concentration of 1,000 copies of input per PCR. Plasmid constructs are described in Materials and Methods. Results are indicated as real-time PCR CT values (± standard deviations).
Neg, negative result.
Panenterhino and Panenterhino/Ge/08 real-time PCRs were then assayed on most of the HRV reference strains. Overall, the sensitivities of the two assays were comparable for all rhinovirus serotypes tested. Enterovirus reference strains were then screened with the three new assays as well as with the previously used HEV-specific real-time PCR. Again, the Panenterhino and the Panenterhino/Ge/08 assays were able to detect all HEV serotypes tested. However, a comparison of threshold cycle (CT) values for each of the 59 strains tested showed that the Entero/Ge/08 combination improved the analytical sensitivity more than the more-generic assays (data not shown). Of note, the two HPeVs tested (HPeV-1 and -2) were not detected by these assays, thus proving that their specificity is restricted to HRV and HEV.
In a last set of experiments following the same extraction and RT procedures, the three new assays were used to screen clinical specimens. The Panenterhino and Panenterhino/Ge/08 assays were tested with specimens from subjects with respiratory symptoms. The validation on clinical specimens was performed on two different types of specimens, as follows: first, on nasopharyngeal samples collected from children with acute otitis media or pneumonia, and second, on lower respiratory tract samples (bronchoalveolar lavage fluid) collected from children with cystic fibrosis (5, 8). For the first study, we compared the results obtained with the Panenterhino PCR and the standard RHP1, RHP2, and HEV on 626 nasopharyngeal samples. Panenterhino proved to be much more sensitive than the previously validated assays for HRV and HEV and showed a positivity rate that was significantly higher (18% versus 7%, respectively), with all RHP1, RHP2, and HEV cases detected by the generic assay. For the second study, we conducted the Panenterhino and Panenterhino/Ge/08 assays in parallel on 157 bronchoalveolar lavage fluid specimens and achieved positivity rates of 42% and 51%, respectively. These results confirm that Panenterhino/Ge/08 is more adapted to circulating HRV strains. This is most probably linked to the inefficient detection of the HRV-C′ strains by the Panenterhino assay. However, the low CT values obtained for the discrepant samples (CT value of more than 39) did not allow further genotyping by sequencing. Similarly, the Entero/Ge/08 assay was used to rescreen 162 cerebrospinal fluid (CSF) specimens sent to our routine laboratories for HEV detection in patients with acute meningoencephalitis symptoms. All these cases have been previously screened with the routinely used HEV assay. Despite an additional cycle of freeze-thaw of the native samples before extraction, five CSF specimens were positive with the new assay versus four with the assay used previously. In one case, we were unable to confirm a previous positivity, which is most probably related to low CT values together with repeated freeze-thaw cycles in samples processed in a routine laboratory.
PNA hybridization probes.
The variety of HRV and HEV serotypes renders impossible the development of antibodies matching all serotypes. In addition, standard in situ hybridization assays require riboprobes with lengths ranging between 100 and 400 nucleotides. As shown in Fig. 1, even the most conserved 5′ NTRs of HRV and HEV do not present conserved stretches of such length. Therefore, we moved to artificially synthesized PNA probes. These probes are characterized by a backbone composed of repeating N-(2-aminoethyl)-glycine units linked by peptide bonds instead of the RNA deoxyribose sugar backbone. Since the backbone of PNA contains no charged phosphate groups, the binding between PNA/RNA strands is stronger than between DNA/RNA or RNA/RNA due to the lack of electrostatic repulsion. This allows the design of very short sequences (18 nucleotides), with high annealing temperatures (55°). In addition, PNAs are not easily recognized by either nucleases or proteases, thus making them resistant to enzyme degradation, and probe stocks can be kept stable for years at −20°C.
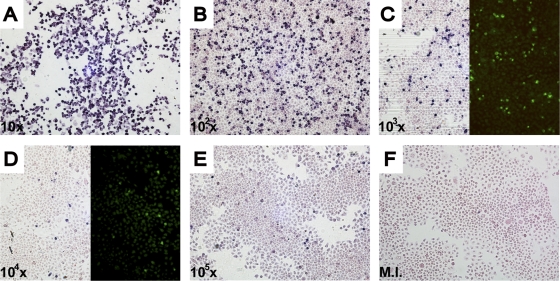
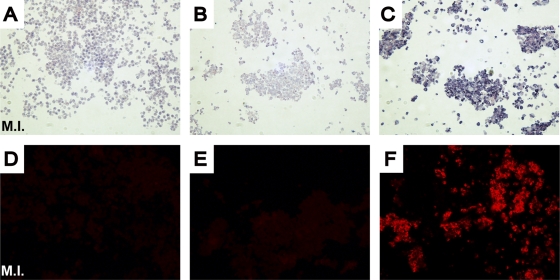
Three different PNA probes were synthesized, the first two specific for HRV-A (A/Ge/08 PNA probe) and HRV-B (B/Ge/08 PNA probe), while the third is more generic, matching all HRV and HEV sequences available (Panenterhino/Ge/08 PNA probe) (Fig. 1; see Materials and Methods). The protocol for this in situ hybridization in cells (see Materials and Methods) is adapted from the PNA ISH detection kit (DakoCytomation). After evaluation of optimal concentration and hybridization temperature, the probes' sensitivities were assessed on cells infected for 16 h with serial dilutions of quantified virus stocks. These sensitivity assays were done for the Panenterhino/Ge/08 PNA probe on HRV-14 (Fig. 3), HRV-94, and EV-71, as well as for the PNA A/Ge/08 and B/Ge/08 probes on HRV-94- and HRV-14-quantified stocks, respectively. All three assays provided similar reproducible results (Fig. 3 and data not shown). HRV-14 detection by immunofluorescence with HRV-14 antibodies conducted in parallel showed similar quantitative detection abilities (Fig. 3C and D). This assay was also conducted successfully on several cell lines (HeLa-OH, Vero, and A549). A panel of HRV and HEV serotypes was tested to determine the specificity of each of the three probes developed. As expected, the PNA probe A/Ge/08 detects HRV-A serotypes (HRV-11, -16, -64, -73) but not HRV-B serotypes (HRV-14, -70, -27, -79, -99). In contrast, the PNA probe B/Ge/08 is able to recognize these HRV-B serotypes but not the HRV-A serotypes. Finally, the Panenterhino/Ge/08 PNA probe gave positive signals on all HRV-A (HRV-64), HRV-B (HRV-14 and -85), and HEV serotypes (CV-A21, E-26, EV-70, and EV-71) tested but did not recognize a control HPeV, HPeV1 (data not shown). In situ hybridization was also successfully applied to formalin-fixed HeLa-OH cells, and fluorescent labeling of positive cells was performed using the Dako Permanent Red detection kit (Fig. 4).
FIG. 3.
Panenterhino/Ge/08 PNA probe sensitivity 16 h postinfection with serial dilutions by 10-fold of a titrated HRV-14 stock (108.25 CCID50/ml). (C and D) Comparison of Panenterhino/Ge/08 PNA probe sensitivity with that of immunofluorescence 16 h postinfection. The dilution factor is shown on the bottom left-hand corner of each panel. M.I., mock infected.
FIG. 4.
In situ hybridization on paraffin-embedded HeLa-OH cells. Visible (A to C) or fluorescent (D to E) labeling of infected cells. Hybridization with the PNA probe A/Ge/08 (A, D, C, and F) or negative control (B, E). The negative control is a random PNA probe (PNA ISH detection kit). Infections were performed with an HRV-64 strain (B, C, E and F), except for mock-infected (M.I.) strains (A, D).
DISCUSSION
New divergent HEV and HRV genotypes have been described and are constantly being identified. In the present study, our goal was to establish a panel of molecular assays adapted to the detection of these viruses. By targeting the 5′ NTR, the most conserved part of the genome, we designed and validated assays that could detect HRV and HEV nucleic acids in clinical specimens by RT-PCR and in cell culture by in situ hybridization. The various constraints related to both the similarities and the diversity of HRV and HEV imply that any 5′ NTR-based PCR assay designed for broad rhinovirus screening will also pick up members of the Enterovirus species. This, together with the fact that a significant number of HEV exhibit a respiratory tropism like all rhinoviruses, led us to propose generic real-time PCR assays detecting all members of the genus that could be used for respiratory screening. In addition, the design of generic assays lowers the risk of missing new emerging strains or still unrecognized variants. Broad HRV and HEV detection by the Panenterhino and Panenterhino/Ge/08 assays was confirmed not only by extensive testing of available human serotypes of the genus but, most importantly, also by testing clinical specimens. Compared to the previously used rhinovirus assays (3), the updated Panenterhino/Ge/08 assay confirmed its ability to detect not only all known rhinoviruses in a sensitive manner but also new HRV-C genotypes, including those with divergent 5′ NTRs. The ability of these assays to target HRV and HEV in a generic manner allowed us to identify new members of the Enterovirus genus (24). This latter point highlights that there is a need for assays that are not restricted to the rhinovirus borders when screening respiratory specimens and that these should include systematically enteroviruses. If necessary, the differentiation between rhino- and enteroviruses should be considered after the screening phase, either by using an additional set of real-time PCR assays, as presented in Table 2, or by sequence analysis. Of note, most subjects screened had significant clinical diseases in the present clinical validation. However, it is important to recognize that when using sensitive nucleic acid detection, HRV can be identified in some instances in subjects with few or no symptoms.
Compared to the previous HEV-specific real-time PCR assay, Entero/Ge/08 showed specificity for HEV that also expanded to the newly discovered EV-104 virus and possibly to other related strains (Table 2). The analytical characteristics of this real-time PCR revealed a significantly higher sensitivity than that of Panenterhino/Ge/08. This illustrates the different trade-offs that need to be considered when designing these assays. Although the more-generic ones provide obvious advantages for respiratory specimens, Enterhino/Ge/08, with its restricted spectrum, was revealed to be probably more appropriate for CSF screening. Given the types of disease considered, acute meningitis and encephalitis, it seems quite reasonable in this case to promote analytical sensitivity.
Diagnostic procedures using cell culture are limited by the expertise and the delay needed to obtain a result. For these reasons, viral culture has a limited place in routine diagnostic microbiology, but there is still a need to consider such assays in reference or research laboratories. In the case of picornaviruses, there are also the additional limitations related to the number of serotypes circulating and the lack of generic detection tools, with each serotype being recognized only by specific antibodies. Here, we took advantage of emerging PNA technologies to design a set of three probes, one specific for rhinovirus A (A/Ge/08 PNA probe), one for rhinovirus B (B/Ge/08 PNA probe), and the third that was supposed to match all known HRV and HEV serotypes (Panenterhino/Ge/08 PNA probe). These probes have the ability to identify rhinoviruses and enteroviruses within 16 h in cell culture. This procedure also has the advantage that it uses standard laboratory techniques and equipment, does not rely on the appearance of a cytopathic effect, does not require a large expertise, and thus could ensure the reproducibility needed. Such an assay presents the additional ability to quantify and assess the kinetics of the infection, which is particularly interesting for the comparison of different genotypes in reverse genetic mutagenesis assays. Finally, it is adapted to formalin-fixed tissues and can be assayed in naturally occurring HRV or HEV infections.
In conclusion, following an extensive validation process, we provide a panel of new and updated real-time PCR assays to detect newly identified rhino- and enterovirus strains. The use of degenerated primers and probes also increase the likelihood to detect divergent variants. We provide, for the first time, a specific, sensitive, and accurate in situ hybridization procedure that has proven efficiency to detect and quantify both rhino- and enteroviruses serotypes early during the viral cycle, thus circumventing the need for specific antibodies or validation based on the apparition of a cytopathic effect.
Acknowledgments
We thank Rosemary Sudan for editorial assistance and Patricia Gindre and Emmanuelle Murith for technical support.
This study was supported by the Swiss National Science Foundation (grant 3200B0-101670 to L.K.), the Department of Medicine of the University Hospitals of Geneva, and the University of Geneva Dean's program for the promotion of women in science (C.T.).
Footnotes
Published ahead of print on 1 April 2009.
REFERENCES
- 1.Arden, K. E., P. McErlean, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2006. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 781232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordey, S., D. Gerlach, T. Junier, E. M. Zdobnov, L. Kaiser, and C. Tapparel. 2008. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA 141568-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deffernez, C., W. Wunderli, Y. Thomas, S. Yerly, L. Perrin, and L. Kaiser. 2004. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J. Clin. Microbiol. 423212-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dufresne, A. T., and M. Gromeier. 2004. A nonpolio enterovirus with respiratory tropism causes poliomyelitis in intercellular adhesion molecule 1 transgenic mice. Proc. Natl. Acad. Sci. USA 10113636-13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilliard, T. N., N. Regamey, J. K. Shute, A. G. Nicholson, E. W. Alton, A. Bush, and J. C. Davies. 2007. Airway remodeling in children with cystic fibrosis. Thorax 621074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junttila, N., N. Leveque, J. P. Kabue, G. Cartet, F. Mushiya, J. J. Muyembe-Tamfum, A. Trompette, B. Lina, L. O. Magnius, J. J. Chomel, and H. Norder. 2007. New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo. J. Med. Virol. 79393-400. [DOI] [PubMed] [Google Scholar]
- 7.Kistler, A., P. C. Avila, S. Rouskin, D. Wang, T. Ward, S. Yagi, D. Schnurr, D. Ganem, J. L. DeRisi, and H. A. Boushey. 2007. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J. Infect. Dis. 196817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kronenberg, A., P. Zucs, S. Droz, and K. Muhlemann. 2006. Distribution and invasiveness of Streptococcus pneumoniae serotypes in Switzerland, a country with low antibiotic selection pressure, from 2001 to 2004. J. Clin. Microbiol. 442032-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamson, D., N. Renwick, V. Kapoor, Z. Liu, G. Palacios, J. Ju, A. Dean, K. St George, T. Briese, and W. I. Lipkin. 2006. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J. Infect. Dis. 1941398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau, S. K., C. C. Yip, H. W. Tsoi, R. A. Lee, L. Y. So, Y. L. Lau, K. H. Chan, P. C. Woo, and K. Y. Yuen. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 453655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, W. M., C. Kiesner, T. Pappas, I. Lee, K. Grindle, T. Jartti, B. Jakiela, R. F. Lemanske, P. A. Shult, and J. E. Gern. 2007. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE 2e966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, X., B. Holloway, R. K. Dare, J. Kuypers, S. Yagi, J. V. Williams, C. B. Hall, and D. D. Erdman. 2008. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46533-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackay, I. M. 2008. Human rhinoviruses: the cold wars resume. J. Clin. Virol. 42297-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McErlean, P., L. A. Shackelton, S. B. Lambert, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2007. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J. Clin. Virol. 3967-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newcombe, N. G., P. Andersson, E. S. Johansson, G. G. Au, A. M. Lindberg, R. D. Barry, and D. R. Shafren. 2003. Cellular receptor interactions of C-cluster human group A coxsackieviruses. J. Gen. Virol. 843041-3050. [DOI] [PubMed] [Google Scholar]
- 16.Norder, H., L. Bjerregaard, L. Magnius, B. Lina, M. Aymard, and J. J. Chomel. 2003. Sequencing of ‘untypable’ enteroviruses reveals two new types, EV-77 and EV-78, within human enterovirus type B and substitutions in the BC loop of the VP1 protein for known types. J. Gen. Virol. 84827-836. [DOI] [PubMed] [Google Scholar]
- 17.Oberste, M. S., K. Maher, S. M. Michele, G. Belliot, M. Uddin, and M. A. Pallansch. 2005. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species human enterovirus A. J. Gen. Virol. 86445-451. [DOI] [PubMed] [Google Scholar]
- 18.Oberste, M. S., K. Maher, D. Schnurr, M. R. Flemister, J. C. Lovchik, H. Peters, W. Sessions, C. Kirk, N. Chatterjee, S. Fuller, J. M. Hanauer, and M. A. Pallansch. 2004. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J. Gen. Virol. 852577-2584. [DOI] [PubMed] [Google Scholar]
- 19.Pulli, T., P. Koskimies, and T. Hyypia. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 21230-38. [DOI] [PubMed] [Google Scholar]
- 20.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 21.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 22.Renwick, N., B. Schweiger, V. Kapoor, Z. Liu, J. Villari, R. Bullmann, R. Miething, T. Briese, and W. I. Lipkin. 2007. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J. Infect. Dis. 1961754-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smura, T., S. Blomqvist, A. Paananen, T. Vuorinen, Z. Sobotova, V. Bubovica, O. Ivanova, T. Hovi, and M. Roivainen. 2007. Enterovirus surveillance reveals proposed new serotypes and provides new insight into enterovirus 5′-untranslated region evolution. J. Gen. Virol. 882520-2526. [DOI] [PubMed] [Google Scholar]
- 24.Tapparel, C., T. Junier, D. Gerlach, S. Van Belle, L. Turin, S. Cordey, K. Mühlemann, N. Regamey, J.-D. Aubert, P. Soccal, P. Eigenmann, E. M. Zdobnov, and L. Kaiser. 2009. New respiratory enterovirus and recombinant rhinoviruses among circulating strains. Emerg. Infect. Dis. 15719-726. [DOI] [PMC free article] [PubMed] [Google Scholar]