Abstract
Since numbers of drug-resistant Mycobacterium tuberculosis strains are on the rise, the simple classification into “susceptible” and “resistant” strains based on susceptibility testing at “critical concentrations” has to be reconsidered. While future studies have to address the correlation of phenotypic resistance levels and treatment outcomes, a prerequisite for corresponding investigations is the ability to exactly determine levels of quantitative drug resistance in clinical M. tuberculosis isolates. Here we have established the conditions for quantitative drug susceptibility testing for first- and second-line agents using MGIT 960 instrumentation and EpiCenter software equipped with the TB eXiST module. In-depth comparative analysis of a range of well-characterized susceptible and resistant clinical isolates has allowed us to propose conditions for testing and to develop criteria for interpretation.
Tuberculosis (TB) is a leading cause of morbidity and mortality worldwide. Developing countries are the most vulnerable, with more than 95% of the cases (32, 33). The present trend is characterized by an alarming emergence of drug resistance (7, 8, 30). Much attention has focused on the burden of multidrug-resistant (MDR) TB, i.e., resistance to the first-line drugs isoniazid and rifampin (rifampicin) (32, 33), and the emergence of extensively drug resistant TB (6, 26). The rise of drug-resistant TB and the increased susceptibility of the human population to TB due to coinfection with human immunodeficiency virus are driving the worldwide TB pandemic and will worsen the situation in the years ahead, with devastating effects in poor countries, whose economies suffer most from this development (19, 20).
In the diagnostic laboratory, testing of mycobacteria for drug susceptibility is substantially different from the general testing procedures used in bacteriology. Rather than determining MICs, a single drug concentration, termed the critical concentration, is usually used to categorize a clinical isolate as susceptible or resistant. This “critical concentration” is more an epidemiological parameter (to distinguish “wild-type” strains from “non-wild-type” strains that are able to grow in the presence of higher drug concentrations [5]) than a clinical cutoff value established to guide treatment decisions (14). With growing knowledge about the mechanisms that underlie drug resistance, it has become evident that drug resistance is multifaceted and that different mutations may lead to different levels of resistance. The acquisition of a resistance mutation leading to a decrease in drug susceptibility should not inevitably exclude an anti-TB drug from a treatment regimen, since low-level resistance does not necessarily imply clinical resistance (3). However, until now, different levels of phenotypic resistance have only rarely been taken into account in the procedures used for in vitro drug susceptibility testing (DST) of mycobacteria (4).
“Critical concentration”-based DST of primary and secondary drugs has been established for the radiometric Bactec 460 instrumentation (Becton Dickinson Microbiology Systems, Sparks, MD) and is considered the “gold standard” in the testing of second-line drugs (22, 25). However, the Bactec 460 system has several drawbacks: (i) it involves the use of sharps and radioisotopes with the need for disposal; (ii) it is only semiautomated; and (iii) it needs considerable hands-on time. The nonradiometric MGIT (mycobacterial growth indicator tube) 960 platform (Becton Dickinsion) has been evaluated extensively for DST of first-line drugs (1, 2, 15, 29) and has recently also been evaluated for second-line DST (16, 25). In contrast to the Bactec 460 instrumentation, the MGIT 960 platform is a fully automated system that uses a fluorescence-quenching-based oxygen sensor for growth detection.
We have previously characterized quantitative drug resistance levels in clinical strains of drug-susceptible and drug-resistant Mycobacterium tuberculosis using radiometric Bactec 460 measurements (28). However, for widespread implementation of quantitative DST in diagnostic mycobacteriology, the technique chosen should be fully automated, compatible with a computerized expert system for interpretation (so as to avoid individual errors and subjectivity), and safe and reliable (e.g., if possible, the system should not use radioactive material nor needles, such as syringes, for inoculation nor needles inside the instrument). To this end, we have subjected a carefully chosen subset of a previously described and well-characterized collection of clinical M. tuberculosis isolates (28) to quantitative measurements of drug susceptibility using the MGIT 960 platform in conjunction with EpiCenter software equipped with the TB eXiST module.
MATERIALS AND METHODS
Susceptibility testing using the Bactec 460 system.
DST for first- and second-line anti-TB drugs was performed using the Bactec 460 system as recommended by the manufacturer (27). A 0.1-ml volume from a positive MGIT tube was inoculated into Bactec 460 12B vials; the final concentrations of drugs are given in Table 1. For the drug-free growth control, the bacterial suspension was diluted 1:100 before inoculation (proportion testing). The vials were incubated at 37°C and read daily until the growth of the control reached a growth index (GI) of >500. Test results were interpreted by comparing the changes in the GI of the growth control with the flasks containing the test drugs by using the standard interpretation procedure (if the GI of the drug is greater than or equal to the GI of the control, the strain is resistant [R]; if the GI of the drug is less than the GI of the control, the strain is susceptible [S]).
TABLE 1.
Concentrations of drugs used for susceptibility testing
| Drug | Concn(s) (mg/liter) used for testing in the following system:
|
|
|---|---|---|
| Bactec 460 | MGIT 960 | |
| Isoniazid | 0.1, 0.4, 1.0, 3.0, 10.0 | 0.1, 0.4, 1.0, 3.0, 10.0 |
| Rifampin | 1.0, 3.0, 10.0, 50.0 | 1.0, 3.0, 10.0, 50.0 |
| Ethambutol | 2.5, 5.0, 12.5, 50.0 | 2.5, 5.0, 12.5, 25.0, 50.0 |
| Streptomycin | 1.0, 10.0, 50.0b | 1.0, 4.0, 20.0b |
| Amikacin | 1.0, 10.0, 50.0 | 1.0, 4.0, 20.0 |
| Ethionamide | 2.5, 12.5a | 2.5, 12.5a |
| Ofloxacin | 2.0, 10.0, 20.0 | 2.0, 10.0, 50.0 |
| Linezolid | 0.4, 4.0 | 1.0, 4.0 |
| Capreomycin | 5.0 | 1.25, 5.0, 25.0 |
Additional testing for selected strains at concentrations of 1.25, 2.5, 5.0, 12.5, and 25.0 mg/liter.
Additional data points were determined for selected strains.
Susceptibility testing using the MGIT 960 system with EpiCenter TB eXiST software.
The MGIT 960 system was used for primary isolation and standard susceptibility testing of first-line drugs (10) as recommended by the manufacturer. MGIT tubes supplemented with 0.8 ml of supplement (MGIT 960 SIRE supplement; Becton Dickinson) were inoculated with 0.1 ml of the drug solution and 0.5 ml of the test strain suspension. For preparation of the drug-free growth control tube, the organism suspension was diluted 1:100 with sterile saline, and then 0.5 ml was inoculated into the tube (proportion testing). For quantitative DST using the MGIT 960 instrumentation, we requested Becton Dickinson to develop custom-designed software with the following characteristics: automated recording of the readings, additional incubation time beyond the time to positivity of the drug-free control, minimization of the number of drug-free control tubes required, graphical representation of the growth unit (GU) value increase and storage of data, and easy handling and documentation. The software is EpiCenter, version 5.53, equipped with the TB eXiST module and available from Becton Dickinson. Table 1 lists the drug concentrations used for comparative analysis. The susceptibility testing sets were placed in the MGIT 960 instrument and continuously monitored using EpiCenter (version 5.53) TB eXiST software.
Results were interpreted as follows. At the time when the GU of the drug-free control tube was >400, if the GU of the drug-containing tube to be compared was ≥100, the strain was R. If the GU of the drug-containing tube was <100, it was incubated for a further 7 days. If it was still <100, the strain was S. If the GU of the drug-containing tube was ≥100 during this further 7 days of incubation after the GU of the drug-free control tube reached >400, the strain was intermediate (I).
Strains and molecular detection of resistance mutations.
The isolates investigated in this analysis represent a subset of 29 clinical M. tuberculosis strains from a previously reported study (28).
For the identification of resistance mutations, the GenoType MTBDRplus assay (Hain Lifescience, Nehren, Germany) was used (11, 17). The GenoType MTBDRplus assay is a reverse hybridization line probe assay designed for rapid detection of rpoB and katG gene mutations. In addition, the strip harbors two wild-type probes covering the promoter region of the inhA gene, and four mutations in this region are targeted using mutated probes (-8T/C, -8T/A, -15C/T, -16A/G). The assay was performed as recommended by the manufacturer. Either the absence of a wild-type probe or a hybridization signal of a mutant probe is an indication of resistance.
For the detection of mutational alterations associated with resistance to ethambutol, amikacin, or streptomycin, i.e., embB position 306 (13), 16S rRNA position 1408 (23), and rpsL positions 42 and 87 (9), PCR-driven gene amplification and nucleic acid sequence determination were applied. Amplified gene fragments were sequenced using the BigDye Terminator cycle-sequencing ready reaction kit (Applied Biosystems Inc., Foster City, CA) and an ABI 3130 DNA genetic analyzer (Applied Biosystems).
RESULTS
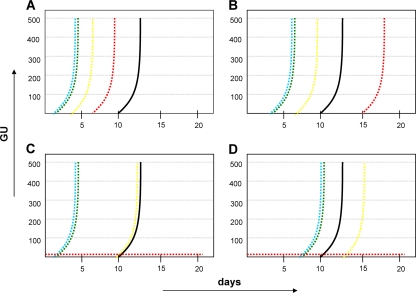
A representative set of well-characterized clinical M. tuberculosis strains, with drug resistance defined at the genotypic level, was used to establish conditions for quantitative DST on the basis of the MGIT 960 instrumentation and EpiCenter, version 5.53, TB eXiST software. A total of 29 clinical isolates of M. tuberculosis, categorized as resistant to one or more first-line drugs on the basis of standard critical-concentration testing, were tested for susceptibility at higher drug concentrations using Bactec 460 and MGIT 960 instrumentation. Table 1 lists the drug concentrations used for comparative analysis. The EpiCenter software allows continuous monitoring of fluorescence, thus enabling a precise assessment of bacterial growth. Continuous growth monitoring, together with an extended incubation period of a further 7 days following the positivity of the drug-free control, facilitated the development of the term “intermediate growth” as an additional characteristic in data interpretation. The phenotypic heterogeneity in isoniazid resistance expression with the katG S315T mutation has been described previously (28) and may serve to illustrate the term “intermediate” (Fig. 1).
FIG. 1.
Phenotypic heterogeneity in the isoniazid resistance of isolates with the katG S315 mutation and the term “intermediate” in MGIT 960 testing with EpiCenter (version 5.53) TB eXiST software. (A) Isolate 177836, resistant at 0.1, 1.0, 3.0, and 10.0 mg/liter; (B) isolate 186137, resistant at 0.1, 1.0, and 3.0 mg/liter and intermediate at 10.0 mg/liter; (C) isolate 186069, resistant at 0.1, 1.0, and 3.0 mg/liter and susceptible at 10.0 mg/liter; (D) isolate 176291, resistant at 0.1 and 1.0 mg/liter, intermediate at 3.0 mg/liter, and susceptible at 10.0 mg/liter. Drug concentrations are represented by colors as follows: blue, 0.1 mg/liter; green, 1.0 mg/liter, yellow, 3.0 mg/liter, red, 10.0 mg/liter; black, drug-free growth control.
To determine the accuracy of quantitative DST, we compared the results of MGIT 960 testing with those of radiometric Bactec 460 testing and related them to the molecular resistance determinants identified. For molecular profiling, the GenoType MTBDRplus assay (Hain Lifescience, Nehren, Germany) was used (rifampin, isoniazid, and ethionamide); in part, mutations were assessed by nucleic acid sequencing (ethambutol, streptomycin, and amikacin).
For isoniazid, we included 11 strains with low-level drug resistance (MICs, ≥0.1 mg/liter and <1 mg/liter) (Table 2) and 18 strains with resistance levels of ≥1 mg/liter (Table 3). All low-level resistant isolates harbored an inhA promoter mutation (C15T). In 14/18 isolates with resistance levels of ≥1 mg/liter, a katG S315T mutation was present. The results obtained with the MGIT 960 platform correlated well with those generated by radiometric Bactec 460 testing over the whole range of drug concentrations tested (Table 3; Fig. 2). The only major discrepancy (S versus R) was observed at a test concentration of 3 mg/liter. Here the Bactec 460 system consistently reported susceptibility for a single isolate, while the MGIT 960 system gave a resistant test result; the two systems gave identical results for this isolate at test concentrations of 1 mg/liter (resistant) and 10 mg/liter (susceptible).
TABLE 2.
Clinical strains with low-level isoniazid resistance
| Strain | Method | Susceptibility to the following drug at the indicated concn (mg/liter):
|
Genetic resistance | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isoniazid
|
Ethionamide
|
|||||||||||
| 0.1 | 0.4 | 1.0 | 3.0 | 10.0 | 1.25 | 2.5 | 5.0 | 12.5 | 25.0 | |||
| TBC 2694 | Bactec 460 | R | S | S | S | S | R | R | NDa | S | S | katG, wtb; inhA, C15T |
| MGIT 960 | R | I | I | S | S | R | R | R | I | S | ||
| TBC 4269 | Bactec 460 | R | S | S | S | S | R | R | R | R | R | katG, wt; inhA, C15T |
| MGIT 960 | R | I | I | S | S | R | R | R | R | R | ||
| NZM 117 | Bactec 460 | R | S | S | S | S | R | R | ND | S | S | katG, wt; inhA, C15T |
| MGIT 960 | R | I | S | S | S | R | R | R | S | S | ||
| NZM 150 | Bactec 460 | R | S | S | S | S | R | R | R | R | ND | katG, wt; inhA, C15T |
| MGIT 960 | R | I | S | S | S | R | R | R | R | I | ||
| NZM 188 | Bactec 460 | R | S | S | S | S | R | R | ND | S | S | katG, wt; inhA, C15T |
| MGIT 960 | R | I | S | S | S | R | R | R | S | S | ||
| TBC 2339 | Bactec 460 | R | S | S | S | S | R | R | ND | S | S | katG, wt; inhA, C15T |
| MGIT 960 | R | S | S | S | S | R | R | R | R | I | ||
| TBC 179320 | Bactec 460 | R | S | S | S | S | R | R | R | R | ND | katG, wt; inhA, C15T |
| MGIT 960 | R | I | S | S | S | R | R | R | R | R | ||
| TBC 179987 | Bactec 460 | R | S | S | S | S | R | R | ND | S | S | katG, wt; inhA, C15T |
| MGIT 960 | R | S | S | S | S | R | R | R | S | S | ||
| NZM 186008 | Bactec 460 | R | S | S | S | S | R | R | ND | S | S | katG, wt; inhA, C15T |
| MGIT 960 | R | S | S | S | S | R | R | R | S | S | ||
| NZM 186016 | Bactec 460 | R | S | S | S | S | R | R | ND | S | S | katG, wt; inhA, C15T |
| MGIT 960 | R | I | S | S | S | R | R | R | S | S | ||
| TBC 176592 | Bactec 460 | R | S | S | S | S | R | S | ND | S | S | katG, wt; inhA, C15T |
| MGIT 960 | R | I | S | S | S | R | R | R | S | S | ||
ND, not done.
wt, wild type.
TABLE 3.
Clinical M. tuberculosis strains with drug resistance: first-line drugs
| Strain | Method | Isoniazid
|
Rifampin
|
Ethambutol
|
Streptomycin
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Susceptibility at the following concn (mg/liter):
|
Genetic resistance | Susceptibility at the following concn (mg/liter):
|
Genetic resistance | Susceptibility at the following concn (mg/liter):
|
Genetic resistance | Susceptibility at the following concn (mg/liter):
|
Genetic resistance | |||||||||||||||||
| 0.1 | 0.4 | 1 | 3 | 10 | 1 | 3 | 10 | 50 | 2.5 | 5 | 12.5 | 25 | 50 | 1 | 4 | 10 | 20 | 50 | ||||||
| NZM 130 | Bactec 460 | R | R | R | R | R | katG, S315T1d; inhA, wtb | R | R | R | R | S531L | R | R | NDa | ND | S | embB codon 306, wt (ATG) | R | ND | S | S | S | rpsL pos.c 42, AAG (wt); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | R | I | R | R | R | R | R | I | S | S | ND | R | S | S | S | S | |||||
| NZM 179 | Bactec 460 | R | R | R | R | R | katG, wt; inhA, C15T | R | R | R | R | S531L plus 2nd mutation | R | R | R | ND | S | embB codon 306, CTG (valine) | R | R | R | R | R | rpsL pos. 42, AGG (Arg); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | R | R | R | R | R | R | R | R | I | S | ND | R | R | R | R | ND | |||||
| NZM 186006 (NZM 154/04) | Bactec 460 | R | R | R | S | S | katG, wt; inhA, wt | R | R | R | R | S531L | R | R | S | ND | S | embB codon 306, ATA (isoleucine) | R | R | R | R | R | rpsL pos. 42, AGG (Arg); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | I | S | R | R | R | R | R | R | S | S | ND | R | R | R | R | ND | |||||
| NZM 186137 (NZM 152/04) | Bactec 460 | R | R | R | R | S | katG, S315T1; inhA, wt | R | R | R | R | S531L | R | R | S | ND | S | embB codon 306, wt (ATG) | R | R | R | R | R | rpsL pos. 42, AGG (Arg); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | R | I | R | R | R | R | R | R | I | S | ND | R | R | R | R | ND | |||||
| TBC 6893 | Bactec 460 | R | R | R | S | S | katG, S315T1; inhA, wt | R | S | S | S | wt | S | S | S | ND | S | embB codon 306, wt (ATG) | R | ND | S | S | S | rpsL pos. 42, AAG (wt); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | I | S | R | S | S | S | S | S | S | S | ND | R | R | ND | S | S | |||||
| TBC 3106 | Bactec 460 | R | R | R | R | S | katG, wt; inhA, wt | R | R | R | R | S531L | R | R | S | ND | S | embB codon 306, wt (ATG) | S | S | S | S | S | ND |
| MGIT 960 | R | R | R | I | S | R | R | R | R | R | R | S | S | ND | S | S | S | S | S | |||||
| NZM 186038 (NZM 3/05) | Bactec 460 | R | R | R | R | S | katG, S315T1; inhA, wt | R | R | R | R | S531L | R | R | S | ND | S | embB codon 306, ATC (isoleucine) | R | R | R | R | R | rpsL pos. 42, AGG (Arg); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | R | I | R | R | R | R | R | R | S | S | ND | R | R | R | R | ND | |||||
| TBC 176291 | Bactec 460 | R | R | R | R | S | katG, S315T1; inhA, wt | R | R | R | R | H526D | R | S | S | ND | S | embB codon 306, wt (ATG) | R | ND | S | S | S | rpsL pos. 42, AAG (wt); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | I | S | R | R | R | R | R | S | S | S | ND | R | S | S | S | S | |||||
| TBC 177836 | Bactec 460 | R | R | R | R | R | katG, S315T1; inhA, wt | R | R | R | R | S531L | R | R | S | ND | S | embB codon 306, wt (ATG) | R | R | R | S | S | rpsL pos. 42, AAG (wt); pos. 87, AGG (Arg) |
| MGIT 960 | R | R | R | R | R | R | R | R | R | R | R | I | S | ND | R | R | R | S | S | |||||
| TBC 181783 | Bactec 460 | R | R | R | S | S | katG, wt; inhA, C15T | R | R | R | R | S531L | R | R | R | ND | S | embB codon 306, GTG (valine) | R | ND | S | S | S | rpsL pos. 42, AAG (wt); pos. 87, AGG (Arg) |
| MGIT 960 | R | R | R | I | S | R | R | R | R | R | R | R | R | S | R | R | ND | I | ND | |||||
| NZM 186069 | Bactec 460 | R | R | R | S | S | katG, S315T1, inhA, wt | R | R | R | R | S531L | R | R | S | ND | S | embB codon 306, ATA (isoleucine) | R | ND | S | S | S | rpsL pos. 42, AAG (wt); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | R | S | R | R | R | R | R | I | S | S | ND | R | S | S | S | S | |||||
| TBC 6890 | Bactec 460 | R | R | R | S | S | katG, S315T1; inhA, wt | R | R | R | R | S531L | S | S | S | ND | S | embB codon 306, wt (ATG) | ND | ND | ND | ND | ND | ND |
| MGIT 960 | R | R | R | I | S | R | ND | ND | ND | S | S | S | S | ND | ND | ND | ND | ND | ND | |||||
| TBC 7392 | Bactec 460 | R | R | R | S | S | katG, S315T1; inhA, wt | S | S | S | S | wt | S | S | S | ND | S | embB codon 306, wt (ATG) | ND | ND | ND | ND | ND | ND |
| MGIT 960 | R | R | R | I | S | S | ND | ND | ND | S | S | S | S | ND | ND | ND | ND | ND | ND | |||||
| TBC 295 | Bactec 460 | R | R | R | R | R | katG, S315T1; inhA, wt | R | R | R | R | S531L | R | S | S | ND | S | embB codon 306, ATA (isoleucine) | R | R | R | R | R | rpsL pos. 42, AGG (Arg); pos. 87, AAG (wt) |
| MGIT 960 | R | R | R | R | R | R | ND | ND | ND | R | I | S | S | ND | ND | ND | ND | ND | ND | |||||
| TBC 179882 | Bactec 460 | R | R | R | R | S | katG, S315T1; inhA, wt | S | S | S | S | wt | S | S | S | ND | S | embB codon 306, wt (ATG) | ND | ND | ND | ND | ND | ND |
| MGIT 960 | R | R | R | R | S | S | ND | ND | ND | S | S | S | S | ND | ND | ND | ND | ND | ND | |||||
| NZM 186073 | Bactec 460 | R | R | R | R | S | katG, S315T1; inhA, wt | S | S | S | S | ND | S | S | S | ND | S | embB codon 306, wt (ATG) | ND | ND | ND | ND | ND | ND |
| MGIT 960 | R | R | R | R | I | S | ND | ND | ND | S | S | S | S | ND | ND | ND | ND | ND | ND | |||||
| NZM 186044 | Bactec 460 | R | R | R | R | R | katG, S315T1; inhA, C15T | ND | ND | ND | ND | ND | R | S | S | ND | S | embB codon 306, ATA (isoleucine) | ND | ND | ND | ND | ND | ND |
| MGIT 960 | R | R | R | R | R | ND | ND | ND | ND | R | S | S | S | ND | ND | ND | ND | ND | ND | |||||
| NZM 178364 | Bactec 460 | R | R | R | R | S | katG, S315T1; inhA, wt | S | S | S | S | wt | S | S | S | ND | S | embB codon 306, wt (ATG) | ND | ND | ND | ND | ND | ND |
| MGIT 960 | R | R | R | R | S | ND | ND | ND | ND | S | S | S | S | ND | ND | ND | ND | ND | ND | |||||
ND, not done.
wt, wild type.
pos., position.
Corresponds to an AGC to ACC substitution.
FIG. 2.
Correlation of Bactec 460 and MGIT 960 susceptibility testing results.
For rifampin, 11 isolates with resistance levels of ≥1 mg/liter were included in the analysis. Complete agreement between the two test systems was observed (Table 3; Fig. 2). Of the 11 isolates tested, 10 showed resistance at >50 mg/liter, and all 10 harbored well-described resistance mutations in the rpoB gene (Table 3). One isolate showed low-level resistance (MIC > 1 mg/liter and < 3 mg/liter) with no detectable rpoB alteration; intermediate test results were not observed.
Ethambutol test results correlated well between the Bactec 460 and MGIT 960 (Table 3; Fig. 2), with three intermediate test results each obtained with the MGIT 960 at 5 mg/liter and at 12.5 mg/liter. Comparison of the test concentration of 25 mg/liter (MGIT 960) with that of 50 mg/liter (Bactec 460) pointed to one discrepancy: the Bactec 460 system reported a sensitive result, while in MGIT 960 testing, the isolate showed resistance at a concentration of 25 mg/liter. Testing of the isolate at a concentration of 50 mg/liter in MGIT 960 gave a susceptible test result, indicating a MIC for the isolate between 25 and 50 mg/liter. Seven of the 12 isolates with resistance levels of ≥2.5 mg/liter showed mutations in embB codon 306, while 5 isolates showed a wild-type (ATG) codon. No resistance of ≥50 mg/liter was found.
Ten isolates with streptomycin resistance levels of ≥1 mg/liter were included in our study. Resistance at >1 mg/liter was consistently detected with both systems (Table 3; Fig. 2). All isolates (n = 4) with resistance levels of ≥50 mg/liter (Bactec 460) or ≥20 mg/liter (MGIT 960) harbored an AAG→AGG mutation at codon 42 of the rpsL gene. Of the strains with lower resistance levels (n = 6), two harbored AAG→AGG mutations at codon 87 of the rpsL gene; in four strains with a low-level-resistant phenotype (≤4 mg/liter), no rpsL mutation was found.
To further evaluate the capacity of the MGIT 960 system for quantitative DST of second-line agents, we determined the resistance profiles of M. tuberculosis isolates for ethionamide, amikacin, ofloxacin, capreomycin, and linezolid.
Nineteen isolates showed ethionamide resistance of ≥2.5 mg/liter in at least one of the two test systems. Of the 19 isolates tested, 2 strains showed deviations at a test concentration of 2.5 mg/liter (Tables 2 and 4): the Bactec 460 system gave a susceptible test result, while resistance was found using the MGIT 960 system. The two strains consistently showed a resistant phenotype at a test concentration of 1.25 mg/liter and susceptibility at a test concentration of 12.5 mg/liter for both test systems. At high drug concentrations, the MGIT 960 system tended to yield more resistant results than the Bactec 460 system, leading to four discrepancies at a test concentration of 12.5 mg/liter (Fig. 2).
TABLE 4.
Clinical M. tuberculosis strains with drug resistance: second line drugs
| Strain | Method | Ethionamide
|
Amikacin
|
Susceptibility to the following drug at the indicated concn (mg/liter):
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Susceptibility at the following concn (mg/liter):
|
Genetic resistance | Susceptibility at the following concn (mg/liter):
|
Genetic resistance | Ofloxacin
|
Linezolid
|
Capreomycin
|
|||||||||||||||||
| 1.25 | 2.5 | 5 | 12.5 | 25 | 1 | 4 | 10 | 20 | 50 | 2 | 10 | 20 | 50 | 0.4 | 1 | 4 | 1.25 | 5 | 25 | ||||
| NZM 130 | Bactec 460 | R | R | NDa | S | S | inhA, wtb | S | ND | S | ND | S | ND | S | S | S | ND | S | ND | S | ND | ND | ND |
| MGIT 960 | R | R | R | R | S | S | S | ND | S | ND | S | S | ND | S | ND | S | S | I | S | S | |||
| NZM 179 | Bactec 460 | R | R | R | R | ND | inhA, C15T | S | ND | S | ND | S | ND | S | S | S | ND | S | ND | S | ND | S | ND |
| MGIT 960 | R | R | R | R | I | S | S | ND | S | ND | S | S | ND | S | ND | S | S | I | S | S | |||
| NZM 186006 (NZM 154/04) | Bactec 460 | ND | ND | ND | ND | ND | inhA, wt | S | ND | S | ND | S | ND | S | S | S | ND | R | ND | S | ND | S | ND |
| MGIT 960 | ND | ND | ND | ND | ND | S | S | ND | S | ND | S | S | ND | S | ND | S | S | I | S | S | |||
| NZM 186137 (NZM 152/04) | Bactec 460 | R | R | ND | S | ND | inhA, wt | R | ND | R | ND | R | rrn, A1408G | S | S | S | ND | R | ND | S | ND | S | ND |
| MGIT 960 | R | R | R | R | R | R | R | ND | R | ND | S | S | ND | S | ND | S | S | R | ND | S | |||
| TBC 6893 | Bactec 460 | S | S | S | S | S | inhA, wt | S | ND | S | ND | S | ND | S | S | S | ND | S | ND | S | ND | ND | ND |
| MGIT 960 | ND | S | S | S | S | S | S | ND | S | ND | S | S | ND | S | ND | S | S | R | S | S | |||
| TBC 3106 | Bactec 460 | S | S | ND | S | S | inhA, wt | S | ND | S | ND | S | ND | S | S | S | ND | S | ND | S | ND | S | ND |
| MGIT 960 | ND | S | ND | S | S | S | S | ND | S | ND | S | S | ND | S | ND | S | S | S | S | S | |||
| NZM 186038 (NZM 3/05) | Bactec 460 | R | R | ND | S | S | inhA, wt | S | ND | S | ND | S | ND | S | S | S | ND | R | ND | S | ND | S | ND |
| MGIT 960 | R | R | R | I | S | S | S | ND | S | ND | S | S | ND | S | ND | S | S | S | S | S | |||
| TBC 176291 | Bactec 460 | ND | ND | ND | ND | ND | inhA, wt | S | ND | S | ND | S | ND | S | S | S | ND | ND | ND | ND | ND | ND | ND |
| MGIT 960 | ND | ND | ND | ND | ND | S | S | ND | S | ND | S | S | ND | S | ND | S | S | S | S | S | |||
| TBC 177836 | Bactec 460 | R | R | ND | S | S | inhA: wt | S | ND | S | ND | S | ND | S | S | S | ND | S | ND | S | ND | S | ND |
| MGIT 960 | R | R | R | R | R | S | S | ND | S | ND | S | S | ND | S | ND | S | S | S | S | S | |||
| TBC 181783 | Bactec 460 | R | R | R | R | ND | inhA: C15T | S | ND | S | ND | S | ND | R | S | S | ND | R | ND | S | ND | S | ND |
| MGIT 960 | R | R | R | R | R | S | S | ND | S | ND | R | S | ND | S | ND | S | S | S | S | S | |||
| NZM 186069 | Bactec 460 | ND | ND | ND | ND | ND | inhA: wt | S | ND | S | ND | S | S | S | S | ND | R | ND | S | ND | S | ND | |
| MGIT 960 | ND | ND | ND | ND | ND | S | S | ND | S | ND | S | S | ND | S | ND | S | S | R | S | S | |||
| TBC 6890 | Bactec 460 | S | S | S | S | S | inhA: wt | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| MGIT 960 | ND | S | S | S | S | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| TBC 7392 | Bactec 460 | ND | ND | ND | ND | ND | inhA: wt | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| MGIT 960 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| TBC 295 | Bactec 460 | R | R | R | R | ND | inhA: wt | S | ND | S | ND | S | ND | S | S | S | S | S | ND | S | ND | ND | ND |
| MGIT 960 | R | R | R | R | R | S | S | ND | S | ND | S | S | S | S | ND | S | S | S | S | S | |||
| TBC 179882 | Bactec 460 | ND | ND | ND | ND | ND | inhA: wt | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| MGIT 960 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| NZM 186073 | Bactec 460 | S | S | S | S | ND | inhA: wt | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| MGIT 960 | S | S | S | S | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| NZM 186044 | Bactec 460 | R | S | S | S | ND | inhA: C15T | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| MGIT 960 | R | R | R | S | S | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| NZM 178364 | Bactec 460 | ND | ND | ND | ND | ND | inhA: wt | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| MGIT 960 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
ND, not done.
wt, wild type.
Of 12 strains tested, one isolate showed high-level resistance to amikacin (≥50 mg/liter). This isolate harbored the typical 16S rRNA mutation A1408G (23). Resistance at all test concentrations for this isolate, as well as susceptibility for the other strains investigated, was consistently determined with both test systems (Table 4).
Twelve strains were available for the comparison of testing for susceptibility to ofloxacin. Both systems consistently gave susceptible test results at a concentration of 2 mg/liter for 11 isolates. One isolate showed resistance at 2 mg/liter and sensitivity at 10 mg/liter (Table 4).
For linezolid and capreomycin, no resistant isolates with MICs above 4 mg/liter and 5 mg/liter, respectively, were present in the collection of strains under investigation. For the isolates studied, both systems consistently revealed sensitive test results at these drug concentrations (Table 4).
DISCUSSION
With the global rise of MDR strains, there is an increasing need to determine susceptibility to first and second-line anti-TB agents exactly. Treatment of patients with drug-resistant TB should be based on reliable and quantitative measures of susceptibility testing, a cornerstone for preventing further amplification of resistance (18) and for optimally exploiting available compounds. Detailed knowledge on quantitative drug resistance may guide empirical treatment of drug-resistant TB, e.g., addressing whether and when to add second-line drugs. A fundamental drawback to this strategy is that even in industrialized countries, only a limited panel of anti-TB drug concentrations is tested, leaving the exact resistance level of clinical M. tuberculosis isolates vestigial. In principle, automatic systems have the potential to meet the challenge of precise determinations of drug resistance levels with reasonable labor input. The MGIT 960 platform has been extensively validated for testing susceptibility to first-line anti-TB agents at critical drug concentrations (2, 24).
Here we systematically evaluated the performance of the MGIT 960 instrumentation for quantitative susceptibility testing of drug-resistant clinical isolates of M. tuberculosis. To investigate the underlying molecular resistance mechanisms, we used direct PCR-mediated sequence analysis of nucleic acids or the GenoType MTBDRplus assay. Resistance to isoniazid or ethionamide is due mainly to alterations in proteins that metabolize the prodrug into its active component, such as KatG or EthA. In contrast, resistance to rifampin, ethambutol, streptomycin, or quinolones is due to mutational alterations in genes encoding the drug target itself, such as rpoB, rpsL, rrs, or gyrA (21, 31).
For the first-line drugs, the quantitative resistance levels determined by the Bactec 460 and MGIT 960 systems were in good agreement. Introduction of the parameter “intermediate,” however, allowed a more-precise determination of resistance in the MGIT 960 system. It turned out that susceptibility testing of isolates with MICs near the test concentration may result in intermediate test results in the MGIT 960 system. The most prominent isoniazid resistance-conferring katG mutation (S315T) is associated with a heterogeneous resistance phenotype, with MICs ranging between 1 and 10 mg/liter.
The need for international standardization of testing of susceptibility to second-line anti-TB agents still remains. We have evaluated the performance of the MGIT 960 system for quantitative testing of susceptibility to the second-line drugs ethionamide, amikacin, ofloxacin, linezolid, and capreomycin. The results with the two test systems show good concordance. However, the conclusions are limited due to the small number of resistant isolates available to us. The drug most frequently affected by Bactec 460-MGIT 960 discrepancies was ethionamide. This is in accordance with previous studies (12, 25), which revealed more resistant results with the MGIT 960 instrumentation than with comparative test systems. Promoter alterations of inhA resulting in ethionamide resistance were observed in 14 of our ethionamide-resistant isolates (Tables 2 and 4). However, ethionamide resistance may also be due to ethA mutations not included in the GenoType MTBDRplus assay.
In general, the MGIT 960 system tended to give resistant test results more frequently than the Bactec 460 system. Medium differences and technical differences in the procedure used for growth measurement may account for this finding. Both instruments determine growth as a measure of metabolic activity. The Bactec 460 instrument measures volatile radioactive CO2 as metabolized from the radioactive precursor palmitic acid; for this purpose, most of the gaseous phase in the vial is removed and replaced. In contrast, the MGIT 960 instrument uses a fluorescence-quenching-based oxygen sensor for growth detection without manipulation of the closed vial. Under conditions of exceedingly slow growth, e.g., near the MIC, the Bactec 460 system would be expected to result in “false” susceptible test results, since it constantly depletes the vial of the little CO2 produced.
Existing procedures for DST of mycobacteria are adequate for screening but require complementation with quantitative DST measures, in particular for those drugs where heterogeneity in phenotypic resistance is present. We have established the conditions for quantitative DST using the MGIT 960 system in combination with EpiCenter software equipped with the TBeXiST module, thus providing a fully automated walk-away system for quantitative DST of M. tuberculosis. This platform allows electronic data management and is compatible with expert systems for interpretation. The MGIT 960 platform in conjunction with the EpiCenter software shows high consistency with Bactec 460 test results over a wide range of concentrations tested for first- and second-line anti-TB drugs. While we note that further studies are needed to address the correlation of phenotypic resistance levels and treatment outcome, we have summarized our recommendations for quantitative DST of M. tuberculosis in Table 5. Widespread implementation of MGIT 960 protocols for quantitative DST should provide standardized data to enable the correlation of results from quantitative DST with clinical outcomes by high-throughput statistical analysis in order to address the issue of phenotypic drug resistance levels and treatment failure. In addition, data sets obtained by an automated standardized procedure based on agreed guidelines provide optimal input for monitoring the epidemiology of resistance at a supranational level.
TABLE 5.
Recommendations for quantitative DST on the basis of the EpiCenter-equipped MGIT 960 system
| Drug | Recommendation | Comment |
|---|---|---|
| Isoniazid | 0.1 mg/liter is used for screening | Mutations in inhA typically lead to R at 0.1 mg/liter and S at 1.0 mg/liter; mutations in katG result in R at 1.0 mg/liter and show varying test results at 10.0 mg/liter |
| Low-level-resistant isolates are R at 0.1 mg/liter and S at 1.0 mg/liter | ||
| R at 10.0 mg/liter indicates that isoniazid is of no clinical use | ||
| Test results of R at 1.0 mg/liter and S at 10.0 mg/liter need further clinical data for interpretation | ||
| 1.0 mg/liter is presumably the most important concn to test | ||
| Rifampin | 1.0 mg/liter is used for screening | Most, if not all, mutations in rpoB are associated with R at >50.0 mg/liter; good correlation between genotype and phenotype |
| 10.0 mg/liter is used to recognize high-level drug resistance | ||
| Ethambutol | 5.0 mg/liter is used for screening | Isolates with mutations in embB are mostly, if not always, susceptible or intermediate at 25.0 mg/liter |
| 25.0 mg/liter is used to recognize high-level drug resistance | ||
| Streptomycin | 1.0 mg/liter is used for screening | Good correlation between genotype and phenotype |
| 20.0 mg/liter is used to recognize high-level drug resistance | ||
| 4.0 mg/liter is used to separate low-level (R at 1.0 mg/liter; S at 4.0 mg/liter) from intermediate (R at 1.0 mg/liter; I or R at 4.0 mg/liter; S at 20.0 mg/liter) resistance | ||
| Amikacin | 1.0 mg/liter is used for screening | |
| 20.0 mg/liter is used to recognize high-level drug resistance | ||
| 4.0 mg/liter is used to separate low-level (R at 1.0 mg/liter; S at 4.0 mg/liter) from intermediate (R at 1.0 mg/liter; I or R at 4.0 mg/liter; S at 20.0 mg/liter) resistance | ||
| Ethionamide | 2.5 mg/liter is used for screening | Mutations in inhA (C15T) may show phenotypic variability: unanimously R at 2.5 and 5.0 mg/liter; I or R at 12.5 mg/liter; S, I, or R at 25.0 mg/liter |
| Additional testing at 12.5 mg/liter | ||
| Ofloxacin | 2.0 mg/liter is used for screening | |
| Additional testing at 10.0 and 50.0 mg/liter | ||
| Capreomycin | 5.0 mg/liter is used for screening | |
| Additional testing at 25.0 mg/liter | ||
| Linezolid | 1.0 mg/liter is used for screening | |
| Additional testing at 4.0 mg/liter |
Acknowledgments
The study was supported in part by grants from the Bundesamt für Gesundheit, Bern, Switzerland. E. C. Böttger has a consultancy agreement with Becton Dickinson.
We thank Susanna Salas for help in writing the manuscript.
Footnotes
Published ahead of print on 1 April 2009.
REFERENCES
- 1.Ädjers-Koskela, K., and M.-L. Katila. 2003. Susceptibility testing with the manual mycobacteria growth indicator tube (MGIT) and the MGIT 960 system provides rapid and reliable verification of multidrug-resistant tuberculosis. J. Clin. Microbiol. 411235-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bemer, P., F. Palicova, S. Rüsch-Gerdes, H. B. Drugeon, and G. E. Pfyffer. 2002. Multicenter evaluation of fully automated BACTEC mycobacteria growth indicator tube 960 system for susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 40150-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttger, E. C. 2006. Mechanisms of mycobacterial drug resistance: implications for treatment and prevention strategies. Chemother. J. 1523-28. [Google Scholar]
- 4.Böttger, E. C., and B. Springer. 2008. Tuberculosis: drug resistance, fitness and strategies for global control. Eur. J. Pediatr. 167141-148. [DOI] [PubMed] [Google Scholar]
- 5.Canetti, G., S. Froman, J. Grosset, P. Hauduroy, M. Langerová, H. T. Mahler, G. Meissner, et al. 1963. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull. W. H. O. 29565-578. [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2006. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs—worldwide, 2000-2004. MMWR Morb. Mortal. Wkly. Rep. 55301-305. [PubMed] [Google Scholar]
- 7.Dorman, S. E., and R. E. Chaisson. 2007. From magic bullets back to the magic mountain: the rise of extensively drug-resistant tuberculosis. Nat. Med. 13295-298. [DOI] [PubMed] [Google Scholar]
- 8.Dye, C., M. A. Espinal, C. J. Watt, C. Mbiaga, and B. G. William. 2002. Worldwide incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 1851197-1202. [DOI] [PubMed] [Google Scholar]
- 9.Finken, M., P. Kirschner, A. Meier, A. Wrede, and E. C. Böttger. 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 91239-1246. [DOI] [PubMed] [Google Scholar]
- 10.Hanna, B. A., A. Ebrahimzadeh, L. B. Elliott, M. A. Morgan, S. M. Novak, S. Rüsch-Gerdes, M. Acio, D. F. Dunbar, T. M. Holmes, C. H. Rexer, C. Savthyakumar, and A. M. Vannier. 1999. Multicenter evaluation of the BACTEC MGIT 960 system for recovery of mycobacteria. J. Clin. Microbiol. 37748-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillemann, D., S. Rüsch-Gerdes, and E. Richter. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 452635-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, T.-S., S. S.-J. Lee, H.-Z. Tu, W.-K. Huang, Y.-S. Chen, C.-K. Huang, S.-R. Wann, H.-H. Lin, and Y.-C. Liu. 2004. Use of MGIT 960 for rapid quantitative measurement of the susceptibility of Mycobacterium tuberculosis complex to ciprofloxacin and ethionamide. J. Antimicrob. Chemother. 53600-603. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, R., A. M. Jordaan, L. Pretorius, E. Engelke, G. van der Spuy, G. Kewley, M. Bosman, P. D. van Helden, R. Warren, and T. C. Victor. 2006. Ethambutol resistance testing by mutation detection. Int. J. Tuberc. Lung. Dis. 1068-73. [PubMed] [Google Scholar]
- 14.Kim, S. J. 2005. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur. Respir. J. 25564-569. [DOI] [PubMed] [Google Scholar]
- 15.Kontos, F., M. Maniati, C. Costopoulos, Z. Gitti, S. Nicolaou, E. Petinaki, S. Anagnostou, I. Tselentis, and A. N. Maniatis. 2004. Evaluation of the fully automated Bactec MGIT 960 system for the susceptibility testing of Mycobacterium tuberculosis to first-line drugs: a multicenter study. J. Microbiol. Methods 56291-294. [DOI] [PubMed] [Google Scholar]
- 16.Krüüner, A., M. D. Yates, and F. A. Drobniewski. 2006. Evaluation of MGIT 960-based antimicrobial susceptibility testing and determination of critical concentration of first- and second-line antimicrobial drugs with clinical drug-resistant strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 44811-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miotto, P., F. Piana, D. M. Cirillo, and G. B. Migliori. 2008. Genotype MTBDRplus: a further step toward rapid identification of drug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 46393-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee, J. S., M. L. Rich, A. R. Socci, J. K. Joseph, F. A. Virú, S. S. Shin, J. J. Furin, M. C. Becerra, D. J. Barry, J. Y. Kim, J. Bayona, P. Farmer, M. C. Smith Fawzi, and K. J. Seung. 2004. Programmes and principles in treatment of multidrug-resistant tuberculosis. Lancet 363474-481. [DOI] [PubMed] [Google Scholar]
- 19.Nunn, P., B. Williams, K. Floyd, C. Dye, G. Elzinga, and M. Raviglione. 2005. Tuberculosis control in the era of HIV. Nat. Rev. Immunol. 5819-826. [DOI] [PubMed] [Google Scholar]
- 20.Ormerod, L. P. 2005. Multidrug-resistant tuberculosis (MDR-TB): epidemiology, prevention and treatment. Br. Med. Bull. 73-7417-24. [DOI] [PubMed] [Google Scholar]
- 21.Parsons, L. M., A. Somoskövi, R. Urbanczik, and M. Salfinger. 2004. Laboratory diagnostic aspects of drug resistant tuberculosis. Front. Biosci. 92086-2105. [DOI] [PubMed] [Google Scholar]
- 22.Pfyffer, G. E., D. A. Bonato, A. Ebrahimzadeh, W. Gross, J. Hotaling, J. Kornblum, A. Laszlo, G. Roberts, M. Salfinger, F. Wittwer, and S. Siddiqi. 1999. Multicenter laboratory validation of susceptibility testing of Mycobacterium tuberculosis against classical second-line and newer antimicrobial drugs by using the radiometric BACTEC 460 technique and the proportion method with solid media. J. Clin. Microbiol. 373179-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prammananan, T., P. Sander, B. A. Brown, K. Frischkorn, G. O. Onyi, Y. Zhang, E. C. Böttger, and R. J. Wallace, Jr. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 1771573-1581. [DOI] [PubMed] [Google Scholar]
- 24.Rüsch-Gerdes, S., C. Domehl, G. Nardi, M. R. Gismondo, H.-M. Welscher, and G. E. Pfyffer. 1999. Multicenter evaluation of the mycobacteria growth indicator tube for testing susceptibility of Mycobacterium tuberculosis to first-line drugs. J. Clin. Microbiol. 3745-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rüsch-Gerdes, S., G. E. Pfyffer, M. Casal, M. Chadwick, and S. Siddiqi. 2006. Multicenter laboratory validation of the BACTEC MGIT 960 technique for testing susceptibilities of Mycobacterium tuberculosis to classical second-line drugs and newer antimicrobials. J. Clin. Microbiol. 44688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah, N. S., A. Wright, G. H. Bai, L. Barrera, F. Boulahbal, N. Martín-Casabona, F. Drobniewski, C. Gilpin, M. Havelková, R. Lepe, R. Lumb, B. Metchock, F. Portaels, M. F. Rodrigues, S. Rüsch-Gerdes, A. Van Deun, V. Vincent, K. Laserson, C. Wells, and J. P. Cegielski. 2007. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg. Infect. Dis. 13380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siddiqui, S. 1996. BACTEC TB system. Product and procedure manual. Becton Dickinson, Sparks, MD.
- 28.Springer, B., R. C. Calligaris-Maibach, C. Ritter, and E. C. Böttger. 2008. Tuberculosis drug resistance in an area of low endemicity in 2004 to 2006: semiquantitative drug susceptibility testing and genotyping. J. Clin. Microbiol. 464064-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tortoli, E., M. Benedetti, A. Fontanelli, and M. T. Simonetti. 2002. Evaluation of automated BACTEC MGIT 960 system for testing susceptibility of Mycobacterium tuberculosis to four major antituberculous drugs: comparison with the radiometric BACTEC 460 TB method and the agar plate method of proportion. J. Clin. Microbiol. 40607-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Rie, A., R. Warren, M. Richardson, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 2000. Classification of drug-resistant tuberculosis in an epidemic area. Lancet 35622-25. [DOI] [PubMed] [Google Scholar]
- 31.Wade, M. M., and Y. Zhang. 2004. Mechanisms of drug resistance in Mycobacterium tuberculosis. Front. Biosci. 9975-994. [DOI] [PubMed] [Google Scholar]
- 32.WHO. 2007. Global tuberculosis control: surveillance, planning, financing. WHO report 2007. WHO/HTM/TB/2007.376. World Health Organization, Geneva, Switzerland.
- 33.WHO/IUATLD, Global Project on Anti-Tuberculosis Drug Resistance Surveillance. 2004. Anti-tuberculosis drug resistance in the world: third global report. WHO/HTM/TB/2004.343. World Health Organization, Geneva, Switzerland.