Abstract
The aim of this study was to develop and evaluate a sensitive method for the simultaneous identification of 14 urogenital potential pathogens. A multiplex PCR-based reverse line blot (mPCR/RLB) assay was developed to detect 14 urogenital pathogens or putative pathogens, namely Trichomonas vaginalis, Streptococcus pneumoniae, Neisseria gonorrhoeae, Chlamydia trachomatis, Ureaplasma parvum, U. urealyticum, Gardnerella vaginalis, Haemophilus influenzae, herpes simplex virus type 1 (HSV1) and HSV2, N. meningitidis, Mycoplasma hominis, M. genitalium, and adenovirus, using two species-specific primer pairs and probes for each. The method was validated using a reference strain or a well-characterized clinical isolate of each target organism and was found to be both sensitive and specific. The limits of detection for the mPCR/RLB assay varied among the 14 target organisms from 4.2 × 10−1 to 7.0 × 10−11 ng/μl of genomic DNA. There were no cross-reactions among any of the probes. This method was used to test 529 first-voided urine specimens from male patients with and without urethritis attending two Sydney sexual health clinics. One or more target species were detected in 193 (36%) subjects. Of 233 positive results, overall 216 (93%) were concordant between mPCR/RLB and a comparator method (culture and/or species-specific PCR), 9 were positive only by mPCR/RLB, and 8 were positive only by the comparator method. The mPCR/RLB method was an accurate, convenient, and inexpensive method for the detection of multiple potential pathogens in first-voided urine specimens from men.
Sexually transmitted infections (STIs) are a major global health problem. Worldwide, an estimated 340 million cases of curable STIs, including chlamydial infection, gonorrhea, trichomoniasis, and syphilis, occur annually, and their incidence is increasing in many parts of the world. In developing countries, their complications rank in the top five disease categories for which adults seek health care (www.who.int/mediacentre/factsheets/fs110/en/). Many STIs cause asymptomatic infection; for example, up to 70% of men and women with gonococcal and/or chlamydial infections are symptom free (www.who.int/mediacentre/factsheets/fs110/en/), which creates the potential for unrecognized transmission with significant implications for both individual and population health.
Urethritis is characterized by discharge and dysuria (37) and is broadly classified as nongonococcal (NGU) or gonococcal. It occurs in both men and women but often is unrecognized in women. Acute NGU is one of the commonest STIs affecting heterosexual men, yet a specific pathogen, most commonly Chlamydia trachomatis, is identified in only 50 to 70% of cases (7). Pelvic inflammatory disease is an important complication of STI in women; C. trachomatis and N. gonorrhoeae commonly are implicated, but often the cause is unknown. Bacterial vaginosis is the commonest cause of vaginal discharge and is associated both with recognized STIs and other genital syndromes (3, 18). Additional epidemiological studies are needed to determine the significance of organisms other than recognized genital pathogens in urethral and vaginal syndromes (7, 13-15). In particular, the pathogenic roles, if any, of the two recently defined human Ureaplasma species (10), U. urealyticum (previously U. urealyticum biovar 2) and U. parvum (previously U. urealyticum biovar 1), and several other genital (32, 43, 44) and respiratory pathogens (20, 30, 33, 42) in NGU are unclear.
The high level of sensitivity of nucleic acid amplification tests, such as PCR, allows the use of less invasive specimen types, including first-voided urine specimens or self-collected vaginal swabs that are unsuitable for less sensitive methods, such as culture and antigen tests (8). This paper describes the development and evaluation of a multiplex PCR-based reverse line blot (mPCR/RLB) assay (19) that can detect any of 14 recognized and potential genital pathogens in urine specimens for use in clinical and epidemiological studies of genital infections.
MATERIALS AND METHODS
Reference strains.
Previously well-characterized clinical isolates of C. trachomatis, N. gonorrhoeae, M. genitalium, herpes simplex virus type 1 (HSV1) and HSV2, adenovirus, T. vaginalis, M. hominis, G. vaginalis, and N. meningitidis, provided by the Centre for Infectious Disease and Microbiology (CIDM) diagnostic laboratory, were used as positive controls. All isolates had been identified according to routine methods (27). In addition, the following organisms were purchased from the American Type Culture Collection (ATCC; Manassas, VA): U. urealyticum (ATCC 27813 and ATCC 27814), U. parvum (ATCC 27818), Haemophilus influenzae (ATCC 10211), and Streptococcus pneumoniae (ATCC 27336).
Clinical specimens.
Five hundred twenty-nine male patients with and without urethral symptoms were enrolled in a study of NGU at Parramatta Sexual Health Clinic (PSHC) and Sydney Sexual Health Centre (SSHC) from November 2006 to September 2007. Men with characteristic gonococcal urethritis, in whom Gram stains of urethral discharge showed gram-negative diplococci, and men who had been treated with antibiotics in the previous 6 weeks were excluded from the study. First-voided urine specimens were collected. Specimens from the SSHC were split, and one portion was sent to the routine diagnostic laboratory serving the clinic for C. trachomatis PCR (Roche COBAS Amplicor). Specimens were stored at 4°C at the clinic and transported in weekly batches, in a cool box, to the Centre for Infectious Diseases and Microbiology (CIDM), where they were stored at 4°C until DNA extraction was performed within 24 h of receipt. Specimens from PSHC were stored under the same conditions until being tested for C. trachomatis using a Roche COBAS Amplicor at the CIDM diagnostic laboratory.
In addition, to assist in the validation of mPCR/RLB results, urethral swabs were collected from all subjects. A Gram-stained smear was examined at the clinic. The swabs were placed in Stuart's transport medium and transported to the CIDM diagnostic laboratory, where cultures for N. gonorrhoeae, aerobic/facultative bacteria (including S. pneumoniae, H. influenzae, N. meningitidis, and G. vaginalis), M. hominis, and Ureaplasma spp. were performed. Swabs were plated on New York City medium, 5% horse blood, chocolate (both in Columbia agar base; Oxoid, Basingstoke, United Kingdom), and A8 mycoplasma (Oxoid, Basingstoke, United Kingdom) agars, incubated for 24 to 48 h in CO2, and identified by microscopic and colony morphology and biochemical and antigen tests (27).
DNA extraction.
The Roche COBAS Amplicor extraction kit (Roche Diagnostics Australia Pty Limited Systems, Australia) was used per the manufacturer's instructions. Briefly, the urine specimens were vortexed thoroughly for 10 s before 500 μl of each specimen was transferred to a tube containing 500 μl of wash buffer. The specimens then were incubated at 37°C for 15 min and centrifuged at 13,000 × g for 5 min. The supernatant was discarded, 250 μl of lysis buffer was added, and after incubation at room temperature for 15 min, 250 μl of specimen diluent was added to the lysate. The tubes then were vortexed and centrifuged for 10 min at 13,000 × g and stored at −70°C.
Primer and probe design.
Two sets of species-specific primers and probes, targeting highly conserved regions, were designed for each organism. The primers and probes used for this assay are shown in Table 1 and in the supplemental material. Primers and probes were designed to have similar physical characteristics to allow simultaneous amplification and hybridization in a multiplex reaction without the loss of sensitivity as follows: melting temperature (Tm), 58 to 65°C; length, 18 to 30 bp; moderate, weak, or no secondary structure; no dimer formation; and amplicon sizes, 80 to 400 bp (19). Some primers were selected from published papers and modified to match the desired characteristics. All probes and primers were checked for specificity against all sequences in GenBank using SeqSearch in the Australian National Genomic Information Services (ANGIS) programs (http://www.angis.org.au). The adenovirus primers used were designed to allow annealing to all 51 known adenovirus types by introducing degenerative base positions (1). Oligonucleotide primers were biotinylated at the 5′ end, and probes had a 5′ amine group and were synthesized by Sigma Aldrich (Sydney, Australia).
TABLE 1.
Oligonucleotide primers and probes developed or modified for the mPCR/RLB assay used in this studya
| Primer/probe nameb | Specificityc (target) | GenBank accession no. | Primer-probe sequenced (5′-3′) | Tme (°C) | Reference or sourceg |
|---|---|---|---|---|---|
| TV-Ap | T. vaginalis (btuB) | 904TGT TGT GAG CTT GAG TGT ACG G883 | 65.1084 | This study | |
| TV-Sp | 916CGA TCT TAA CCA CCT TGT TTC C945 | 63.31923 | 14 (mod) | ||
| NGpSb | N. gonorrhoeae (CP) | M10316 | 3249TGC TGT TTC AAG TCG TCC AG3268 | 64.06359 | This study |
| NGpAp | 3317GAT AGT CAT AGC AGG GCT GTT C3296 | 61.5549 | This study | ||
| NGpSp | 3452CCG TAA CGT CTC TAA GTC TGC TT3474 | 62.51156 | This study | ||
| NGpAb | 3503CGA AGC CGC CAG CAT AGA GC3484 | 71.22094 | This study | ||
| NG16Sb | N. gonorrhoeae (ITS) | AF223396 | 404CCA AAA CTT AAC AAA TGA AAG CAA G428 | 63.41 | This study |
| NG1S | 453TGA TTT GCG AAG TAG AAT AAC G474 | 60.64 | This study | ||
| NG2A2 | 456ATC AAA ATA AGC TGC TAA AAA CAG433 | 59.41 | This study | ||
| NGITSAb | 490TGT TAA AGA TCG ATG CGT CGT472 | 64.33 | This study | ||
| CT24b | C. trachomatis (CP) | X06707 | 840GGG ATT CCT GTA ACA ACA AGT CAG G864 | 67.33045 | This study |
| CTS1p | 865TTG CGC ATA ATT TTA GGC TTG885 | 63.59178 | This study | ||
| CTA2p | 1021ACA CTT TGT CTC GAT GAA AGA CA999 | 62.57137 | This study | ||
| CT27b | 1047CCT CTT CCC CAG AAC AAT AAG AAC AC1022 | 67.37814 | This study | ||
| UP-Sb | U. parvum (ureB) | AF085731 | 637GAT CAC ATT TTC ACT TGT TTG AAG TG662 | 64.37767 | 23 (mod) |
| UP-Ap | 702CTT CAT TTC CTT TTT CAT CAA AAA ATA C675 | 63.43446 | This study | ||
| UP-Sp | 688AAA AAG GAA ATG AAG ATA AAG AAC G712 | 61.40322 | 23 (mod) | ||
| UP-Ab | 735AAC GTC GTC CAT AAG CAA CTT TG713 | 65.8875 | 23 (mod) | ||
| UU-Ap | U. urealyticum (ureB) | 705CTT CAT TTC CTT TTT CAT CAA AAA ATA C678 | 63.43446 | This study | |
| UU-Sp | 691AAA AAG GAA ATG AAG ATA AAG AAC G715 | 61.40322 | 23 (mod) | ||
| UU-Ab | 739AAA CGA CGT CCA TAA GCA ACT TTA716 | 64.34317 | 23 (mod) | ||
| GV-Ap | G. vaginalis (ITS) | 416TCC TGT CTA CCA AGG CAT CC397 | 63.97078 | This study | |
| GV-Sp | 632CGT GTG ATA ACC GTC AGG TG651 | 64.06355 | This study | ||
| HSV1-Sp | HSV1 (gD) | 545CGT TTG AGA CCG CCG GCA562 | 73.04224 | This study | |
| HSV2-Sp | 572CCT TCG AGA CCG CGG GTA589 | 68.53278 | This study | ||
| NM-Sb | N. meningiditis (porA) | AY319969 | 929GCT TCG GTA ATG CAG TTC CA948 | 64.94242 | 17 (mod) |
| NM-Ap | 1010CTG GTA TTT TCG CCT TTT TTA C989 | 17 (mod) | |||
| NM-Sp | 953TCA GCT ATG CCC ATG GTT970 | This study | |||
| MH-Ap | M. hominis (gap) | 770CTG AAT AAA CAA CTG TTT TAA CAC CTT CGCT740 | 68.7016 | 2 (mod) | |
| MH-Sp | 702CAG GTG CTA AAA AGG TGT TTA TTA CTG CT730 | 66.55416 | This study | ||
| MgPAa-Ap | M. genitalium (mgpA) | 1463TAT CAT ACC TTC TGA TTG CAA AGT1445 | 60.39573 | This study | |
| MgPa-Sp | 1473CGG TAG AGC TTT ATA TGA TAT TAA CTT AGC1502 | 61.46476 | This study | ||
| AdVdeSb | Adenovirus (hexon) | GCC SCA RTG GKC WTA CAT GCA CAT Cf | 69.2 | 1 (mod) | |
| AdVdeAp | CCY ACR GCC AGI GTR WAI CGM RCY TTG TA | 68.16371 | 1 (mod) | ||
| AdVdeSp | GCC CGY GCM ACI GAI ACS TAC TTC | 63.76665 | 1 (mod) | ||
| AdVdeAb | CAG CAC SCC ICG RAT GTC AAA | 62.80243 | 1 (mod) |
In addition to primers specifically designed or modified for this study, published primers were used without modification for several targets. Details are shown in the supplemental material.
The suffix b indicates a biotin-labeled primer, and p indicates an amine-labeled probe. An A indicates antisense, and S indicates sense.
Abbreviations: CP, cryptic plasmid; ITS region, intergenic spacer region; gD, glycoprotein D.
Numbers represent the base positions at which the primer/probe sequence starts and finishes (starting at point 1 of the corresponding gene sequence in GenBank).
Melting temperatures were provided by the primer synthesizer (Sigma-Aldrich).
S = G+C; R = A+G; K = G+T; W = A+T; Y = C+T; M = A+C; I = inosine.
Some primers were modified (mod) from the published primers.
mPCR amplification.
mPCR amplification was performed using a 25-μl reaction mixture containing 10 μl template DNA, 0.075 μl of each forward (100 pmol μl−1) and reverse (100 pmol μl−1) primer, 1.25 μl deoxynucleoside triphosphates (0.125 mM of each deoxynucleoside triphosphate), 2.5 μl 10× buffer (Qiagen), 3.0 μl 25 mM MgCl2 (final concentration, 3.0 mM), 0.2 μl Qiagen HotStar Taq polymerase (5 U μl−1), and water to 25 μl. The thermal profile involved initial denaturation for 15 min at 95°C, 40 cycles of 30 s at 94°C, 55°C for 30 s, and 72°C for 90 s, and a final extension for 10 min at 72°C, followed by a hold at 22°C. Inhibition controls were not included in the assay.
RLB assay.
The RLB assay was performed as previously described (19). Briefly, probes were labeled and fixed to the membrane in various concentrations (0.6 to 10.8 pmol/μl) to determine the optimal conditions. Each PCR product was denatured and immediately chilled on ice. Hybridization was performed at 60°C for 60 min. The washed membrane was incubated in peroxidase-labeled streptavidin conjugate (Roche, Germany) at 42°C for 60 min. The membrane then was incubated in chemiluminescence blotting substrate (ECL direct system; Roche) for 2 min and covered with Hyperfilm X-ray film (Amersham). The film was exposed for 5 min.
sPCR.
Single PCRs (sPCRs), using different targets from those used in the mPCR/RLB assay, were used as comparator methods for T. vaginalis, HSV1 and HSV2, M. genitalium, and adenovirus to confirm the specificity of the mPCR targets. The oligonucleotide primers used for sPCR are shown in Table 2. sPCRs for the other nine pathogens, using the same primers as those used in the mPCR, were performed when culture and mPCR/RLB results were discrepant. The same primers were used to confirm results, as sPCR generally is more sensitive than mPCR and the possibility of cross-reactions, which can occur in mPCR, is avoided.
TABLE 2.
Oligonucleotide primers used for sPCR
| Primer name | Specificity (target) | GenBank accession no. | Primer sequencea (5′-3′) | Tmb (°C) | Reference |
|---|---|---|---|---|---|
| Tv1 | T. vaginalis 18S rRNA gene | U17510 | 874TAA TGG CAG AAT CTT TGG AG894 | 59.2 | 25 |
| Tv2 | 1185GAA CTT TAA CCG AAG GAC TTC1165 | 58.3 | |||
| HSVPolA1 | HSV DNA polymerase | ATC ATC TAC CGC GAC ACG GACT | 68.8 | 49 | |
| HSVPolA2 | TCC ACG CCC TTG ATG AGC ATC T | 72.0 | |||
| MG16-45F | M. genitalium 16S rRNA gene | X77334 | 45TAC ATG CAA GTC GAT CGG AAG TAG C69 | 68.8 | 16 |
| MG16-447R | 469AAA CTC CAG CCA TTG CCT GCT AG447 | 69.2 | |||
| AD1 | Adenovirus hexon gene | U20821 | CTG ATG TAC TAC AAC AGC ACT GGC AAC ATG GG | 76.1 | 36 |
| AD2 | GCG TTG CGG TGG TGG TTA AAT GGG TTT ACG TTG TCC AT | 83.4 | |||
| GV1 | G. vaginalis | LO8167 | 364TTA CTG GTG TAT CAC TGT AAG G385 | 55.8 | 51 |
| 16S-23S rRNA gene | |||||
| GV2 | 23S | 695CCG TCA CAG GCT GAA CAG T677 | 64.1 |
Numbers represent the base positions at which the primer/probe sequence starts and finishes (starting at point 1 of the corresponding gene sequence in GenBank).
Melting temperatures were provided by the primer synthesizer (Sigma-Aldrich).
sPCRs were performed using a 25-μl reaction mixture containing 10 μl template DNA, 0.25 μl of each forward (100 pmol μl−1) and reverse (100 pmol μl−1) primer, 1.25 μl deoxynucleoside triphosphates (0.125 mM of each), 2.5 μl 10× buffer (Qiagen), 3.0 μl 25 mM MgCl2 (final concentration, 3.0 mM), 0.2 μl Qiagen HotStar Taq polymerase (5 U μl−1), and water to 25 μl. The thermal profile involved initial denaturation for 15 min at 95°C, 40 cycles of 30s at 94°C, 55°C for 30s, and 72°C for 90 s, and a final extension for 10 min at 72°C, followed by a hold at 22°C.
Plasmid construction.
Reference strains of all species were amplified using primers (Table 1; also see the supplemental material) targeting species-specific genes. Amplified products were inserted into a pGEM-T Easy vector system (Promega) and transformed into competent Escherichia coli cells JM109 according to the manufacturer's instructions. The Luria-Bertani-ampicillin-5-bromo-4-chloro-3-indolyl-â-d-galactopyranoside-isopropyl-β-d-thiogalactopyranoside plates were screened for positive clones and subcultured. The clones then were extracted. DNA concentrations were determined using UV spectrometry.
Analytical sensitivity.
The analytical sensitivity of the mPCR/RLB assay was estimated using a series of 10-fold dilutions of plasmid templates of all species to determine the lowest limit of detection, which was expressed as nanograms per microliter of DNA in the last sample positive in the dilution series for each organism.
The results of mPCR/RLB assays were compared to those of the Roche COBAS Amplicor PCR for C. trachomatis in the same specimens (performed by routine diagnostic laboratories serving both clinics) and with those of urethral swab cultures for N. gonorrhoeae, Ureaplasma spp., M. hominis, and aerobic/facultative bacteria, including S. pneumoniae, H. influenzae, and G. vaginalis. Final results, after repeating any tests that were discrepant, were accepted as true positives if (i) both RLB probes were positive or one was positive and the result was confirmed by sPCR; (ii) the culture was positive; and/or (iii) sPCR using an alternative target was positive.
Data analysis.
Data analysis was carried out using SPSS software (version 15.0; SPSS Inc., Chicago, IL). Univariate analysis was performed using Fisher's exact test and the chi-squared test for categorical variables. A 95% confidence interval was used.
RESULTS
Testing and validation with clinical isolates and reference strains of target organisms showed that each target species was amplified, hybridized, and correctly identified by the mPCR/RLB assay (Fig. 1). Initially nonspecific binding during hybridization was observed, which was eliminated by adjusting reaction conditions and components. Final probe concentrations used to label the RLB membrane were 10.8 pmol/μl for HSV2, 5.4 pmol/μl for S. pneumoniae and adenovirus, 0.6 pmol/μl for N. gonorrhoeae, and 1.8 pmol/μl for all other species. In all cases both probes gave positive results in the RLB if target DNA was present in the sample.
FIG. 1.
mPCR/RLB results using reference strains. Lanes: 1 and 2, T. vaginalis; 3 and 4, S. pneumoniae; 5 and 6, N. gonorrhoeae; 7 and 8, C. trachomatis; 9 and 10, U. parvum; 11 and 12, U. urealyticum; 13 and 14, G. vaginalis; 15 and 16, H. influenzae; 17 and 18, HSV1; 19 and 20, HSV2; 21 and 22, N. meningitidis; 23, blank; 24, M. hominis; 25 and 26, M. genitalium; and 27 and 28, adenovirus type 1.
Analytical sensitivity of mPCR/RLB and comparison to alternative methods.
The limit of detection by mPCR/RLB ranged from 4.2 × 10−1 to 7.0 × 10−11 ng/μl for different species. Results for mPCR/RLB and the comparator method for each target species (except HSV2, which was not detected in any specimens) are shown in Table 3. Of a total of 233 positive results, 211 (90%) were concordant in mPCR/RLB and comparator methods on initial testing; 14 were positive in mPCR/RLB only (of which 10 were resolved by repeating the comparator method); and 7 were positive in the comparator method only.
TABLE 3.
Comparison of results of mPCR/RLB and comparator methods in detection of genital infection/colonization with 14 recognized or putative genital pathogens
| Organism | Limit of detectiona (ng/μl) | No. positively detected by mPCR/RLBb | Comparator methodc (target) | No. positively detected by comparator method |
|---|---|---|---|---|
| T. vaginalis | 1.5 × 10−9 | 1 | sPCR (18S rRNA gene) | 1 |
| S. pneumoniae | 7.4 × 10−1 | 1 | Culture | 1 |
| N. gonorrhoeae | 4.3 × 10−2 | 7 | Culture and Amplicor PCR | 2 (+5)d |
| C. trachomatis | 7.0 × 10−11 | 55 | Amplicor PCR | 50 (+5)d |
| Ureaplasma spp.e | 7.8 × 10−9 | 84 | Culture | 86 |
| G. vaginalis | 7.8 × 10−3 | 3 | sPCR (16-23S rRNA gene) | 3 |
| H. influenzae | 7.0 × 10−8 | 31 | Culture | 35 |
| HSV1 | 4.3 × 10−8 | 8 | sPCR (pol) | 5 |
| N. meningitides | 8.2 × 10−8 | 2 | Culture | 1 |
| M. hominis | 5.5 × 10−1 | 15 | Culture | 16 |
| M. genitalium | 2.1 × 10−9 | 15 | sPCR (16S rRNA gene) | 15 |
| Adenovirus | 6.3 × 10−7 | 4 | sPCR (hexon gene) | 4 |
For mPCR/RLB.
sPCR, using the same primers as those for mPCR, was performed on specimens with discrepant mPCR/RLB and culture results. In all cases the mPCR/RLB and sPCR results were concordant.
Comparator methods were either the culture of urethral swab collected at the same time as first-voided urine specimen or sPCR on the same urine DNA extract as that used for mPCR/RLB, using a different, species-specific target (except for adenoviruses, for which the same hexon gene target was used).
Of the 7 and 55 specimens positive by mPCR/RLB for N. gonorrhoeae and C. trachomatis, respectively, only 2 and 50 were positive initially in the Roche Amplicor PCR; all were positive on retesting.
Urethral specimens were cultured for ureaplasmas, but isolates were not speciated. Ureaplasma spp. identified in the mPCR/RLB-positive specimens are shown in Fig. 2.
Of the 55 specimens in which C. trachomatis was detected using the mPCR/RLB method, 5 (10%) initially were negative using the Roche COBAS Amplicor PCR. However, on retesting, C. trachomatis was detected in all five specimens by the Amplicor PCR. All specimens that initially were positive with the Amplicor PCR also were positive with mPCR/RLB. Although subjects with clinical or microscopic evidence of gonorrhea on presentation were excluded, seven had positive tests for gonorrhea in the mPCR/RLB test and were culture positive. Only two of these specimens were positive for N. gonorrhoeae using the Roche COBAS Amplicor PCR when tested initially, but all were positive on retesting.
Ureaplasma spp. were identified in 86 urethral swab specimens by culture but in only 84 urine specimens by mPCR/RLB (U. parvum, 31; U. urealyticum, 53). All mPCR/RLB results were confirmed by species-specific sPCR.
Clinical specimens.
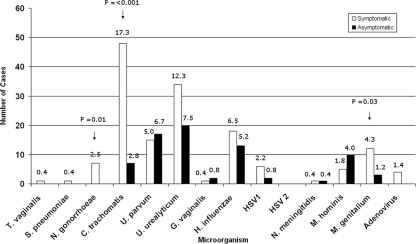
The age of the 529 subjects ranged from 19 to 76 years (mean, 37 years; median, 35 years). One or more pathogens or putative pathogens were identified in 193 (36%) men, including 136 of 277 (49%) men with urethritis symptoms and 57 of 252 (23%) asymptomatic men. Two or more target organisms were identified in 30 men (16 symptomatic and 14 asymptomatic) (Table 4). Figure 2 shows the numbers and percentages of specimens in which each organism was detected by symptom status. A simple comparison of results in symptomatic and asymptomatic men showed that C. trachomatis (48/277 and 7/252, respectively; P < 0.001), N. gonorrhoeae (7/277 and 0/252, respectively; P = 0.01), and M. genitalium (12/277 and 3/252, respectively; P = 0.03) were detected significantly more frequently in men with symptoms. There were no significant differences in detection rates between symptomatic and asymptomatic men for any other pathogens. A detailed analysis of clinical and epidemiological data will be reported separately (D. Couldwell, unpublished data).
TABLE 4.
Mixed genital infections/colonization with target organisms detected by mPCR/RLB assay from 30 men with and without urethral symptoms
| Symptom(s)a | Organisms detectedb | No. of cases |
|---|---|---|
| Yes | NG, HI | 1 |
| CT, NG | 2 | |
| CT, MG | 1 | |
| CT, UU | 1 | |
| CT, HSV1, UP | 1 | |
| CT, NM | 1 | |
| CT, HI | 1 | |
| MG, UU | 3 | |
| MG, UP, GV | 1 | |
| MG, HI | 1 | |
| UU, MH | 1 | |
| UP, ADV | 1 | |
| UP, MH | 1 | |
| Total | 16 | |
| No | CT, MH | 2 |
| UU, MG, MH | 1 | |
| UU, MG, HI, GV | 2 | |
| UU, GV | 1 | |
| UU, HI | 1 | |
| UU, MH | 1 | |
| HSV1, HI | 1 | |
| HSV1, UP | 1 | |
| UP, MH | 4 | |
| Total | 14 |
A symptomatic man was a patient who had urethral symptoms (dysuria, urethral discomfort, or urethral discharge), and an asymptomatic man was a patient who presented with no symptoms.
Abbreviations: CT, C. trachomatis; NG, N. gonorrhoeae; HI, H. influenzae; UU, U. urealyticum; GV, G. vaginalis; UP, U. parvum; MH, M. hominis; MG, M. genitalium; ADV, adenovirus; and NM, N. meningitidis.
FIG. 2.
Results of mPCR/RLB on first-voided urine specimens for 14 target urogenital organisms from men with and without symptoms of urethritis. Numbers on bars indicate the percentages of subjects with positive results for each target organism. Denominators are 277 for symptomatic men and 252 for asymptomatic men; the y axis shows the number of positive specimens. The arrows indicate organisms detected significantly more frequently in men with urethral symptoms than in men without symptoms (P ≤ 0.05).
DISCUSSION
We have developed an mPCR/RLB hybridization assay that permits the reliable, simultaneous detection of 14 known or potential urogenital pathogens, many of which are difficult to identify by other methods. We have used mPCR/RLB previously to identify multiple pathogens in respiratory specimens and blood cultures (45, 46). Others (40) have used gel-based mPCR to identify Ureaplasma spp., M. genitalium, and M. hominis in first-voided urine samples, and there is a recent report of an mPCR to detect 16 pathogens using a microplate assay (25). mPCR/RLB potentially is applicable to routine diagnosis, can be modified to add or delete targets, and is particularly suitable for epidemiological studies to examine the roles of putative pathogens in genital syndromes. It is more practicable and less expensive than microarray technology.
Overall, 75 target organisms were detected in 57 of 252 asymptomatic men, and 158 were detected in 136 of 277 men with symptoms; multiple organisms were identified in approximately equal numbers of men with and without symptoms. They included two mixed infections with N. gonorrhoeae and C. trachomatis, which is not uncommon and probably results from simultaneous transmission (25, 31). Both are well-established genital pathogens, whether or not they cause symptoms, and M. genitalium also has been implicated in NGU (7, 24). In this study, all three were significantly associated with the presence of urethral symptoms (P < 0.05). The rate of the detection of M. genitalium was similar to that reported by others (5, 25, 40), and more widespread testing for it in patients with NGU has been advocated (6).
The roles of the other organisms or combinations of organisms targeted in this study are uncertain, since many commonly are found among the normal genital flora. Providing further evidence for their roles in urethritis was the aim of the clinical component of this study (of which the results will be reported separately). There were differences between symptomatic and asymptomatic men in the rates of the detection of HSV1, adenovirus, and U. urealyticum as in other studies (7, 50), but the numbers were small and overall differences did not reach statistical significance.
The comparison of mPCR/RLB results to those of alternative detection methods showed very good correlation. Several organisms were detected in very few (<5) specimens (T. vaginalis, S. pneumoniae, G. vaginalis, N. meningitidis, and adenovirus), but results agreed in all but one (one culture negative and mPCR/RLB positive for N. meningitidis). mPCR/RLB identified N. gonorrhoeae, HSV1, and M. genitalium in all specimens that were positive by comparator methods and HSV1 in three additional specimens. It did not detect M. hominis in one and Ureaplasma spp. in two urine specimens from men whose urethral swabs were culture positive. These specimens were from men attending SSHC, where urine specimens were stored at 4°C for several days before being processed, which may have reduced the sensitivity of mPCR/RLB compared to that of the culture of urethral swabs, which were stored at room temperature in Stuart's transport medium. The refrigeration of specimens for several days had no apparent effect on the detection of other pathogens. Stellrecht et al. (40), using urine and swabs for PCR, recorded sensitivities similar to those of culture for Ureaplasma spp., M. genitalium, and M. hominis, and similar sensitivities have been reported by others for other species (2, 12, 23, 40, 50). Nevertheless, these results suggest that specimens should be processed for PCR as soon as possible after collection and, if they cannot be tested immediately, stored as DNA extracts.
False-negative mPCR/RLB results also may have resulted from the prolonged storage of DNA extracts (up to 18 months at −20°C) prior to testing, which can affect DNA quality and PCR efficiency (28), or from PCR inhibitors in urine (9, 34).
Initial false-negative results for Roche COBAS Amplicor PCR for N. gonorrhoeae and C. trachomatis (some from both diagnostic laboratories performing routine testing for the two clinics) were positive on retesting. These results reflect the real-life pitfalls of diagnostic testing, even by reputable laboratories using generally reliable assays.
A significant limitation of this study, in common with other studies of new, potentially more sensitive tests, was that there was no single gold standard for the analysis of mPCR/RLB. We chose to culture, where practicable, a different specimen (urethral swab) for target bacteria, for which urine would have been inappropriate, to identify men with urogenital colonization/infection. To optimize the reliability of the mPCR/RLB, we measured limits of detection, quantitatively, using cloned targets; confirmed all positive results using culture or sPCRs; designed primers and probes based on targets used in well-established sPCR methods, which had been shown to be specific; and avoided contamination by the use of appropriate negative and no-DNA controls. There were relatively few discrepancies between methods, and we believe that the decision to regard any confirmed positive result as a true positive for the purposes of comparison was justified. Nevertheless, we cannot exclude the possibility that a small number of mPCR/RLB results were false positives. Even in this relatively large number of subjects, most target organisms were identified too infrequently to calculate accurate sensitivities and predictive values.
This study confirmed the sensitivity and specificity of the mPCR/RLB assay for the detection of a wide range of potential urogenital pathogens in first-voided urine specimens. However, cross-reactivity can occur if primers and probes are not designed correctly, and the optimization of reaction components and conditions is required to produce a stable system without nonspecific reactions. The advantages of mPCR/RLB are that it can simultaneously test up to ∼40 specimens for up to ∼40 target genes in a single reaction, and it could be used for a variety of specimens other than urine, including cervical smears collected for cytology (48) or self-collected vaginal swabs. Given the high frequency of the multiple species identified, we believe that such an approach could contribute to an effective public health response to STIs.
Supplementary Material
Acknowledgments
We thank Vitali Sintchenko and Heather Gidding for assistance in planning the study. Victor Weixiong provided some of the primer/probe designs. We also thank the virology staff at CIDM for performing all DNA extractions, Bin Wang from the Westmead Millennium Institute for assistance with cloning experiments, and clinical research teams at SSHC and PSHC for the collection of urine samples and clinical data.
Footnotes
Published ahead of print on 8 April 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Allard, A., B. Albinsson, and G. Wadell. 2001. Rapid typing of human adenovirus by general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baczynska, A., H. F. Svenstrup, J. Fedder, S. Birkelund, and G. Christiansen. 2004. Development of real-time PCR for detection of Mycoplasma hominis. BMC Microbiol. 351-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, J. V., C. Farquhar, C. Owen, and P. Mangtani. 2004. Sexually transmitted infections in women who have sex with women. Sex. Transm. Infect. 80244-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Bjornelius, E., P. Lidbrink, and J. J. Jensen. 2000. Mycoplasma genitalium in nongonococcal urethritis—a study in Swedish male STD patients. Int. J. STD AIDS 11292-296. [DOI] [PubMed] [Google Scholar]
- 6.Bowden, F. J., S. N. Tabrizi, S. M. Garland, and C. K. Fairley. 2002. Sexually transmitted infections: new diagnostic approaches and treatments. Med. J. Aust. 176551-557. [DOI] [PubMed] [Google Scholar]
- 7.Bradshaw, C. S., S. N. Tabrizi, T. R. H. Read, S. M. Garland, C. A. Hopkins, L. M. Moss, and C. K. Fairley. 2006. Etiologies of non-gonococcal urethritis: bacteria, viruses, and the association with orogenital exposure. J. Infect. Dis. 193336-345. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M. Y. 2005. Changes in testing methods for genital Chlamydia trachomatis in New South Wales, Australia. Sex. Health 2251-253. [DOI] [PubMed] [Google Scholar]
- 9.Chong, S., D. Jang, X. I. Song, J. Mahony, A. Petrich, P. Barriga, and M. Chernesky. 2003. Specimen processing and concentration of Chlamydia trachomatis added can influence false negative rates in the LCx assay but not in the APTIMA Combo 2 assay when testing for inhibitors. J. Clin. Microbiol. 41778-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deguchi, T., T. Yoshida, T. Miyazawa, M. Yasuda, H. Tamaki, H. Ishiko, and S. Maeda. 2004. Association of Ureaplasma urealyticum (biovar 2) with nongonoccocal urethritis. Sex Trans. Dis. 31192-195. [DOI] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Falk, L., and J. S. Jensen. 2005. Signs and symptoms of urethritis and cervicitis among women with or without Mycoplasma genitalium or Chlamydia trachomatis infection. Sex Trans. Infect. 8173-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagman, M., L. Forslin, H. Moi, and D. Danielsson. 1991. Neisseria meningitidis in specimens from urogenital sites. Sex Trans. Dis. 18228-232. [DOI] [PubMed] [Google Scholar]
- 14.Hardick, J., S. Yang, S. Lin, D. Duncan, and C. Gaydos. 2003. Use of the Roche LightCycler instrument in a real-time PCR for Trichomonas vaginalis in urine samples from females and males. J. Clin. Microbiol. 125619-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann, A. A., and P. Elsner. 1988. Urethritis caused by Neisseria meningitidis group B: a case report. Sex. Trans. Dis. 15150-151. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, J. S., B. E. D. Birthe, and P. Lidbrink. 2004. Use of TaqMan 5′ nuclease real-time PCR for quantitative detection of Mycoplasma genitalium DNA in males with and without urethritis who were attendees at a sexually transmitted disease clinic. J. Clin. Microbiol. 42683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordens, Z. J., and J. E. Heckels. 2005. A novel porA-based real time PCR for detection of meningococcal carriage. J. Med. Microbiol. 54463-466. [DOI] [PubMed] [Google Scholar]
- 18.Keane, F. E. A., B. J. Thomas, L. Whitaker, A. Renton, and D. Taylor-Robinson. 1997. An association between non-gonococcal urethritis and bacterial vaginosis and the implications for patients and their sexual partners. Genitourin. Med. 73373-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong, F., and G. L. Gilbert. 2006. Multiplex PCR-based reverse line blot hybridisation assay (mPCR/RLB)—a practical epidemiological and diagnostic tool. Nat. Protoc. 12668-2680. [DOI] [PubMed] [Google Scholar]
- 20.Koroglu, M., Y. Yakupogullari, and F. Aydogan. 2007. A case of urethritis due to Streptococcus pneumoniae. Sex. Trans. Dis. 341040. [DOI] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Reference deleted.
- 23.Mallard, K., K. Schopfer, and T. Bodmer. 2005. Development of real-time PCR for the differential detection and quantification of Ureaplasma urealyticum and Ureaplasma parvum. J. Clin. Microbiol. 6013-19. [DOI] [PubMed] [Google Scholar]
- 24.Manhart, L. E., K. K. Holmes, and J. P. Hughes. 2007. Mycoplasma genitalium among young adults in the United States: an emerging sexually transmitted infection. Am. J. Public Health 971118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masue, N., T. Deguchi, S. Yokoi, T. Yamada, K. Ohkusu, and T. Ezaki. 2007. System for simultaneous detection of 16 pathogens related to urethritis to diagnose mixed infection. Int. J. Urol. 1439-42. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Murray, P. R., E. J. Baron, J. Jorgensen, M. A. Pfaller, and M. L. Landry (eds.). 2007. Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 28.Ng, D. P. K., D. Koh, S. G. L. Choo, V. Ng, and Q. Fu. 2004. Effect of storage conditions on the extraction of PCR-quality genomic DNA from saliva. Clin. Chim. Acta 343191-194. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Noble, R. C. 1985. Colonisation of the urethra with Streptococcus pneumoniae: a case report. Genitourin. Med. 61345-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oriel, J. D. 1975. Infection with chlamydia group A in men with urethritis due to N. gonorrhoeae. J. Infect. Dis. 131376-382. [DOI] [PubMed] [Google Scholar]
- 32.Povlsen, K., E. Bjornelius, P. Lidbrink, and I. Lind. 2002. Relationship of Ureaplasma urealyticum biovar 2 to non-gonococcal urethritis. Eur. J. Clin. Microbiol. Infect. Dis. 2197-101. [DOI] [PubMed] [Google Scholar]
- 33.Quentin, R., J. M. Mellovet, and P. Y. Sizaret. 1989. Typing of urogenital, maternal and neonatal isolates of Haemophilus influenzae and Haemophilus parainfluenzae in correlation with clinical source of isolation and evidence for a genital specificity of H. influenzae biotype IV. J. Clin. Microbiol. 272286-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauter, C., M. Mueller, I. Diterich, S. Zeller, D. Hassler, T. Meergans, and T. Hartung. 2005. Critical evaluation of urine-based PCR assay for diagnosis of Lyme borreliosis. Clin. Diagn. Lab. Immun. 12910-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Sarantis, H. J. G., M. Brown, M. Petric, and R. Tellier. 2004. Comprehensive detection and serotyping of human adenovirus by PCR and sequencing. J. Clin. Microbiol. 423963-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scholes, D., A. Stergachis, F. E. Heidrich, H. Andrilla, K. K. Holmes, and W. E. Stamm. 1996. Prevention of pelvic inflammatory disease by screening for cervical chlamydial infection. N. Engl. J. Med. 3341362-1366. [DOI] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Reference deleted.
- 40.Stellrecht, K. A., A. M. Woron, N. G. Mishrik, and R. A. Venezia. 2004. Comparison of multiplex PCR assay with culture for detection of genital mycoplasmas. J. Clin. Microbiol. 421528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Sturm, S. A. W. 1986. Haemophilus influenzae and Haemophilus parainfluenzae in nongonococcal urethritis. J. Infect. Dis. 153165-167. [DOI] [PubMed] [Google Scholar]
- 43.Taylor-Robinson, D. 2007. The role of mycoplasmas in pregnancy outcomes. Best Pract. Res. Clin. Obstet. Gynaecol. 21425-438. [DOI] [PubMed] [Google Scholar]
- 44.Taylor-Robinson, D., and W. M. McCormack. 1980. The genital mycoplasmas (the first of two parts). N. Engl. J. Med. 3021003-1010. [DOI] [PubMed] [Google Scholar]
- 45.Wang, H., F. Kong, P. Jelfs, G. James, and G. L. Gilbert. 2004. Simultaneous detection and identification of common cell culture contaminant and pathogenic mollicutes strains by reverse line blot hybridization. Appl. Environ. Microbiol. 701483-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Y., F. Kong, G. L. Gilbert, M. Brown, W. Gao, S. Yu, and Y. Yang. 2008. Use of a multiplex PCR-based reverse line blot (mPCR/RLB) hybridisation assay for the rapid identification of bacterial pathogens. Clin. Microbiol. Infect. 14155-160. [DOI] [PubMed] [Google Scholar]
- 47.Reference deleted.
- 48.Xiong, L., F. Kong, H. Zhou, and G. L. Gilbert. 2006. Use of PCR and reverse line blot hybridization assay for simultaneous detection and serovar identification of Chlamydia trachomatis. J. Clin. Microbiol. 441413-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto, L. J., D. G. Tedder, R. Ashley, and M. J. Levin. 1991. Herpes simplex virus DNA in cerebrospinal fluid of a patient with mollaret's meningitis. N. Engl. J. Med. 3251082-1085. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida, T., T. Deguchi, S.-T. Meda, Y. Kubota, M. Tamaki, S. Yokoi, M. Yasuda, and H. Ishiko. 2007. Quantitative detection of Ureaplasma parvum (biovar 1) and Ureaplasma urealyticum (biovar 2) in urine specimens from men with and without urethritis by real-time polymerase chain reaction. Sex. Trans. Dis. 34416-419. [DOI] [PubMed] [Google Scholar]
- 51.Zariffard, M. R., M. Saifuddin, B. E. Sha, and G. T. Spear. 2002. Detection of bacterial vaginosis-related organisms by real-time PCR for Lactobacilli, Gardnerella vaginalis and Mycoplasma hominis. FEMS Immun. Med. Microbiol. 34277-281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.