Abstract
Periodontitis is one of the most common chronic inflammatory diseases. A number of putative bacterial pathogens have been associated with the disease and are used as diagnostic markers. In the present study, we compared the prevalence of oral bacterial species in the subgingival biofilm of generalized aggressive periodontitis (GAP) (n = 44) and chronic periodontitis (CP) (n = 46) patients with that of a periodontitis-resistant control group (PR) (n = 21). The control group consisted of subjects at least 65 years of age with only minimal or no periodontitis and no history of periodontal treatment. A total of 555 samples from 111 subjects were included in this study. The samples were analyzed by PCR of 16S rRNA gene fragments and subsequent dot blot hybridization using oligonucleotide probes specific for Aggregatibacter (Actinobacillus) actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythia, a Treponema denticola-like phylogroup (Treponema phylogroup II), Treponema lecithinolyticum, Campylobacter rectus, Fusobacterium spp., and Fusobacterium nucleatum, as well as Capnocytophaga ochracea. Our data confirm a high prevalence of the putative periodontal pathogens P. gingivalis, P. intermedia, and T. forsythia in the periodontitis groups. However, these species were also frequently detected in the PR group. For most of the species tested, the prevalence was more associated with increased probing depth than with the subject group. T. lecithinolyticum was the only periodontopathogenic species showing significant differences both between GAP and CP patients and between GAP patients and PR subjects. C. ochracea was associated with the PR subjects, regardless of the probing depth. These results indicate that T. lecithinolyticum may be a diagnostic marker for GAP and C. ochracea for periodontal health. They also suggest that current presumptions of the association of specific bacteria with periodontal health and disease require further evaluation.
Periodontitis is a chronic inflammatory disease of infectious origin leading to destruction of tooth-supporting tissues and is the major cause of tooth loss in adults. However, all patients are not equally susceptible to periodontitis. In addition to the microbial challenge, other factors, such as genetics, environment, and host factors, play a role in the pathogenesis of this disease (24). The most common form is chronic periodontitis, which is characterized by a slow, gradual loss of periodontal attachment (3). In contrast to this form, rapid destruction of periodontal attachment is evident in aggressive periodontitis. Generalized aggressive periodontitis usually affects young adults under the age of 30, with attachment loss occurring in pronounced episodes of tissue destruction (2).
Many studies have been performed to evaluate the composition of the subgingival biofilm and identify key periodontal pathogens by both cultivation and molecular methods. More than 700 different species have been identified in the oral cavity, many of which are yet to be cultivated (1, 25). Of these species, only a small number are suspected periodontal pathogens, such as Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. This group of bacteria, characterized as the “red complex,” was highly associated with periodontal tissue destruction (28). Other bacteria, such as Prevotella intermedia, Campylobacter rectus, Fusobacterium species, Peptostreptococcus micros, and various spirochetes, have been implicated in the development of periodontitis (33). Aggressive periodontitis has been postulated to be frequently associated with Aggregatibacter actinomycetemcomitans and P. gingivalis (2). Localized aggressive periodontitis in particular seems to be characterized by specific infection with A. actinomycetemcomitans (27, 34), whereas chronic periodontitis is rather a mixed bacterial infection, not associated with any specific microorganism.
In the present study, we reevaluated the association of putative periodontal pathogens in patients with generalized aggressive or chronic periodontitis versus species in a periodontitis-resistant control group to identify species which are incompatible with periodontal health. As a control group, we chose a population of older adults with only minimal or no periodontitis and no history of periodontal treatment, considering these subjects resistant to periodontitis. We analyzed the presence of A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia, a T. denticola-like phylogroup (Treponema phylogroup II), Treponema lecithinolyticum, C. rectus, Fusobacterium spp., and Fusobacterium nucleatum, as well as Capnocytophaga ochracea. We also examined whether the presence or absence of these species was related to pocket depth.
MATERIALS AND METHODS
Clinical samples.
Patients diagnosed with chronic (CP) or generalized aggressive periodontitis (GAP) according to the criteria of the 1999 International Workshop for Classification of Periodontal Disease and Conditions (3) were included in this study. Patients with GAP (n = 44) and with CP (n = 46) had been referred for periodontal treatment to the Departments of Periodontology at the University Hospital Charité Berlin and the University of Würzburg, respectively. A periodontitis-resistant (PR) group of 21 subjects 65 years of age or older with minimal or no periodontitis recruited from a private practice in Berlin served as the control group. Clinical criteria for patient selection are presented in Table 1. All subjects were previously untreated. Exclusion criteria for all subjects were chronic systemic disease or anti-inflammatory or antimicrobial therapy within the last 6 months; pregnant or lactating women were also excluded. Demographics of all subjects are presented in Table 2.
TABLE 1.
Clinical criteria for patient selection
| Group | Criterion(a) for inclusion |
|---|---|
| PR control | Age of ≥65 yr |
| ≥20 natural teeth | |
| Probing depth at any site of ≤5 mm | |
| Clinical attachment loss at any site of ≤2 mm | |
| CP | ≥4-mm probing depth at ≥30% of residual teeth |
| GAP | Disease onset estimated at <30 yr based on clinical examination, past radiographs, and/or interview |
| ≥6-mm probing pocket depth at >3 permanent teeth other than first molars and incisors |
TABLE 2.
Patient demographics
| Characteristic | Value for groupa
|
||
|---|---|---|---|
| GAP | CP | PR | |
| Age (yr) ± SD | 34.4 ± 6.5 | 55.2 ± 11.2 | 66.6 ± 1.5 |
| Gender [no. (%)] | |||
| Male | 19 (43.2) | 21 (45.7) | 8 (38.1) |
| Female | 25 (56.8) | 25 (54.3) | 13 (61.9) |
| Smoker [no. (%)] | |||
| Current | 17 (38.6) | 15 (32.6) | NDb |
| Former | 4 (9.1) | 4 (8.7) | ND |
| Never | 23 (52.3) | 27 (58.7) | ND |
| Patient samples | |||
| Mean PDc (mm) ± SD | 7.5 ± 2.9 | 5.2 ± 2.4 | 3.7 ± 0.9 |
For GAP, n = 44; for CP, n = 46; for PR, n = 21.
ND, not determined.
PD, probing depth.
A total of 555 samples from 111 subjects were included in this study after informed consent was obtained. For each subject, subgingival plaque samples were taken from the four deepest periodontal pockets and if present from an additional healthy control site with a probing depth of ≤3 mm by insertion of three sterile paper points (ISO 35; Becht, Offenburg, Germany) after removal of supragingival plaque. In the GAP group all sample sites with the deepest periodontal pockets showed a probing depth of ≥6 mm and in the PR group a probing depth of <6 mm. The paper points were removed after 10 s, placed in 1 ml of reduced transport fluid (30) containing 25% glucose, transferred to the laboratory, and processed immediately.
DNA extraction and amplification.
PCR amplification of the subgingival plaque samples was performed as described earlier (20). Briefly, aliquots (100 μl) of each specimen were centrifuged at 13,000 × g for 10 min in a Labofuge 400 R instrument (Heraeus, Germany). The pellets were resuspended in 100 μl lysis buffer (5). No further purification of nucleic acids was performed, and 1 μl of the bulk DNA was used for in vitro amplification by PCR (final reaction volume, 100 μl) in a thermal cycler (Trioblock, Biometra, Germany), using 30 cycles of denaturation (1 min, 95°C), annealing (1 min, 56°C), and extension (1 min, 72°C). The broad-range bacterial primers TPU1 (5′-AGA GTT TGA TCM TGG CTC AG-3′) (corresponding to positions 8 to 27 in the Escherichia coli 16S rRNA gene) (4) and RTU3 (5′-GWA TTA CCG CGG CKG CTG-3′) (corresponding to complementary positions 519 to 536 in E. coli 16S rRNA) (4) were used for 16S rRNA gene amplification. Successful amplification was verified by agarose gel electrophoresis.
Oligonucleotide probes.
Species-specific, genus-specific, or phylotype-specific oligonucleotide probes (16 to 29 bases) as 16S rRNA/DNA-directed probes were used to detect Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythia, Treponema group II (TRE II) (Treponema denticola-like), Treponema lecithinolyticum, Campylobacter rectus, Capnocytophaga ochracea, Fusobacterium spp., and F. nucleatum.
Previously published oligonucleotides were reevaluated. In order to assess the specificity, the target sequences were compared to 16S rRNA entries of prokaryotes in the EMBL and GenBank databases, accessible (as of July 2002) by using the software program BLASTN of the Husar program package (version 4.0; Heidelberg Unix Sequence Analysis Resources; DKFZ, Heidelberg, Germany). All probes were checked with the program OLIGO 4.0 for their use in a hybridization assay. Oligonucleotide probe sequences and references are shown in Table 3. More details on the oligonucleotide probes are available at probeBase (16).
TABLE 3.
Oligonucleotide probes used for dot blot hybridization
| Target species/genus | Probe name as deposited in probeBase | Probe sequence (5′-3′) | Reference |
|---|---|---|---|
| A. actinomycetemcomitans | ACAC | TCC ATA AGA CAG ATT C | Sunde et al., 2003 (29) |
| P. gingivalis | POGI | CAA TAC TCG TAT CGC CCG TTA TTC | Sunde et al., 2003 (29) |
| P. intermedia | PRIN | CTT TAC TCC CCA ACA AAA GCA GTT TAC AA | Sunde et al., 2003 (29) |
| T. forsythia | B(T)AFO | CGT ATC TCA TTT TAT TCC CCT GTA | Sunde et al., 2003 (29) |
| T. denticola-like (TRE II) | TRE II | GCT CCT TTC CTC ATT TAC CTT TAT | Moter et al., 1998 (20) |
| T. lecithinolyticum | TLEC | CAC TCT CAG AAA GGA GCA AGC TCC | Moter et al., 2006 (21) |
| C. rectus | CARE | TTA ACT TAT GTA AAG AAG | This study |
| C. ochracea | CAOC | TCG GGC TAT CCC CCA GTG AAA GGC AGA T | This study |
| Fusobacterium spp.a | FUSO | CTA ATG GGA CGC AAA GCT CTC | Sunde et al., 2003 (29) |
| F. nucleatum F. periodonticum | FUNU | ATG TTG TCC CTA V(GCA)CT GTG AGG C | This study |
The sequence of the genus Fusobacterium-specific probe matches those of F. nucleatum, F. necrophorum, F. mortiferum, F. simiae, F. gonidiaformans, F. alocis, F. varium, F. russii, F. ulcerans, F. periodonticum, F. perfoetens, F. equinum, F. naviforme, and F. canifelinum.
Bacterial strains.
Bacterial strains were used as positive and negative controls (Fig. 1). The identities of target bacteria and closely related species were verified by 16S rRNA gene sequencing or biochemical tests using the rapid ID32A system (bioMérieux, Marcy-l'Etoile, France). PCR analysis of the reference bacteria was performed with cell pellets collected from culture as described for the subgingival plaque samples.
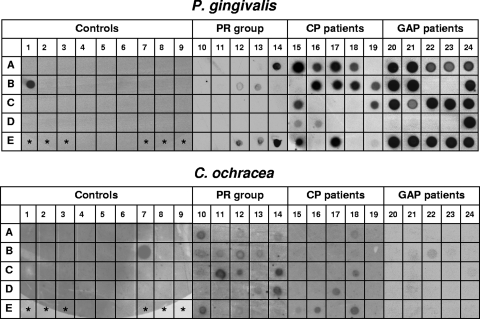
FIG. 1.
Dot blot hybridizations of identical membranes with probes for P. gingivalis and C. ochracea. In columns 1 to 9, PCR products of the following strains were applied as controls: Actinobacillus actinomycetemcomitans ATCC 43718 (A1); Actinobacillus actinomycetemcomitans ATCC 33384 (A2); Actinobacillus actinomycetemcomitans serotype a (A3); Leptotrichia buccalis MCCM 00448 (A4); Pasteurella haemolytica ATCC 33396 (A5); Haemophilus influenzae ATCC 33391 (A6); Haemophilus influenzae clinical isolate (A7); Haemophilus aphrophilus NCTC 55906 (A8); Haemophilus paraphrophilus ATCC 29241 (A9); Porphyromonas gingivalis ATCC 33277 (B1); Prevotella intermedia ATCC 25611 (B2); Porphyromonas assacharolyticus ATCC 25260 (B3); Prevotella nigrescens NCTC 9336 (B4); Prevotella oralis MCCM 00684 (B5); Prevotella buccalis ATCC 33690 (B6); Capnocytophaga ochracea ATCC 27872 (B7); Capnocytophaga sputigena ATCC 33612 (B8); Capnocytophaga gingivalis ATCC 33624 (B9); Campylobacter rectus ATCC 33238 (C1); Campylobacter concisus ATCC 33237 (C2); Bacteroides gracilis ATCC 33236 (C3); Bacteroides fragilis ATCC 25285 (C4); Eikenella corrodens CCUG 2138 (C5); Kingella kingae ATCC 23330 (C6); Veillonella parvula ATCC 10790 (C7); Veillonella dispar ATCC 17748 (C8); Klebsiella pneumoniae ATCC 23357 (C9); Fusobacterium nucleatum ATCC 25586 (D1); Flavobacterium odoratum MCCM 02932 (D2); Neisseria lactamica ATCC 23970 (D3); Streptococcus mutans ATCC 35668 (D4); Streptococcus intermedius ATCC 27335 (D5); Actinomyces viscosus ATCC 15987 (D6); Actinomyces israelii ATCC 10048 (D7); Eubacterium lentum ATCC 25559 (D8); Selenomonas sp. clinical strain (D9); Fusobacterium simiae CCUG 16798 (E4); Fusobacterium periodonticum CCUG 14345 (E5); and Fusobacterium necrophorum NCTC 25286 (E6). Asterisks indicate empty fields without PCR product. In columns 10 to 14, 15 to 19, and 20 to 24, PCR products from subgingival plaque samples of PR subjects, CP patients, and GAP patients (five patients each) were applied, respectively. Each column represents four deep pockets plus one control site from a single patient. For an accurate analysis, the samples from each patient were spotted on the membrane in a random order. Samples were considered positive if the dot was clearly visible above the background level of the negative controls.
Dot blot hybridization.
Amplified DNA was spotted onto nylon membranes, and dot blot hybridization was carried out as described earlier (20). Briefly, an aliquot (1 μl) of heat-denatured PCR product was applied on nylon membranes (Hybond N; Amersham, Buckinghamshire, Great Britain) and fixed by UV cross-linking.
PCR products of 42 amplified DNA samples of oral and extraoral bacterial strains served as controls to ensure stringent hybridization conditions in all dot blot hybridizations. These included the respective species as positive controls and the phylogenetically closest relatives at the probe binding sites. Furthermore, if the probes TRE II or TLEC were used, a control membrane with amplified DNA samples from either recombinant clones retrieved from the original oral treponema 16S rRNA gene library (5) or known cultivable treponemes was included as a control in all dot blot hybridizations as published previously (21).
Probes were labeled with digoxigenin-ddUTP (Boehringer, Mannheim, Germany) and detected by chemiluminescence according to the manufacturer's recommendations. All hybridizations were performed at 54°C. Stringency washes were optimized for each probe by varying the washing temperature (40°C to 64°C) and the washing buffer (containing 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-0.2% sodium dodecyl sulfate [SDS] or 0.1× SSC-0.1% SDS). X-ray films were exposed to the membranes for 2 to 12 h. After stripping with 0.2 N NaOH with 0.1% SDS (stripping buffer), identical membranes were used for multiple hybridization experiments with the probes mentioned above.
Statistical analysis.
Statistical evaluation was performed for descriptive and for inferential purposes. The exact chi-square tests with Bonferroni's correction for multiple comparisons were applied to compare the prevalences of the target bacteria between the three groups. A subject was regarded as positive for a certain genospecies if the organism was detected in at least one sample. The Bonferroni-corrected Mann-Whitney U tests, as well as tests for a difference in proportions using the central limit theorem, were used to examine differences in the numbers of sites per patient positive for the target species. Analysis of variance with Bonferroni's correction for multiple comparisons in the post hoc tests was performed for determination of differences between the groups with respect to target bacteria at different probing depths. The mean percentages of positive sites were derived by averaging the positive sites of each species within a subject and then across subjects in the clinical groups. The presence of bacteria at various probing depths was evaluated by logistic regression analysis adjusted for clustering on the subjects. For all statistical tests, P values less than 0.05 were considered significant.
RESULTS
PCR amplification was successful for all subgingival plaque specimens. In dot blot hybridization, none of the negative controls showed a signal. This indicated that no carryover of amplified material occurred (data not shown). Using optimized hybridization and washing conditions for each oligonucleotide probe, only PCR products of the appropriate target species were detected by the specific probes on the control membranes. No cross-reactions were observed.
Prevalences of target bacteria in subject groups.
All target species were detected in subgingival plaque samples from both periodontitis groups and the PR group. Obvious differences were observed between the patient groups regarding signal intensities of the dot blot hybridizations, suggesting a higher number of periodontal pathogens in the GAP patients (Fig. 1). However, since PCR-amplified DNA was used for hybridization experiments, the signal intensity was not quantitatively analyzed.
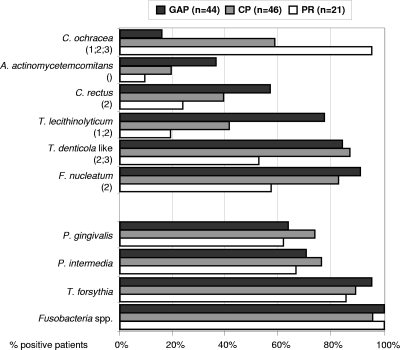
The prevalences of the target species in the different groups are shown in Fig. 2. The number of individuals positive for a given species or phylotype varied considerably between the groups. Except for C. ochracea, the prevalence of most genospecies was highest in the GAP group, followed by the CP and PR groups (Fig. 2). Significant differences between the groups were found for T. lecithinolyticum (P = 0.001, GAP versus CP; P < 0.001, GAP versus PR), TRE II (T. denticola-like) (P = 0.007, GAP versus PR; P = 0.002, CP versus PR), F. nucleatum (P = 0.001, GAP versus PR), and C. rectus (P = 0.013, GAP versus PR). A. actinomycetemcomitans showed an overall significant difference (P = 0.038) but no significant differences between the groups in the post hoc test.
FIG. 2.
Prevalence of target species in the GAP patients, CP patients, and PR subjects as determined by dot blot hybridizations using oligonucleotide probes. A patient was regarded as positive if at least one sample was positive. Numbers in parentheses indicate statistical significances between the groups: 1, GAP versus CP; 2, GAP versus PR; and 3, CP versus PR, as determined by chi-square analysis with Bonferroni's correction for multiple comparisons. Empty parentheses (), as for A. actinomycetemcomitans, indicate an overall significance but no significant differences between the groups in the post hoc test results.
C. ochracea was detected significantly more often in PR subjects than in the two patient groups and significantly more often in CP patients than in GAP patients (P < 0.001, GAP versus CP and PR; P = 0.002, CP versus PR).
Number of positive sites per patient.
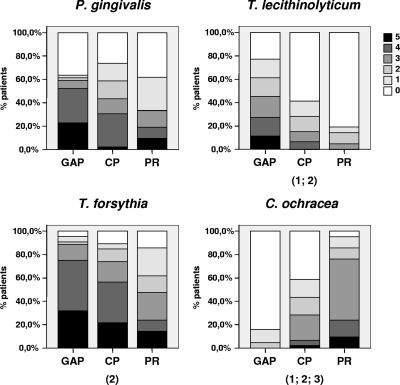
To semiquantitatively assess the presence of the different species, we compared the number of positive sites per patient. Most putative periodontal pathogens were detected in more sites in both periodontitis groups, with numbers being highest in the GAP group (Fig. 3). The differences between the groups were statistically significant for T. lecithinolyticum (P < 0.001, GAP versus CP and PR), TRE II (T. denticola-like) (P < 0.001, GAP versus PR and CP versus PR; data not shown), F. nucleatum (P < 0.001, GAP versus PR; P = 0.006, CP versus PR; data not shown), T. forsythia (P = 0.001, GAP versus PR), Fusobacteria spp. (P = 0.010, GAP versus PR; data not shown), and C. rectus (P = 0.009, GAP versus PR; data not shown). C. ochracea showed significantly more positive sites in the PR group than in the two periodontitis groups (P < 0.001, GAP versus CP and PR and CP versus PR) (Fig. 3).
FIG. 3.
Percentage of patients with 1, 2, 3, 4, or 5 of the sites colonized by target bacteria as revealed by dot blot analysis. Numbers in parentheses indicate statistical significances between the groups: 1, GAP versus CP; 2, GAP versus PR; and 3, CP versus PR, as determined with the Mann-Whitney U test. P values were adjusted for multiple comparisons (Bonferroni's correction).
Presence of target bacteria depending on probing depths.
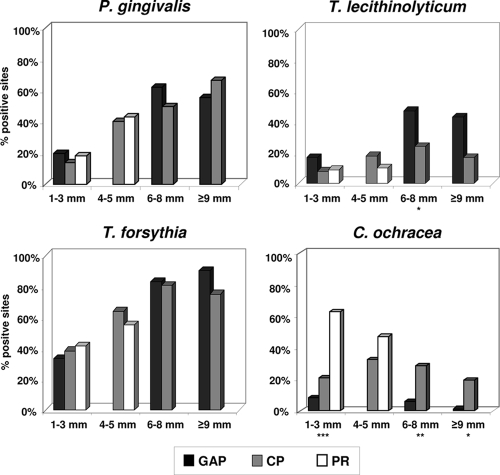
The question arises of whether the greater prevalence of various bacteria in the periodontitis groups is due merely to the deeper pockets found in advanced periodontitis, resulting in more bacterial mass in the paper point sample. To evaluate the relationship between the existence of certain periodontal species and disease severity, logistic regression analysis adjusted for clustering on the subjects, as well as comparison of mean positive samples from pockets of different probing depths, was performed. The probability of occurrence of C. ochracea and A. actinomycetemcomitans showed no significant correlation with the probing depth. For all other putative periodontal pathogens, there was a positive correlation which was statistically significant (P ≤ 0.001).
According to the inclusion criteria, the four deepest pockets had probing depths of ≥6 mm in the GAP group and <6 mm in the PR group. Comparison of mean positive samples was performed for all clinical groups only at probing depths of ≤3 mm. At probing depths of 4 to 5 mm, comparison was done only for the CP and PR groups, and at probing depths of 6 to 8 mm and ≥9 mm, only for the two periodontitis groups. At sites of 4 to 5 mm, the putative periodontal pathogens TRE II (T. denticola-like) and F. nucleatum were found significantly more often in CP patients than in PR subjects (P < 0.001 and P = 0.027, respectively; data not shown). At sites of 6 to 8 mm, T. lecithinolyticum, A. actinomycetemcomitans (data not shown), and C. rectus (data not shown) were detected significantly more often for GAP patients than for CP patients (P = 0.013, 0.005, and 0.044, respectively) (Fig. 4). In contrast, C. ochracea was detected significantly more often in PR subjects than in the two patient groups at sites of ≤3 mm (P < 0.001) (Fig. 4). At sites of 6 to 8 mm and ≥9 mm, the differences were statistically significant between CP and GAP patients (P = 0.003 and 0.011, respectively) (Fig. 4).
FIG. 4.
Percentage of positive samples at different probing depths. According to the inclusion criteria, the four deepest pockets showed a probing depth of ≥6 mm in the GAP group and <6 mm in the PR group. Therefore, only the CP patient group and the PR group were compared at probing depths of 4 to 5 mm, and only the two periodontitis groups were compared at probing depths of 6 to 8 mm and ≥9 mm. Asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001) indicate significant differences as determined by analysis of variance. The mean percentages of positive sites were derived by averaging the positive sites of each species within a subject and then across subjects in the clinical groups.
DISCUSSION
It is widely accepted that the etiology of periodontitis is polymicrobial in nature (23). Worsening or improvement of periodontal status is accompanied by a shift in the bacterial composition of subgingival plaque (14). It has therefore been suggested that microbial testing can be used for diagnosis and to optimize periodontal therapy and assess its outcome, especially when treatment with antimicrobial drugs is considered. However, this strategy may be confounded, since initiation and progression of periodontal disease are influenced by the interaction of myriad genetic, environmental, host, and microbial factors (22, 24, 26, 32). Further, molecular studies reveal an unexpectedly high diversity of microorganisms whose relevance for initiation and progression of disease still remains to be investigated. Nevertheless, current microbiological testing mainly involves the classical suspected oral pathogens.
In the present study, we examined the prevalences of 10 periodontal bacterial species in 2 patient groups with advanced chronic or generalized aggressive periodontitis, as well as a periodontitis-resistant control group. We chose PCR of 16S rRNA gene fragments and subsequent dot blot hybridization as the detection method. Although quantitative analysis cannot be accurately performed with this approach, signal intensities in GAP samples appear much stronger than those in CP samples, even more so than those in PR samples (Fig. 1). In a semiquantitative approach, we compared the percentages of positive sites per patient. As expected, most putative periodontal pathogens were found more often in the periodontitis groups, especially the GAP group, than in the PR group (Fig. 3). The only significant differences in microbial prevalences between GAP and CP patients were seen for T. lecithinolyticum (Fig. 2).
Three widely accepted periodontal pathogens, T. forsythia, P. gingivalis, and P. intermedia, indeed showed no differences in prevalence between the three groups. This has also been shown by Kumar et al. (14) using quantitative 16S cloning and sequencing. These authors could not detect any significant association of T. forsythia and P. gingivalis with disease. However, these species represent only a small percentage of the total bacterial species, and the open-ended approach of their study may not be geared to detect an association of these species with disease. In another study, Kumar et al. demonstrated a significant association of both T. forsythia and P. gingivalis, as well as T. denticola, with chronic periodontitis, using PCR amplification of 16S rRNA genes (13). T. forsythia, P. gingivalis, and T. denticola are members of the so-called “red complex” based on checkerboard DNA-DNA hybridization of 13,000 plaque samples from 185 subjects (28). These three species are considered to be highly associated with advanced periodontitis. In the present study, the high prevalences of these species were confirmed in the periodontitis groups, but the PR group also showed very high prevalence rates (95%, 89%, and 86% for T. forsythia, 64%, 74%, and 62% for P. gingivalis, and 70%, 76%, and 67% for P. intermedia in GAP, CP, and PR groups, respectively, Fig. 2). The high prevalences of P. gingivalis and T. forsythia found here are similar to those in other studies examining aggressive periodontitis patients (12). Griffen and coworkers (8) found a comparable prevalence of P. gingivalis in periodontitis subjects using nested PCR analysis of samples from all teeth. However, the prevalence of P. gingivalis was significantly lower in age-matched healthy subjects. Although the mean age was higher, other inclusion criteria for the healthy control group were similar to those used in the present study. The authors concluded that P. gingivalis is highly associated with periodontitis, in accordance with results in other studies.
Most previous studies of the association of specific bacteria with periodontitis did not examine the influence of probing depth. We questioned whether the detection of a bacterial species may be related more to the depth of the sampled pocket than to a certain diagnosis. For most species in our study, we found that the probing depth had a much greater impact on the occurrence of the species than did the diagnosis. The prevalence of P. gingivalis was highly associated (P < 0.001) with pocket depth, as revealed by logistic regression analysis adjusted for clustering on the subjects.
Treponemes have previously been associated with periodontitis, and T. denticola especially has been suggested as a diagnostic marker (28). In the present investigation, treponemes were more prevalent and were found in more sites in the periodontitis groups than in the control group. This was the case for both for T. lecithinolyticum and TRE II species, which include T. denticola. However, the difference was greatest for T. lecithinolyticum, as we have previously shown (21). Only T. lecithinolyticum showed significant differences between the two periodontitis groups. The differences between CP patients and the PR control group were not statistically significant, which again were highly significant for TRE II. This confirms the results of Kumar et al. (13), who compared the microbiota of chronic periodontitis patients with age-matched controls and detected T. denticola and to a lesser extent T. lecithinolyticum for significantly more diseased patients than controls.
A. actinomycetemcomitans has been closely associated with aggressive periodontitis, especially localized forms (previously known as localized juvenile periodontitis) (7, 34). It has been shown that the prevalence of A. actinomycetemcomitans in severe or refractory periodontitis seems to be inversely related to age. (26). However, Umeda et al. (32) found a positive correlation between age and the prevalence of A. actinomycetemcomitans in the subgingival space or saliva, using 16S rRNA PCR analysis. In the present study, the prevalence of A. actinomycetemcomitans is highest in the GAP patients, who are also the youngest, and lowest in the PR subjects, who are the oldest. The differences between the groups were statistically significant (Fig. 2). Logistic regression analysis showed that A. actinomycetemcomitans was the only “classical” putative periodontal pathogen tested whose presence did not correlate with probing depth. This is contrary to findings of Mombelli et al. (19) that the presence of A. actinomycetemcomitans is significantly associated with probing depth. However, these workers used culturing techniques that may not have detected low levels of the organism in shallow pockets. The detection frequency of A. actinomycetemcomitans was rather low for all subjects (36%, 20%, and 10% in GAP, CP, and PR subjects, respectively), which is in accordance with results in other studies analyzing samples from aggressive periodontitis patients (11, 12). Using DNA probe analysis of the four deepest pockets of patients, Haffajee and Socransky (9) found a 38% false-negative detection rate for species with low prevalences, such as A. actinomycetemcomitans. This finding questions the use of A. actinomycetemcomitans as an appropriate diagnostic marker.
In contrast to the suspected periodontal pathogens noted above, C. ochracea was significantly more prevalent and showed significantly more positive sites per patient in the control group than in the periodontitis groups. It was also significantly more prevalent in the CP than in the GAP patient group, independent of probing depth. This is consistent with results in other studies, which found that high levels of C. ochracea were related to a lower risk of disease progression (6, 10), suggesting that C. ochracea can be considered beneficial to the host. The detection of such beneficial species may be as important as detection of periodontal pathogens, and it has been suggested that recolonization of periodontal pockets with beneficial bacteria following scaling and root planing may be a useful clinical strategy (31). This approach has been studied in the gastrointestinal tract by use of probiotics or microbial replacement therapy (17) and deserves further investigation in the treatment of periodontitis.
Microbial testing has been advocated both for periodontal diagnosis and to discriminate between chronic and aggressive forms of periodontitis in order to better tailor the treatment approach. The results of this study indicate that testing for bacteria which are now presently targeted may not be sufficient. Most commercial tests include such putative pathogens as A. actinomycetemcomitans, P. gingivalis, T. forsythia, T. denticola, Fusobacterium spp., and P. intermedia, which were included in this study. None of these organisms showed significant differences in prevalence between the two periodontitis groups. Their presence correlated with probing depth rather than diagnosis. The association of certain putative pathogens with severe periodontitis shown in many studies may be explained in part by deeper pockets in these patient groups. This view is consistent with the conclusion of Mombelli et al. (18) that the presence or absence of A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythia, or C. rectus cannot discriminate between subjects with aggressive or chronic periodontitis. Listgarten and Loomer (15) also questioned whether microbial identification should be considered as a strategy in the management of patients with periodontitis. They concluded that there was no strong evidence supporting the benefit of microbial testing, partly because of a lack of standardization among diagnostic laboratories.
In summary, the results of the present study indicate that currently recognized periodontal pathogens, such as P. gingivalis, P. intermedia, and T. forsythia, can be frequently detected in PR subjects and in both GAP and CP patients. However, the bacterial load seems to be lower in PR subjects, suggesting that these species are opportunistic pathogens. TRE II species (including T. denticola) are highly associated with GAP and CP patients compared to PR subjects, whereas T. lecithinolyticum was the only species showing significant differences both between GAP and CP patients and between GAP patients and PR subjects. Colonization by C. ochracea appears beneficial to the host, since it is associated with a stable periodontal condition, as evidenced by a high prevalence and frequency, independent of pocket depth, in subjects resistant to periodontitis. Therefore, we suggest T. lecithinolyticum be considered a diagnostic marker for GAP and C. ochracea an indicator of periodontal health.
Acknowledgments
We thank Cindy Hefenbrock and Jork Schneiderheinze for excellent technical assistance. We further thank Clarence L. Trummel, University of Connecticut Health Center, Farmington, CT, for his helpful comments and for reviewing the manuscript.
The species were obtained from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) and CCUG (Culture Collection University of Göteborg) and were kindly provided by R. Mutters (Marburg, Germany), G. Conrads (Aachen, Germany), C. Wyss (Zürich, Switzerland), A. Mombelli (Bern, Switzerland), and E. Esdorn (Charité, Berlin, Germany). The DNA of Tannerella forsythia was kindly provided by I. Olson, Institute of Oral Biology, Oslo University.
This study was supported by the Deutsche Forschungsgemeinschaft (to B.R., L.E.-N., and J.-P.B.; GRK 325), a grant (01KI9318) from the Bundesministerium für Bildung und Forschung (to U.B.G.), a Körber European Research Award (to U.B.G.), and a Rahel-Hirsch grant (to A.M.).
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 435721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Academy of Periodontology. 2000. Parameter on aggressive periodontitis. J. Periodontol. 71867-869. [DOI] [PubMed] [Google Scholar]
- 3.Armitage, G. C. 1999. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 41-6. [DOI] [PubMed] [Google Scholar]
- 4.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 754801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Göbel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 621889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dzink, J. L., S. S. Socransky, and A. D. Haffajee. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15316-323. [DOI] [PubMed] [Google Scholar]
- 7.Fine, D. H., K. Markowitz, D. Furgang, K. Fairlie, J. Ferrandiz, C. Nasri, M. McKiernan, and J. Gunsolley. 2007. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 453859-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffen, A. L., M. R. Becker, S. R. Lyons, M. L. Moeschberger, and E. J. Leys. 1998. Prevalence of Porphyromonas gingivalis and periodontal health status. J. Clin. Microbiol. 363239-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haffajee, A. D., and S. S. Socransky. 1992. Effect of sampling strategy on the false-negative rate for detection of selected subgingival species. Oral Microbiol. Immunol. 757-59. [DOI] [PubMed] [Google Scholar]
- 10.Haffajee, A. D., S. S. Socransky, C. Smith, and S. Dibart. 1991. Relation of baseline microbial parameters to future periodontal attachment loss. J. Clin. Periodontol. 18744-750. [DOI] [PubMed] [Google Scholar]
- 11.Kamma, J. J., A. Contreras, and J. Slots. 2001. Herpes viruses and periodontopathic bacteria in early-onset periodontitis. J. Clin. Periodontol. 28879-885. [DOI] [PubMed] [Google Scholar]
- 12.Kamma, J. J., M. Nakou, R. Gmur, and P. C. Baehni. 2004. Microbiological profile of early onset/aggressive periodontitis patients. Oral Microbiol. Immunol. 19314-321. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82338-344. [DOI] [PubMed] [Google Scholar]
- 14.Kumar, P. S., E. J. Leys, J. M. Bryk, F. J. Martinez, M. L. Moeschberger, and A. L. Griffen. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 443665-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Listgarten, M. A., and P. M. Loomer. 2003. Microbial identification in the management of periodontal diseases. A systematic review. Ann. Periodontol. 8182-192. [DOI] [PubMed] [Google Scholar]
- 16.Loy, A., F. Maixner, M. Wagner, and M. Horn. 2007. probeBase—an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 35D800-D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madden, J. A., S. F. Plummer, J. Tang, I. Garaiova, N. T. Plummer, M. Herbison, J. O. Hunter, T. Shimada, L. Cheng, and T. Shirakawa. 2005. Effect of probiotics on preventing disruption of the intestinal microflora following antibiotic therapy: a double-blind, placebo-controlled pilot study. Int. Immunopharmacol. 51091-1097. [DOI] [PubMed] [Google Scholar]
- 18.Mombelli, A., F. Casagni, and P. N. Madianos. 2002. Can presence or absence of periodontal pathogens distinguish between subjects with chronic and aggressive periodontitis? A systematic review. J. Clin. Periodontol. 29(Suppl. 3)10-21; discussion, 37-38. [DOI] [PubMed] [Google Scholar]
- 19.Mombelli, A., R. Gmur, C. Gobbi, and N. P. Lang. 1994. Actinobacillus actinomycetemcomitans in adult periodontitis. I. Topographic distribution before and after treatment. J. Periodontol. 65820-826. [DOI] [PubMed] [Google Scholar]
- 20.Moter, A., C. Hoenig, B. K. Choi, B. Riep, and U. B. Göbel. 1998. Molecular epidemiology of oral treponemes associated with periodontal disease. J. Clin. Microbiol. 361399-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moter, A., B. Riep, V. Haban, K. Heuner, G. Siebert, M. Berning, C. Wyss, B. Ehmke, T. F. Flemmig, and U. B. Göbel. 2006. Molecular epidemiology of oral treponemes in patients with periodontitis and in periodontitis-resistant subjects. J. Clin. Microbiol. 443078-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nibali, L., D. R. Ready, M. Parkar, P. M. Brett, M. Wilson, M. S. Tonetti, and G. S. Griffiths. 2007. Gene polymorphisms and the prevalence of key periodontal pathogens. J. Dent. Res. 86416-420. [DOI] [PubMed] [Google Scholar]
- 23.Nishihara, T., and T. Koseki. 2004. Microbial etiology of periodontitis. Periodontol. 2000 3614-26. [DOI] [PubMed] [Google Scholar]
- 24.Page, R. C., and K. S. Kornman. 1997. The pathogenesis of human periodontitis: an introduction. Periodontol. 2000 149-11. [DOI] [PubMed] [Google Scholar]
- 25.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 1833770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodenburg, J. P., A. J. van Winkelhoff, E. G. Winkel, R. J. Goene, F. Abbas, and J. de Graff. 1990. Occurrence of Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in severe periodontitis in relation to age and treatment history. J. Clin. Periodontol. 17392-399. [DOI] [PubMed] [Google Scholar]
- 27.Slots, J., H. S. Reynolds, and R. J. Genco. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological Investig. Infect. Immun. 291013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25134-144. [DOI] [PubMed] [Google Scholar]
- 29.Sunde, P. T., I. Olsen, U. B. Göbel, D. Theegarten, S. Winter, G. J. Debelian, L. Tronstad, and A. Moter. 2003. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology 1491095-1102. [DOI] [PubMed] [Google Scholar]
- 30.Syed, S. A., and W. J. Loesche. 1972. Survival of human dental plaque flora in various transport media. Appl. Microbiol. 24638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teughels, W., M. G. Newman, W. Coucke, A. D. Haffajee, H. C. Van Der Mei, S. K. Haake, E. Schepers, J. J. Cassiman, J. Van Eldere, D. van Steenberghe, and M. Quirynen. 2007. Guiding periodontal pocket recolonization: a proof of concept. J. Dent. Res. 861078-1082. [DOI] [PubMed] [Google Scholar]
- 32.Umeda, M., C. Chen, I. Bakker, A. Contreras, J. L. Morrison, and J. Slots. 1998. Risk indicators for harboring periodontal pathogens. J. Periodontol. 691111-1118. [DOI] [PubMed] [Google Scholar]
- 33.van Winkelhoff, A. J., B. G. Loos, W. A. van der Reijden, and U. van der Velden. 2002. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J. Clin. Periodontol. 291023-1028. [DOI] [PubMed] [Google Scholar]
- 34.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 121-20. [DOI] [PubMed] [Google Scholar]