Abstract
MicroRNA (miRNA)-binding site polymorphisms that could contribute to disease risk and prognosis are rapidly being identified and investigated as this genetic variation may have a potentially profound impact on human health. A recently described variant allele in the KRAS 3′ untranslated region that arises in the let-7 miRNA complementary site (KRAS-LCS6) and leads to increased KRAS expression in lung cancer was examined for its association with the occurrence of head and neck squamous cell carcinoma (HNSCC). We examined the prevalence of the KRAS-LCS6 variant allele in a population-based case–control study of HNSCC to determine if this KRAS-LCS6 genotype was associated with disease occurrence and patient survival. Although the KRAS-LCS6 variant genotype was not associated with the overall risk of HNSCC, cases with the KRAS-LCS6 variant genotype had significantly reduced survival [hazard ratio (HR), 1.6; 95% confidence interval (CI), 1.0–2.5] in models controlled for confounders of survival. This risk was greatest in cases of oral cavity carcinoma (HR, 2.7; 95% CI, 1.4–5.3). These data demonstrate that cases with the KRAS-LCS6 variant have significantly reduced survival time and suggest that this variant may alter the phenotype or therapeutic response of this disease.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the eighth most common cancer in men in the USA, with ∼45 000 new cases diagnosed in both sexes each year, resulting in over 11 000 deaths (1). There are three major etiologic factors that contribute to HNSCC risk and prognosis: tobacco use, alcohol consumption and human papillomavirus (HPV) infection (2,3). In addition, genetic variation has been shown to modify the risk of disease (4,5) and in certain instances has been associated with patient survival (4,6). The majority of the current literature has employed a biologically based candidate gene approach to investigate the association of normal genetic variation with HNSCC. Emerging studies are focusing both on employing genome-wide platforms and investigating phenotypically well-characterized genetic variation. The latter strategy now includes microRNA (miRNA)-related single nucleotide polymorphisms (SNPs) which can occur in miRNA genes themselves, or in their target sequences (7,8). MiRNAs are short non-coding RNAs which prevent translation of their target genes by binding the highly evolutionarily conserved 3′ untranslated regions (UTRs) of mRNAs (9). The highly critical role miRNAs play in gene regulation is widely accepted, and altered expression of miRNAs in human cancers has been well documented (10). Recently, critical examinations of miRNA expression profiles in HNSCC have underlined the importance of miRNA expression alterations in HNSCC tumorigenesis (11,12).
The biological relevance of miRNA-binding site SNPs in influencing the risk of human diseases such as cancer has recently been reviewed by Chen et al. (7). An example of a miRNA-binding site SNP that has been shown to alter disease risk is a let-7 complementary site SNP in the 3′ UTR of KRAS (13). The let-7 family of miRNAs has been shown to regulate KRAS (14). A member of the family of RAS oncogenes that are well-characterized GTPases, KRAS is activated by somatic mutation in many human cancer types (15). Activation of the KRAS proto-oncogene via mutation in adenocarcinomas of the lung, colon and pancreas is well documented (16). In contrast, similar mutations of KRAS in squamous cell carcinomas are relatively rare (15,16) although the growth-promoting character of KRAS activation (associated with companion modes of activation of this and related pathways) is almost certainly a feature of most solid tumors and, in fact, amplification of KRAS has been reported to promote growth of HNSCC (17). A recent study of non-small cell lung cancer (NSCLC) indicated that individuals with <40 pack-years smoked and any variant allele in the KRAS let-7 complementary site SNP (KRAS-LCS6) had a significantly increased risk of disease (13). Further, these authors characterized the function of the KRAS-LCS6 SNP, determining that the variant allele was associated with both increased KRAS expression and decreased let-7 levels (13). Work in another lab has shown that low levels of let-7a-2 correlate with poor survival in lung adenocarcinoma (18). We hypothesized that activation of the KRAS pathway might be accomplished in squamous cell carcinomas via action of this normal variant and thereby associated with susceptibility to this disease and with patient outcome.
Here, we have examined this hypothesis in a population-based case–control study of HNSCC. Further, we examined the potential association of the KRAS-LCS6 SNP with disease outcome, defined as overall patient survival.
Materials and methods
Study population
The study population has been described previously (5,19). Briefly, incident cases of HNSCC were identified from nine medical facilities in the Boston metropolitan area, with the histological classification of malignancy ascertained from the pathology report of the participating hospitals and confirmed by an independent study pathologist. Population-based controls were drawn from the same greater Boston population and matched to cases by gender, age (±3 years) and town of residence using the Massachusetts town lists. All cases and controls enrolled in the study provided written informed consent as approved by the institutional review boards of the participating institutions. Survival time was determined for cases using publicly available databases.
DNA isolation and genotyping
DNA was extracted from whole blood or buccal cells with the QIAamp DNA mini kit according to the manufacturer's protocol (Qiagen, Valencia, CA). Genotyping of the KRAS let-7 microRNA binding site SNP was done using Taqman® allelic discrimination (Applied Biosystems, Foster City, CA) with a custom-designed primer probe set—forward primer: GCCAGGCTGGTCTCGAA, reverse primer: CTGAATAAATGAGTTCTGCAAAACAGGTT, reporter sequence 1: CTCAAGTGATTCACCCAC-VIC and reporter sequence 2: CAAGTGATGCACCCAC-FAM. Genotyping was performed in a blinded fashion, appropriate controls were included in each run and ∼10% of samples were duplicated in a coded fashion as quality assurance with >95% concordance observed between replicates.
Statistical analysis
Data were analyzed using the SAS software, and all P-values represent two-sided statistical tests. Tests for Hardy–Weinberg equilibrium were conducted. Unconditional logistic regression was used to calculate adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for the association of KRAS let-7 microRNA binding site SNP genotype and HNSCC risk adjusting for age, sex, HPV16 status, alcohol consumption and tobacco smoking pack-years. For analyses by tumor location, cases were grouped according to International Classification of Diseases-9 code, with oral cancer encompassing International Classification of Diseases-9 141–145, pharyngeal cancers 146–149 and laryngeal cancers 161. As homozygous variant and heterozygous genotypes had similar ORs and in order to improve power for examination of the rare variant allele, these groups were combined with homozygous wild-type as the referent. Patient survival was examined using Cox proportional hazards modeling to control for variables related to patient survival. These survival probability models included variables representing the combined homozygous variant and heterozygous genotypes (any variant) and were controlled for patient age (in decades) and tumor stage (1 or 2 versus 3 or 4). Normalized signal data resulting from miRNA microarray were averaged within tumor sites (oral and pharyngeal) for each let-7 homolog. MiRNA expression data were subjected to a two-sided, unpaired, Student's t-test assuming equal means and unequal variance to determine differences in expression between tumor sites.
Results
Table I describes the characteristics of the study population. The mean age, distribution of gender and distribution of race between cases and controls were similar. An increased relative risk of HNSCC with increasing pack-years smoked was observed: subjects in the highest quartile had an OR of 3.6 (95% CI, 2.5–5.2), and this relationship has been described previously in our study population (4,5,19). Specifically, compared with oral and pharyngeal cancers, the magnitude of increased laryngeal cancer risk for heavy tobacco users was greatly elevated (OR, 21.2; 95% CI, 7.9–57.0). In addition, relative to subjects who drank two or fewer drinks per week on average, individuals in the heaviest drinking quartile (>15 drinks per week) had a significantly increased risk of HNSCC overall (OR, 2.5; 95% CI, 1.7–3.7), though this association did not hold for risk of laryngeal cancer. Further, as previously reported (3), HPV16 seropositive individuals had a significantly increased risk of HNSCC (OR, 4.2; 95% CI, 2.9–6.0), with risk of pharyngeal tumors being the greatest (OR, 8.1; 95% CI, 5.0–13.2).
Table I.
Participant characteristics of HNSCC cases and controls genotyped for KRAS-LCS6
| Characteristic | All study participants |
Oral cancer (n = 283) |
Pharyngeal cancer (n = 132) |
Laryngeal cancer (n = 98) |
||
| Cases (n = 513), n (%) | Controls (n = 597), n (%) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | |
| Age (years), mean (SD) | 59.7 (11.5) | 61.0 (11.4) | ||||
| Gender | ||||||
| Female | 135 (26.3) | 165 (27.6) | ||||
| Male | 378 (73.7) | 432 (72.4) | ||||
| Race | ||||||
| White | 468 (91.2) | 544 (91.1) | ||||
| Non-white | 45 (8.8) | 53 (8.9) | ||||
| Tobacco (lifetime pack-years)a | ||||||
| ≤1 | 104 (20.2) | 224 (37.5) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) |
| >1 to ≤8 | 45 (8.8) | 75 (12.6) | 1.3 (0.8–2.0) | 0.8 (0.5–1.3) | 0.8 (0.3–1.9) | 3.6 (1.1–12.5) |
| >8 to ≤34 | 120 (23.4) | 155 (26.0) | 1.5 (1.1–2.2) | 1.1 (0.7–1.9) | 1.1 (0.6–2.1) | 6.7 (2.4–18.7) |
| >34 | 244 (47.6) | 143 (24.9) | 3.6 (2.5–5.2) | 2.9 (1.8–4.7) | 3.6 (2.0–6.5) | 21.2 (7.9–57.0) |
| Alcohol use (lifetime average drinks/week)a | ||||||
| ≤2 | 82 (16.0) | 146 (24.5) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) |
| >2 to ≤6 | 80 (15.6) | 169 (28.3) | 0.8 (0.5–1.2) | 0.8 (0.5–1.3) | 0.8 (0.4–1.7) | 0.8 (0.4–1.7) |
| >6 to ≤15 | 90 (17.5) | 137 (22.9) | 1.1 (0.7–1.6) | 1.1 (0.7–1.9) | 1.2 (0.6–2.5) | 0.8 (0.3–1.7) |
| >15 | 261 (50.9) | 145 (24.3) | 2.5 (1.7–3.7) | 2.9 (1.8–4.7) | 2.2 (1.1–4.3) | 1.6 (0.8–3.2) |
| HPV16 seropositivityb | ||||||
| No | 370 (72.1) | 482 (89) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) |
| Yes | 143 (27.9) | 59 (11) | 4.2 (2.9–6.0) | 3.0 (2.0–4.5) | 8.1 (5.0–13.2) | 2.2 (1.2–4.2) |
| KRAS-LCS6 genotype | ||||||
| Wt/Wt | 413 (80.5) | 500 (83.7) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) | 1.0 (reference) |
| Wt/Var + Var/Var | 100 (19.5) | 97 (16.3) | 1.3 (0.9–1.8) | 1.2 (0.8–1.8) | 1.5 (0.9–2.5) | 1.2 (0.6–2.1) |
Models are controlled for all variables in tables. SD, standard deviation.
Tobacco pack-years and average drinks per week based on quartile distribution in controls.
Seurm HPV16 antibody status was available for 1024 subjects, remaining subjects were coded as missing and included in the model.
We examined the association of the variant genotype of a KRAS 3′ UTR let-7 miRNA-binding site SNP (KRAS-LCS6) with case status. Genotype frequencies were within Hardy–Weinberg equilibrium. In models controlling for potential confounders, we did not observe an association between presence of the KRAS-LCS6 variant allele and risk of HNSCC or an association with specific sites of this disease (Table I). In addition, the estimate of risk of HNSCC among KRAS-LCS6 variant allele carriers was not modified after stratifying on smoking pack-years, alcohol consumption or HPV16 seropositivity (data not shown).
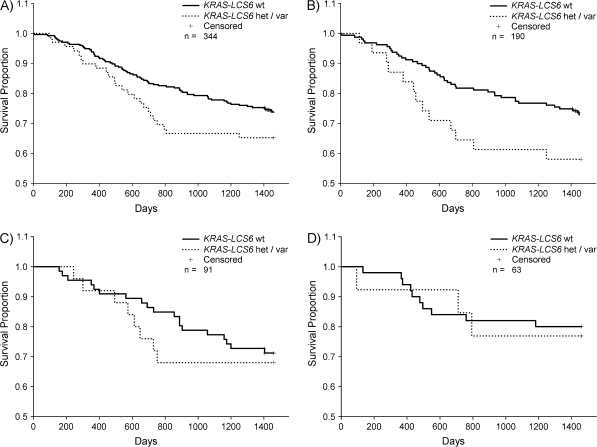
Next, we examined the association of the KRAS-LCS6 variant genotype with disease survival. Figure 1 shows the Kaplan–Meier survival probability plots stratified by KRAS-LCS6 genotype for all cases and for specific tumor locations. Using a multivariate Cox proportional hazards model to control for potential confounders of the association of the variant allele with disease survival, revealed that cases carrying any KRAS-LCS6 variant allele had a significantly increased hazard ratio (HR, 1.6; 95% CI, 1.0–2.5) (Table II). To determine if this effect varied by tumor location, we stratified the analyses by tumor site. Cases of oral cancer with any variant allele at KRAS-LCS6 had significantly reduced survival (HR, 2.7; 95% CI, 1.4–5.3) (Table II). Pharyngeal tumors (HR, 1.4; 95% CI, 0.6–3.4) and laryngeal tumors (HR, 3.1; 95% CI, 0.7–13.4) did not demonstrate any significant survival association based on KRAS-LCS6 genotype.
Fig. 1.
Kaplan–Meier survival probability plots stratified by KRAS-LCS6 genotype for head and neck tumors with available staging data. Survival time is defined as time from diagnosis to death or last known follow-up where crosses represent censored values. The log-rank method was used to test for a difference between strata. Solid lines represent cases with wild-type KRAS-LCS6 genotype; dotted lines represent cases carrying any KRAS-LCS6 variant alleles. (A) All tumor sites, n = 344, P = 0.20. (B) Oral cancers, n = 190, P = 0.06. (C) Pharyngeal cancers, n = 91, P = 0.65. (D) Laryngeal cancers, n = 63, P = 0.83.
Table II.
Cases of HNSCC carrying the KRAS-LCS6 variant allele have significantly reduced survival
| Covariate | All HNSCC cases |
Oral cancer cases |
||||
| Total n = 344, n (%) | HR (95% CI) | P-value | Total n = 190, n (%) | HR (95% CI) | P-value | |
| Age | ||||||
| ≤50 | 77 (22.4) | 1.0 (reference) | 50 (26.3) | 1.0 (reference) | ||
| >50 to ≤60 | 111 (32.3) | 1.1 (0.6–2.2) | 0.73 | 62 (32.6) | 1.4 (0.7–3.0) | 0.38 |
| >60 to ≤70 | 106 (30.8) | 1.8 (0.9–3.4) | 0.09 | 48 (25.3) | 1.3 (0.6–2.9) | 0.51 |
| >70 | 50 (14.5) | 3.5 (1.8–6.9) | <0.001 | 30 (15.8) | 3.3 (1.4–7.4) | <0.005 |
| Gender | ||||||
| Female | 85 (24.7) | 1.0 (reference) | 54 (28.4) | 1.0 (reference) | ||
| Male | 259 (75.3) | 1.5 (0.9–2.5) | 0.16 | 136 (71.6) | 1.6 (0.8–3.4) | 0.20 |
| Race | ||||||
| White | 312 (90.7) | 1.0 (reference) | 177 (93.2) | 1.0 (reference) | ||
| Non-white | 32 (9.3) | 1.2 (0.6–2.1) | 0.63 | 13 (6.8) | 0.8 (0.3–2.1) | 0.62 |
| Tobaccoa | ||||||
| ≤1 | 68 (19.8) | 1.0 (reference) | 44 (23.2) | 1.0 (reference) | ||
| >1 to ≤8 | 29 (8.4) | 0.2 (0.1–0.9) | <0.04 | 20 (10.5) | 0.3 (0.1–1.2) | 0.08 |
| >8 to ≤34 | 86 (25.0) | 1.1 (0.6–2.1) | 0.73 | 48 (25.3) | 0.9 (0.4–2.0) | 0.78 |
| >34 | 161 (46.8) | 1.5 (0.8–2.7) | 0.20 | 78 (41.0) | 1.4 (0.6–3.1) | 0.39 |
| Alcohol usea | ||||||
| ≤2 | 55 (16.0) | 1.0 (reference) | 29 (15.3) | 1.0 (reference) | ||
| >2 to ≤6 | 55 (16.0) | 0.8 (0.4–1.7) | 0.49 | 32 (16.8) | 0.7 (0.2–2.0) | 0.46 |
| >6 to ≤15 | 57 (16.6) | 0.6 (0.3–1.3) | 0.23 | 33 (17.4) | 0.9 (0.3–2.4) | 0.77 |
| >15 | 177 (51.4) | 0.8 (0.5–1.5) | 0.54 | 96 (50.5) | 1.1 (0.4–2.9) | 0.84 |
| HPV16 seropositivity | ||||||
| No | 241 (70.1) | 1.0 (reference) | 143 (75.3) | 1.0 (reference) | ||
| Yes | 103 (29.9) | 0.6 (0.4–1.0) | <0.05 | 47 (24.7) | 0.7 (0.4–1.3) | 0.26 |
| Stage | ||||||
| 1 or 2 | 97 (28.2) | 1.0 (reference) | 55 (29.0) | 1.0 (reference) | ||
| 3 or 4 | 247 (71.8) | 2.3 (1.4–3.8) | <0.002 | 135 (71.0) | 3.4 (1.7–6.9) | <0.001 |
| KRAS-LCS6 genotype | ||||||
| Wt/Wt | 275 (80.0) | 1.0 (reference) | 159 (83.7) | 1.0 (reference) | ||
| Wt/Var + Var/Var | 69 (20.0) | 1.6 (1.0–2.5) | <0.04 | 31 (16.3) | 2.5 (1.3–4.9) | <0.01 |
Models are controlled for all variables in tables.
Tobacco pack-years and average drinks per week based on quartile distribution in controls.
While increased KRAS expression is a known functional consequence of variant KRAS-LCS6 genotype, decreased levels of let-7 expression have also been associated with presence of the variant allele (13). Hence, to provide confirmation that the differences that we observed in survival were associated with differences in the tumor phenotype; we first investigated whether differential expression of let-7 was associated with KRAS-LCS6 genotype. MiRNA microarray data were available for a subset of tumors (array data Gene Expression Omnibus accession #GSE11163; tumors with genotype data n = 14). We first investigated the expression of let-7 levels using data detailed in Avissar et al. (20). Consistent with the literature, compared with tumors from KRAS-LCS6 wild-type individuals, tumors from variant allele carriers had significantly reduced expression of three let-7 miRNAs (Table III). Next, we examined these tumors for site-specific let-7 miRNA expression levels and found that four let-7 miRNAs had significantly reduced expression in oral tumors compared with pharyngeal tumors (Table IV). Since three oral tumors (no pharyngeal tumors) were from patients carrying a variant allele, we next restricted to oral tumors and found no significant difference in let-7 expression levels by KRAS-LCS6 genotype (data not shown).
Table III.
Tumors from patients harboring a variant allele at KRAS-LCS6 have lower let-7 family miRNAs expression levels compared with tumors from wild-type individuals
| miRNA | Mean (SD) by KRAS-LCS6 genotype |
P-value* | |
| Wild-type (n = 11) | Any variant allele (n = 3) | ||
| let_7a | 14.1 (0.48) | 13.5 (0.28) | 0.05 |
| let_7b | 13.6 (0.42) | 13.0 (0.43) | 0.10 |
| let_7c | 13.6 (0.44) | 12.9 (0.28) | 0.02 |
| let_7d | 12.2 (0.43) | 11.7 (0.23) | 0.02 |
| let_7e | 9.3 (0.73) | 8.8 (0.54) | 0.26 |
| let_7f | 11.1 (0.50) | 10.7 (0.38) | 0.16 |
| let_7g | 10.5 (0.50) | 9.8 (0.65) | 0.21 |
| let_7i | 11.3 (0.04) | 10.3 (0.50) | 0.07 |
SD, standard deviation.
Two-sided unpaired t-test.
Table IV.
Let-7 family miRNA expression is lower in oral tumors compared with pharyngeal tumors
| miRNA | Mean (SD) by tumor site |
P-value* | |
| Oral (n = 10) | Pharyngeal (n = 4) | ||
| let_7a | 13.7 (0.19) | 14.7 (0.65) | 0.02 |
| let_7b | 13.3 (0.29) | 13.7 (0.73) | 0.18 |
| let_7c | 13.1 (0.18) | 13.8 (0.67) | 0.02 |
| let_7d | 11.8 (0.12) | 12.4 (0.52) | 0.005 |
| let_7e | 9.0 (0.76) | 9.7 (0.55) | 0.15 |
| let_7f | 10.2 (0.18) | 11.4 (0.56) | 0.003 |
| let_7g | 10.1 (0.34) | 10.6 (0.63) | 0.08 |
| let_7i | 10.9 (0.45) | 11.4 (0.51) | 0.10 |
SD, standard deviation.
Two-sided unpaired t-test.
Finally, since the KRAS-LCS6 variant genotype is known to result in increased KRAS levels (13), we examined existing mRNA expression array data on 16 tumors with available patient blood and genotyping data (B.C. Christensen, C.M. Marsit, K.T. Kelsey, unpublished data). Tumors from patients carrying the KRAS-LCS6 variant allele (n = 5) had increased KRAS mRNA relative to wild-type individuals (n = 11) and this observation approached statistical significance (P = 0.10).
Discussion
The importance of miRNA and miRNA-binding site polymorphisms (mirSNPs) in disease risk and prognosis are rapidly being defined and investigated in human cancers (7,8). We investigated the prevalence of a recently characterized variant allele in the 3′ UTR of KRAS which inhibits downregulation by its cognate miRNA let-7 in a population-based case–control study of HNSCC. Previous work has demonstrated that KRAS is regulated by the let-7 family of miRNAs (14), and characterized the associations between the KRAS-LCS6 variant allele and increased KRAS expression, as well as reduced let-7 expression (13). Importantly, we found that presence of any KRAS-LCS6 variant allele was significantly associated with poor prognosis in HNSCC, and that prognosis was worst among cases of oral cancer.
Aberrant activation of KRAS via somatic mutation is a well-recognized cancer-related alteration identified in many types of human tumors (21). More specifically, KRAS mutation in NSCLC is significantly more prevalent in adenocarcinomas than in squamous tumors (22), and a paucity of KRAS mutations have been observed in other squamous tumors such as HNSCC. In one review, 19% of NSCLCs had KRAS mutation compared with only 3% of HNSCCs (16). An increase in KRAS expression associated with the KRAS-LCS6 variant allele could therefore be a surrogate for KRAS activation via somatic mutation. Evidence for the hypothesis that the KRAS-LCS6 variant allele may substitute for KRAS mutation comes from the observed increase in the risk of NSCLC in patients carrying the KRAS-LCS6 variant allele who are low to moderate smokers (13). However, we did not find any association between the KRAS-LCS6 variant allele and risk of HNSCC. Further, risk was not modified when stratifying on exposure to tobacco, alcohol or HPV16 seropositivity. Although our finding suggests that KRAS-LCS6 variant allele-associated KRAS activation does not confer an increased risk of HNSCC, cases of HNSCC carrying the KRAS-LCS6 variant allele had significantly reduced survival compared with wild-type individuals. We also found independent contributions of HPV16 seropositive status and tumor stage to survival, and although tobacco and alcohol increase risk of HNSCC, we did not observe independent survival effects for either of these exposures.
In the context of no increased risk of HNSCC for individuals with the variant allele, our observation of significantly reduced survival among HNSCC cases carrying the KRAS-LCS6 variant allele suggests that the variant phenotype may be associated with tumor progression rather than initiation. Studies of KRAS expression in HNSCC have indicated that amplified KRAS promotes the growth of HNSCC (17), and presence of K-ras protein in HNSCC tumors has been associated with late-stage tumors and increased tumor size (23). We observed that both increased KRAS expression and reduced levels of let-7 expression were associated with presence of the KRAS-LCS6 variant allele, and these findings are consistent with those of Chin et al. (13). In addition, we found reduced let-7 miRNA levels in oral tumors relative to pharyngeal tumors. Although this is in part due to three of the oral tumors being variant allele carriers, restricting the analysis to oral tumors did not reveal an effect for KRAS-LCS6 genotype on let-7 expression, suggesting that variant allele status alone does not fully account for the observed differences in let-7 expression levels by tumor site. Instead, tumors from different sites may have differential let-7 miRNA expression due to normal cell of origin differences in miRNA expression profiles.
Not surprisingly, different cell types have been shown to have different miRNA expression profiles (24). Therefore, the lack of disease risk for KRAS-LCS6 variant allele carriers may be explained by a better tolerance of normal head and neck epithelium to variant allele-associated phenotypic alterations. However, in the context of an evolving tumor, the KRAS-LCS6 variant allele phenotype (increased KRAS and reduced let-7 expression) may differentially contribute to tumor progression in oral cavity tumors relative to tumors from other head and neck sites. As normal oral and pharyngeal epithelium are different cell types with specific transcriptomes, alterations in let-7 family miRNA expression will impact expression of miRNA target transcripts in a cell-type-specific way. Hence, differential dysregulation of miRNA targets could explain the more aggressive phenotype associated with reduced survival in oral cancer relative to head and neck tumors from other sites. Further support for this hypothesis comes from Tokumaru et al. (25) who demonstrate that let-7 downregulates dicer, a critical component of the miRNA processing machinery. In fact, reduced let-7 levels have been shown to result in both increased dicer expression and altered miRNA expression patterns (25). Slight alterations in dicer expression could result in widespread dysregulation of miRNA activity. Since we observed no difference in the KRAS-LCS6 variant allele frequency between tumor sites, alterations in KRAS and let-7 expression may simply have a more profound effect in cancers of oral epithelium due to cell-type-specific patterns of miRNA expression and their regulatory target transcripts. Indeed, individual miRNAs are thought to potentially regulate dozens, if not hundreds of mRNAs (26). However, since different cell types have different mRNA expression profiles, altered expression of a single miRNA could have profoundly different impacts due to cell-type-specific differences in the constitutively expressed target transcripts.
Our study suggests that patients with HNSCC carrying the KRAS-LCS6 variant allele have significantly reduced survival compared with wild-type individuals. Notably, response to treatment is unlikely to affect KRAS-LCS6-related reduced survival as these patients undergo standardized tumor stage-specific treatment modalities, and tumor stage is controlled for in our survival analysis. These findings were tumor site specific with cases of oral cancer having the worst prognosis, whereas cases of laryngeal and pharyngeal carcinoma with the variant allele did not have an altered survival pattern. Further studies are necessary to confirm these findings and to determine the mechanism through which the KRAS-LCS6 variant genotype alters the phenotype of these tumors. In addition, our findings suggest that patients with tumors of the oral cavity carrying the KRAS-LCS6 variant allele may benefit from the development of small RNA-based therapeutics.
Funding
NCI (R01CA078609 and R01CA100679); NIEHS (T32ES007272).
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papillomavirus
- HR
hazard ratio
- miRNA
microRNA
- NSCLC
non-small cell lung cancer
- OR
odds ratio
- SNP
single nucleotide polymorphism
- UTR
untranslated region
References
- 1.Jemal A, et al. Cancer statistics, 2007. CA Cancer J. Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Blot WJ, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48:3282–3287. [PubMed] [Google Scholar]
- 3.Furniss CS, et al. Human papillomavirus 16 and head and neck squamous cell carcinoma. Int. J. Cancer. 2007;120:2386–2392. doi: 10.1002/ijc.22633. [DOI] [PubMed] [Google Scholar]
- 4.Marsit CJ, et al. A genotype-phenotype examination of cyclin D1 on risk and outcome of squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2008;14:2371–2377. doi: 10.1158/1078-0432.CCR-07-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters ES, et al. Glutathione S-transferase polymorphisms and the synergy of alcohol and tobacco in oral, pharyngeal, and laryngeal carcinoma. Cancer Epidemiol. Biomarkers Prev. 2006;15:2196–2202. doi: 10.1158/1055-9965.EPI-06-0503. [DOI] [PubMed] [Google Scholar]
- 6.Hopkins J, et al. Genetic polymorphisms and head and neck cancer outcomes: a review. Cancer Epidemiol. Biomarkers Prev. 2008;17:490–499. doi: 10.1158/1055-9965.EPI-07-2714. [DOI] [PubMed] [Google Scholar]
- 7.Chen K, et al. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 8.Saunders MA, et al. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl Acad. Sci. USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, et al. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Chang SS, et al. MicroRNA alterations in head and neck squamous cell carcinoma. Int. J. Cancer. 2008;123:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramdas L, et al. miRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissue. Head Neck. 2009;31:642–654. doi: 10.1002/hed.21017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin LJ, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Rodenhuis S, et al. Clinical significance of ras oncogene activation in human lung cancer. Cancer Res. 1992;52:2665s–2669s. [PubMed] [Google Scholar]
- 16.Lea IA, et al. Genetic pathways and mutation profiles of human cancers: site- and exposure-specific patterns. Carcinogenesis. 2007;28:1851–8. doi: 10.1093/carcin/bgm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoa M, et al. Amplification of wild-type K-ras promotes growth of head and neck squamous cell carcinoma. Cancer Res. 2002;62:7154–7156. [PubMed] [Google Scholar]
- 18.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Hsiung DT, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 20.Avissar M, et al. MicroRNA expression ratios are preditive of head and neck squamous cell carcinoma. Clin. Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbacid M. ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 22.Keohavong P, et al. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin. Cancer Res. 1996;2:411–418. [PubMed] [Google Scholar]
- 23.McDonald JS, et al. Immunohistochemical detection of the H-ras, K-ras, and N-ras oncogenes in squamous cell carcinoma of the head and neck. J. Oral Pathol. Med. 1994;23:342–346. doi: 10.1111/j.1600-0714.1994.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 24.Mattick JS, et al. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005 doi: 10.1093/hmg/ddi101. 14 Spec No 1, R121–R32. [DOI] [PubMed] [Google Scholar]
- 25.Tokumaru S, et al. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 26.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]