Abstract
Loss of NF-E2-related factor 2 (Nrf2) signaling increases susceptibility to acute toxicity, inflammation and carcinogenesis in mice due to the inability to mount adaptive responses. In contrast, disruption of Keap1 (a cytoplasmic modifier of Nrf2 turnover) protects against these stresses in mice, although inactivating mutations in Keap1 have been identified recently in some human cancers. Global characterization of Nrf2 activation is important to exploit this pathway for chemoprevention in healthy, yet at-risk individuals and also to elucidate the consequences of hijacking the pathway in Keap1-mutant human cancers. Liver-targeted conditional Keap1-null, Albumin-Cre:Keap1(flox/−) (CKO) mice provide a model of genetic activation of Nrf2 signaling. By coupling global gene expression analysis of CKO mice with analysis of pharmacologic activation using the synthetic oleanane triterpenoid 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im), we are able to gain insight into pathways affected by Nrf2 activation. CDDO-Im is an extremely potent activator of Nrf2 signaling. CKO mice were used to identify genes modulated by genetic activation of Nrf2 signaling. The CKO response was compared with hepatic global gene expression changes in wild-type mice treated with CDDO-Im at a maximal Nrf2 activating dose. The results show that genetic and pharmacologic activation of Nrf2 signaling modulates pathways beyond detoxication and cytoprotection, with the largest cluster of genes associated with lipid metabolism. Genetic activation of Nrf2 results in much larger numbers of detoxication and lipid metabolism gene changes. Additionally, analysis of pharmacologic activation suggests that Nrf2 is the primary mediator of CDDO-Im activity, though other cell-signaling targets are also modulated following an oral dose of 30 μmol/kg.
Introduction
The NF-E2-related factor 2 (Nrf2)-signaling pathway plays a crucial role in mediating a cytoprotective response that protects against a wide variety of carcinogenic and toxic insults. The transcription factor Nrf2 is bound to Keap1 in the cytoplasm, resulting in Keap1-facilitated proteasomal degradation of Nrf2 (1). Chemical inducers (triterpenoids, dithiolethiones, isothiocyanates and phenolic antioxidants) or endogenous stimuli (electrophilic stress and oxidative stress) can activate Nrf2 signaling by modifying reactive cysteines in Keap1, causing dissociation of Nrf2 or impaired proteasomal degradation of Nrf2 (2). This results in translocation of Nrf2 to the nucleus, where it accumulates, binds and activates the antioxidant response element found in the regulatory domain of many cytoprotective genes. Activation of these cytoprotective genes results in a broad prosurvival adaptive response, which includes gene products involved in conjugation/detoxication, antioxidative response, proteasome function, molecular chaperones and inhibition of inflammation (3).
Studies using Nrf2-null mice have shown that loss of Nrf2 signaling increases susceptibility to acute toxicity, inflammation and carcinogenesis. Nrf2-null mice are more susceptible to acetaminophen-induced hepatotoxicity (4) and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced toxicity in the striatum of the brain (5). Inflammatory injury of the colon caused by dextran sulfate treatment is increased in Nrf2-deficient mice compared with wild-type (6). Nrf2-deficient mice are more susceptible in a variety of chemical-induced carcinogenesis models. For example, Nrf2-null mice develop a higher burden of gastric neoplasia following treatment with benzo[a]pyrene compared with wild-type mice (7). Furthermore, Nrf2 is essential for the chemopreventive actions of sulforaphane and oltipraz in this model and others. Oltipraz and sulforaphane significantly reduce the multiplicity of gastric neoplasia in wild-type mice, but these protective actions are lost in Nrf2-deficient mice (7,8). Sulforaphane also decreases skin tumor incidence in wild-type but not in Nrf2-deficient mice (9).
Synthetic oleanane triterpenoids have recently been shown to be extremely potent inducers of Nrf2 signaling in vivo (10). Although originally developed as anti-inflammatory agents, rodent studies have shown that this class of triterpenoids provides chemoprotection against several disease states through induction of Nrf2-regulated genes. For example, 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im) is an effective chemopreventive agent against aflatoxin-induced hepatic tumorigenesis, with pronounced protection at oral doses as low as 1 μmol/kg body wt and complete inhibition of tumorigenesis at 100 μmol/kg body wt. Protection is achieved through induction of Nrf2-regulated hepatic detoxification and cytoprotective genes, resulting in reduced aflatoxin–DNA adduct formation and inhibition of tumorigenesis (11). This class of triterpenoids has also been shown to reduce the number, size and severity of lung tumors induced by vinyl carbamate (12). Furthermore, protection is not limited to carcinogenesis; treatment with triterpenoids protects against cigarette smoke-induced emphysema and cardiac dysfunction through activation of Nrf2 signaling (13). The broad tissue distribution of these agents combined with potent activation of Nrf2-regulated cytoprotective responses make synthetic oleanane triterpenoids ideal agents for chemoprevention.
Nrf2 signaling can be constitutively activated by deletion of Keap1. Furthermore, tissue-specific disruption of Keap1 in mice has been shown to protect against toxic insults such as methylmercury (14) and acetaminophen (15), as well as acute inflammatory liver injury (16). In contrast, inactivating mutations in Keap1 have been identified recently in some human cancers, including non-small cell lung cancer (17) and breast cancer (18). Activation of Nrf2 signaling in tumor cells may provide a survival advantage and promote resistance to chemotherapy. Global gene expression changes induced by alteration of Nrf2 signaling have not yet been fully characterized. Global characterization of Nrf2 activation is important to exploit this pathway for chemoprevention in healthy, yet at-risk individuals and also to elucidate the consequences of hijacking the pathway in Keap1-mutant human cancers. Our study addresses this issue by coupling global gene expression analysis of genetic Nrf2 activation in liver-targeted conditional Keap1-null, Albumin-Cre:Keap1(flox/−) (CKO) mice with analysis of pharmacologic activation using the extremely potent Nrf2 inducer CDDO-Im (10,11,19,20). CKO mice are ideal models for genetic activation of Nrf2 signaling, as it is well established that modification of Keap1 function primarily results in changes in Nrf2 activity. For example, Keap1-null mice die just prior to weaning due to hyperkeratosis of the forestomach and esophagus related to Nrf2-regulated changes in several squamous epithelial genes; breeding Keap1-null mice with Nrf2-deficient mice reverses this phenotype (21). In addition, early studies of RNA transcripts increased in the CKO mouse (compared with wild-type) show that the vast majority of genes modulated were either known Nrf2 targets or contained an antioxidant response element in their regulatory region (15).
The comparisons used in our study provide a unique opportunity to gain insight into pathways modulated by differing modes of Nrf2 activation. In addition, we are able to broadly evaluate the pharmacodynamic action of CDDO-Im in order to identify the predominant molecular pathways contributing to the chemopreventive action of CDDO-Im. The results described in this study can also be used to consider how CDDO-Im or other Nrf2 inducers could best be used for cancer chemoprevention.
Materials and methods
Chemicals
CDDO-Im was synthesized as described previously (22).
Animals and treatment
All experiments were approved by the Johns Hopkins University Animal Care and Use Committee. Mice were fed AIN-76A-purified diet without ethoxyquin. Male 9-week-old mice were used for all experiments. Liver-targeted conditional Keap1-null mice (CKO, Alb-Cre:Keap1flox/−) were generated by crossing Alb-Cre:Keap1+/− mice with Keap1flox/− mice. Alb-Cre:Keap1+/− mice, Keap1flox/− mice and Alb-Cre:Keap1flox/+ (genetic control wild-type, Albumin-Cre:Keap1(flox/+) [WT]) mice were generated on a C57BL/6J background (15). While this targeting strategy results in complete disruption of Keap1 in hepatocytes, these animals exhibit hypomorphic expression of Keap1 in other cell types (our unpublished results). Genotypes were assigned using polymerase chain reaction (PCR) analysis of tail genomic DNA and later confirmed using hepatic genomic DNA. Nrf2-disrupted mice were generated from inbred Nrf2-heterozygous mice on a C57BL/6J background (23). For gene expression analyses, mice were gavaged with a single dose of 30 μmol CDDO-Im/kg body wt or vehicle consisting of 10% Cremophor EL (Sigma–Aldrich, St Louis, MO), 10% dimethyl sulfoxide and 80% phosphate-buffered saline and sacrificed 6 h later. This dose of CDDO-Im is known to be a maximal Nrf2-activating dose (10) that also provides protection against tumorigenesis (11).
Nqo1 transcript measurement
Total RNA was isolated from individual snap frozen livers using Versagene RNA purification kit (Gentra Systems, Minneapolis, MN) and complementary DNA (cDNA) was synthesized using iSCRIPT cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Gene expression measurements were performed using TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA) and iQ Supermix (Bio-Rad). Gene expression measurements were normalized to the endogenous reference gene Gapdh, which was not modulated. Fold-change values for gene expression data from real-time quantitative PCR were determined using the 2−ΔΔCt relative quantification method as published (24).
Microarray preparation
Total RNA was isolated as above. RNA quality was evaluated using an Agilent Bioanalyzer 2100 (Applied Biosystems). Purified RNA was used for double-stranded cDNA synthesis. Double-stranded cDNA synthesis was performed using the Superscript Double-Stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA). Complementary RNA synthesis was then performed using the Bioarray High Yield RNA Transcript Labeling Kit (Enzo, Farmingdale, NY). Both cDNA and complementary RNA were purified using the Affymetrix GeneChip Sample Cleanup Module (Affymetrix, Santa Clara, CA). Complementary RNA was used for the fragmentation reaction. The entire fragmentation product was then hybridized to the Affymetrix Mouse Genome 430 2.0 array chip for 18 h. The chips were removed, washed and stained using an Affymetrix GeneChip Fluidics Station 400. The chips were then scanned using an Affymetrix GeneChip Scanner 3000 7G.
Microarray data analysis
The CEL files (raw data files) were background corrected using the ‘bg.adjust.gcrma’ function of ‘gcrma’ v.2.12.1 (25) and then base 2 logarithm transformed. The transformed data were quantile normalized using the ‘normalize.AffyBatch.quantiles’ function of affy v.1.8.1 (26) as implemented in R v.2.7.0. Each probe set was modeled independently using a linear fixed effects model adapted from (27) to account for fixed treatment, genotype and probe affinity effects. The linear fixed effects model and filtering procedures are described in detail in the supplementary data (available at Carcinogenesis Online). Briefly, present and absent calls were used to filter for probe sets that had at least two present calls in any three replicates of any treatment or genotype groups. A 1.7-fold-change threshold was also used. A false discovery rate procedure (28) was applied to correct for multiple hypothesis testing. Each P-value was assigned a corresponding q-value to identify differentially expressed probe sets with high statistical significance at a 5% false discovery rate. Several genes were selected from the resulting gene lists for validation by real-time PCR using TaqMan Gene Expression Assays (Applied Biosystems). Microarray data discussed in this publication have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus and are accessible through Gene Expression Omnibus Series accession number GSE15633.
Biological questions and gene lists
Comparisons were designed to address several biological questions, as described below.
Which genes are modulated by genetic activation of Nrf2 signaling through deletion of Keap1?
Genetic activation: CKO versus WT.
Which genes are modulated by pharmacologic activation using CDDO-Im?
Pharmacologic activation: WT + CDDO-Im versus WT + vehicle.
Which genes are common to both genetic and pharmacologic activation of Nrf2 signaling?
Common: Modulated in both CKO versus WT and WT + CDDO-Im versus WT + vehicle. Gene expression changes are Keap1–Nrf2-dependent and CDDO-Im modulated.
Which genes are uniquely modulated by genetic activation in CKO mice?
Unique to genetic activation: Modulated only in CKO versus WT. Gene expression changes are Keap1–Nrf2-dependent, but not CDDO-Im modulated.
Which genes are uniquely modulated by pharmacologic activation using CDDO-Im?
Unique to pharmacologic activation: Modulated only in WT + CDDO-Im versus WT + vehicle. Gene expression changes are CDDO-Im modulated, but Keap1–Nrf2 independent.
Which genes can CDDO-Im modulate above and beyond the state of Nrf2 hyperactivation in the liver of CKO mice?
CKO + CDDO-Im versus CKO + vehicle.
The gene lists in this experiment were produced based on the linear model analysis. Depending on the biological questions, different statistical criteria were applied to generate the answers. This procedure is described in detail in supplementary data (available at Carcinogenesis Online).
Functional category analysis
In order to uncover the similar and unique biological processes and molecular functions among the genes, the Affymetrix probe identification numbers and their corresponding fold-change values were uploaded into Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood, CA, www.ingenuity.com). Effective use of Ingenuity Pathways Analysis requires a smaller input gene list (in our experience, <500 genes is optimal). For this reason, a more stringent q-value cutoff of 10−10 was used for this analysis.
Determination of CDDO-Im concentration in tissues
Liver samples were homogenized on ice in acetonitrile. The homogenate was vortexed and sonicated and then centrifuged at 20 000g for 10 min. The supernatant was diluted 1:1 with 20 mM ammonium acetate pH 7.4. Diluted buffered acetonitrile extracts were recentrifuged at 20 000g for 5 min, and supernatants transferred to Waters 0.8 ml Total Recovery Vials.
Sample vials were placed in a Waters 2695 HPLC and held at 5°C prior to loading on a Waters XTerra MS C18 5 μm column protected by a Waters 2.1 × 10 mm Guard Column. Sample (25–100 μl) was loaded, with initial column conditions 46% acetonitrile, 10 mM ammonium acetate pH 7.4. The column was subjected to an 8 min gradient from 46 to 94% acetonitrile and then washed with 94% acetonitrile for 6 min before loading the next sample. CDDO-Im was detected using a Waters ZQ mass spectrometer with an electrospray ionization probe. Analysis and quantitation were carried out using the Waters MassLynx software package. Quantitation was against standard curves generated by adding dilutions of compound to acetonitrile extract from control liver.
Measurement of tissue lipid levels
Liver (100 mg) was homogenized in 1 ml of buffer containing 18 mM Tris, pH 7.5, 300 mM mannitol, 50 mM EGTA and 0.1 mM phenylmethylsulfonyl fluoride. Five hundred microliters of homogenate was mixed with 4 ml chloroform:methanol (2:1) and incubated overnight at room temperature with occasional shaking. Then, 1 ml of dH2O was added, samples were vortexed and centrifuged at 3000g for 5 min. The lower lipid phase was removed and dried under nitrogen gas. Lipid pellets were dissolved in a mixture of isopropanol and Triton X-100 (4:1). Triglyceride and cholesterol levels were measured using Vet ACE chemistry system (Alfa Wassermann, West Caldwell, NJ). Free fatty acid levels were determined according to manufacturer's instructions (Roche Diagnostics, Penzberg, Germany). Longer treatment periods were used to evaluate CDDO-Im effects on liver lipid content. Mice were treated with 30 μmol CDDO-Im/kg body wt on Monday, Wednesday and Friday and sacrificed after 1 week or 3 months of treatment.
Results
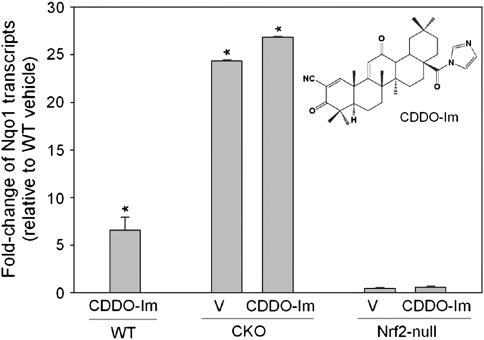
Activation of Nrf2 signaling through deletion of Keap1 (genetic activation) and maximal pharmacologic activation using CDDO-Im (structure shown in Figure 1, inset) results in increased hepatic RNA transcripts of a prototypic Nrf2 target gene nicotinamide adenine dinucleotide phosphate dehydrogenase, quinone 1 (Nqo1). The presence of a functional antioxidant response element has been confirmed in the regulatory region of mouse Nqo1 (29). Earlier studies established that treatment of wild-type mice with a single dose of 30 μmol CDDO-Im/kg body wt produced maximal induction of Nqo1 transcripts in liver (10). This dose has also been shown to provide protection against aflatoxin-induced hepatic tumorigenesis in rats (11). In the present study, mice were treated with a single dose of 30 μmol CDDO-Im/kg body wt by gavage and sacrificed 6 h later. Treatment with CDDO-Im results in a 6.5-fold increase in RNA transcripts of Nqo1 in WT mice. Genetic activation in liver-targeted CKO mice results in 24.4-fold induction of Nqo1. When CKO mice are treated with CDDO-Im Nqo1 transcripts are only slightly increased to 26.8-fold. Nqo1 is not induced in Nrf2-null mice, confirming Nrf2 dependence. The fold changes in Nqo1 transcripts illustrate the differences between genetic and pharmacologic activation of Nrf2 signaling. As shown here, genetic activation can result in a much larger magnitude of transcript induction. While pharmacologic activation with CDDO-Im results in peak Nrf2 transcriptional activation by 6–9 h that returns to near baseline by 24 h (10), genetic activation results in induction that is constitutively sustained. This results in differing degrees and duration of cumulative Nrf2 activation. In addition, it is important to note that the 6 h time point used to evaluate pharmacologic activation was also chosen to minimize the inclusion of secondary or downstream gene expression changes induced by CDDO-Im.
Fig. 1.
Modulation of Nrf2 target gene, Nqo1, by pharmacologic (CDDO-Im) and genetic (conditional Keap1 null, CKO) activation. Lack of Nqo1 transcript induction in Nrf2-null mice confirms Nrf2 dependence. Bars indicate mean ± SEM, n = 4–5/group, *P < 0.05 versus vehicle (V)-treated WT. Inset, chemical structure of CDDO-Im.
Hepatic global gene expression was studied using microarray to compare the overall gene expression signatures modulated by pharmacologic or genetic activation of Nrf2 signaling. Gene expression changes modulated by genetic activation were defined by comparing CKO mice with WT mice (n = 3 mice/group for all comparisons). Pharmacologic activation was evaluated in WT mice 6 h following gavage with vehicle or 30 μmol CDDO-Im/kg body wt. Resulting gene lists for genetic and pharmacologic activation were compared to identify the gene changes common to both, as well as genes unique to each mode of activation. In addition, because previous studies in Nrf2-null mice have suggested that CDDO-Im can modulate Nrf2-independent genes [(11) and our unpublished observations], CKO mice were treated with vehicle or 30 μmol CDDO-Im/kg body wt to determine if CDDO-Im can modulate additional genes in the state of constitutive Nrf2 hyperactivation (suggesting that these changes are Keap1–Nrf2 independent). The overall experimental design for generating these gene lists as well as their interpretation is shown in Figure 2. Genetic and pharmacologic activation of Nrf2 signaling results in overlapping, but distinct gene expression changes. Using a stringent q-value cutoff for pathway analysis, ∼400 genes were identified that are modulated by genetic activation in CKO mice. Of these, 196 are unique to genetic activation and 174 are also modulated by CDDO-Im. CDDO-Im treatment uniquely modulates 84 genes in WT mice. Surprisingly, very little overlap occurs between genes that are modulated by CDDO-Im in CKO mice and the Keap1–Nrf2 independent, CDDO-Im-modulated gene list. One possible explanation would be altered metabolism and transport of CDDO-Im in the liver of CKO mice resulting in a reduced tissue concentration of CDDO-Im. CDDO-Im levels were determined in liver tissue from the mice used in the microarray experiment. Surprisingly, the concentration of CDDO-Im is actually increased in the liver of CKO mice compared with WT mice (11.5 μM compared with 2.2 μM). Both concentrations far exceed the picomolar concentrations required for induction of Nqo1 in cultured hepatocytes (19). The small number of genes modulated by CDDO-Im in CKO mice may reflect secondary or compensatory effects.
Fig. 2.
Experimental design diagram and interpretation of gene changes in each group. The number of genes modulated in each group is indicated using most stringent filtering criteria (q-value < 10−10), n = 3/group. WT; genetic control mice Alb-Cre:Keap1(flox/+), CKO; conditional Keap1-null Alb-Cre:Keap1(flox/−).
Validation was performed by comparing gene expression values determined by real-time PCR and microarray. Representative genes were selected from different functional categories including xenobiotic metabolism (Nqo1, Gclc), lipid metabolism (Fasn), cell signaling (Bmp6, Ddit4) and cell cycle regulation (Ccnd1, Cdkn1a, Gadd45g). Pearson correlation analysis was performed between the microarray and real-time PCR results and confirmed a strong correlation (R = 0.889, P < 0.0001).
Overlapping gene expression changes result from genetic and pharmacologic Nrf2 activation
Gene changes common to genetic and pharmacologic activation of Nrf2 were evaluated to identify the primary biological functions and pathways associated with these genes. Ingenuity Pathways Analysis clustered these gene changes into several functional groups. The five most significant functional categories, in descending order, were lipid metabolism, molecular transport, carbohydrate metabolism, cell signaling and xenobiotic metabolism (Table I, along with examples from each category). Identification of known Nrf2-dependent genes and biological functions provides internal validation for this approach (3,30).
Table I.
Changes common to genetic and pharmacologic activation of Nrf2 signaling
| Fold change |
|||
| Pharmacologic | Genetic | ||
| Lipid metabolism | |||
| Sterol regulatory element-binding factor-1 | Srebf1 | −5.65 | −2.43 |
| Lipase, endothelial | Lipg | −5.63 | –4.12 |
| Fatty acid synthase | Fasn | –5.04 | –2.54 |
| Acetyl-coenzyme A carboxylase alpha | Acaca | –4.70 | –1.82 |
| ELOVL family member 6, elongation of long chain fatty acids (yeast) | Elovl6 | –4.56 | –2.66 |
| ATP citrate lyase | Acly | –3.68 | –3.15 |
| Adipose differentiation-related protein | Adfp | –1.93 | –1.79 |
| Molecular transport | |||
| Solute carrier family 22 (organic anion transporter), member 7 | Slc22a7 | –1.71 | –4.35 |
| CD36 antigen | Cd36 | 1.73 | 8.49 |
| ATP-binding cassette, subfamily D (ALD), member 1 | Abcd1 | 1.87 | 1.77 |
| Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | Slc11a1 | 1.88 | 1.91 |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 5 | Abcc5 | 2.47 | 37.91 |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 4 | Abcc4 | 2.58 | 81.02 |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 1 | Abcc1 | 3.66 | 1.86 |
| Solute carrier family 2 (facilitated glucose transporter), member 1 | Slc2a1 | 4.00 | 1.75 |
| Carbohydrate metabolism | |||
| Pyruvate kinase liver and red blood cell | Pklr | –4.30 | –2.54 |
| Glycerol phosphate dehydrogenase 2, mitochondrial | Gpd2 | –4.06 | –2.07 |
| Phosphogluconate dehydrogenase | Pgd | 2.00 | 2.24 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 3C | Ppp1r3c | 2.02 | 2.68 |
| UDP-glucose dehydrogenase | Ugdh | 2.18 | 2.15 |
| UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 | B3galt1 | 2.21 | 1.71 |
| Cell signaling | |||
| Ubiquitin-specific peptidase 7 | Usp7 | –2.76 | –1.73 |
| Jun oncogene | Jun | –2.22 | –3.20 |
| Inhibitor of kappaB kinase gamma | Ikbkg | 2.34 | 1.82 |
| Ubiquitin specific peptidase 2 | Usp2 | 2.50 | 1.74 |
| Dual specificity phosphatase 4 | Dusp4 | 2.91 | 2.13 |
| Myelocytomatosis oncogene | Myc | 3.09 | 2.94 |
| Xenobiotic metabolism | |||
| Glutathione S-transferase, mu 4 | Gstm4 | 2.15 | 4.42 |
| Glutathione reductase 1 | Gsr | 2.42 | 2.18 |
| Glutamate-cysteine ligase, catalytic subunit | Gclc | 2.90 | 2.38 |
| Glutathione S-transferase, theta 3 | Gstt3 | 2.99 | 5.14 |
| NAD(P)H dehydrogenase, quinone 1 | Nqo1 | 12.83 | 38.88 |
All values reported are statistically significant (q-value < 10−10), as described in Materials and Methods. NAD(P)H, nicotinamide adenine dinucleotide phosphate.
Xenobiotic metabolism is an exceptionally well-characterized function of Nrf2 signaling. Genetic and pharmacologic activation of Nrf2 signaling result in upregulation of known Nrf2 target genes in this group, including glutathione S-transferases, glutathione biosynthesis genes and Nqo1.
Molecular transport changes have also been shown previously to be regulated by Nrf2 signaling. In this study, molecular transport genes were modulated by Keap1 deletion and CDDO-Im treatment. Both solute carrier family members and adenosine triphosphate (ATP)-binding cassette transporters were modulated, with the majority being upregulated.
Unexpectedly, the largest group of overlapping gene changes was associated with lipid metabolism. Genes in this group are primarily downregulated and related to lipid and fatty acid biosynthesis. In addition, sterol regulatory element-binding transcription factor 1 (Srebf1) is significantly downregulated by genetic and pharmacologic activation of Nrf2 signaling and is a key regulator of many fatty acid synthesis genes.
Carbohydrate metabolism is also modulated by genetic and pharmacologic activation of Nrf2 signaling. These gene expression changes are related to several different aspects of carbohydrate metabolism, including glycolysis, the pentose phosphate pathway, regeneration of nicotinamide adenine dinucleotide for use in glycolysis and fatty acid oxidation and glycosaminoglycan biosynthesis. These changes are associated with different functions in carbohydrate metabolism, making the overall effect unclear.
Gene expression changes involved in signal transduction were identified following genetic and pharmacologic activation of Nrf2 signaling. However, these changes do not cluster into any one specific pathway. Jun and Myc oncogene transcripts were modulated. In addition, transcript levels of inhibitor of kappaB kinase gamma were increased. Other examples of signaling-related transcript changes include ubiquitin-specific peptidases 2 and 7, as well as dual specificity phosphatase 4.
Further analysis of gene changes common to genetic and pharmacologic activation of Nrf2 signaling identified several cell cycle and DNA damage response genes that have not been previously identified as Nrf2 target genes. For example, cyclin-dependent kinase inhibitor 1A (p21) was increased 3.6- and 2.1-fold by genetic and pharmacologic activation, respectively. In addition, cyclin D1 (−3.1- and −2.5-fold), DNA damage-inducible transcript 4 (10.4- and 8.0-fold) and growth arrest and DNA damage-inducible 45 gamma (3.8- and 2.4-fold) were modulated by genetic and pharmacologic activation of Nrf2 signaling. It is important to note that the genes associated with DNA damage are not well characterized and may in fact play a part in oxidative stress response (31).
Genetic activation of Nrf2 signaling results in unique gene expression changes
Genetic activation of Nrf2 signaling resulted in gene expression changes that did not occur through pharmacologic activation with CDDO-Im. While there are ∼200 genes uniquely modulated in CKO mice, many of these genes have functions associated with the same biological function categories identified as common to genetic and pharmacologic activation, namely lipid metabolism, molecular transport, xenobiotic metabolism and carbohydrate metabolism (Table II). This suggests that although similar biological functions are altered, genetic activation results in an amplified response. Many of these genes can be validated as Nrf2 dependent based on previous literature (3,30,32,33). It is possible that a few of the Keap1-dependent gene expression changes may be Nrf2 independent. However, identification of concordant changes in biological functions between genetic and pharmacologic manipulation, together with the knowledge that most of the Keap1-dependent gene changes are known to be Nrf2 dependent, suggests that this effect is minimal.
Table II.
Changes unique to genetic activation of Nrf2 signaling
| Fold change | ||
| Genetic | ||
| Lipid metabolism | ||
| Fatty acid desaturase 2 | Fads2 | –3.23 |
| 3-Hydroxy-3-methylglutaryl-coenzyme A synthase 2 | Hmgcs2 | –2.41 |
| Fatty acid-binding protein 2, intestinal | Fabp2 | –2.14 |
| Elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 | Elovl2 | –2.00 |
| Peroxisome proliferator-activated receptor alpha | Ppara | –1.95 |
| Lipase, hepatic | Lipc | –1.89 |
| Fatty acid-binding protein 1, liver | Fabp1 | –1.83 |
| Peroxisome proliferator-activated receptor gamma | Pparg | 2.87 |
| Molecular transport | ||
| Solute carrier family 3, member 1 | Slc3a1 | −4.91 |
| ATP-binding cassette, subfamily G (WHITE), member 5 | Abcg5 | 1.99 |
| Solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | Slc1a1 | 6.39 |
| Solute carrier family 22 (organic anion/cation transporter), member 12 | Slc22a12 | 31.90 |
| Xenobiotic metabolism | ||
| Cytochrome P450, family 3, subfamily a, polypeptide 11 | Cyp3a11 | −2.52 |
| Glutathione S-transferase, mu 2 | Gstm2 | 4.28 |
| Cytochrome P450, family 2, subfamily b, polypeptide 10 | Cyp2b10 | 7.38 |
| Flavin containing monooxygenase-3 | Fmo3 | 16.26 |
| Cytochrome P450, family 2, subfamily b, polypeptide 9 | Cyp2b9 | 20.89 |
| Carbohydrate metabolism | ||
| Arginine vasopressin receptor 1A | Avpr1a | –6.66 |
| Insulin-like growth factor-1 | Igf1 | –2.57 |
| UDP-glucose pyrophosphorylase-2 | Ugp2 | 2.13 |
| Phosphorylase kinase beta | Phkb | 2.40 |
| ST3 beta-galactoside alpha-2,3-sialyltransferase-6 | St3gal6 | 9.50 |
| Protein synthesis | ||
| Eukaryotic translation initiation factor 2-alpha kinase-2 | Eif2ak2 | –2.28 |
| Eukaryotic translation initiation factor 4E-binding protein-1 | Eif4ebp1 | 1.91 |
| Eukaryotic translation initiation factor 4E member-3 | Eif4e3 | 4.12 |
All values reported are statistically significant (q-value < 10−10), as described in Materials and Methods.
The known Nrf2-regulated functions of xenobiotic metabolism and molecular transport were also significantly modulated by gene expression changes unique to CKO mice. Xenobiotic metabolism changes include cytochrome P450s, flavin-containing monooxygenase-3 and glutathione S-transferase mu 2. Molecular transport changes include 13 different members of the solute carrier family and three ATP-binding cassette transporter family members. Examples include the glutamate transporter Slc1a1, an organic ion transporter Slc22a12, and Abcg5, a critical mediator of biliary cholesterol secretion.
Lipid metabolism genes are the largest group of changes unique to CKO mice. These genes are again primarily associated with downregulation of lipid metabolism and biosynthesis. In addition, peroxisome proliferator-activated receptors alpha and gamma are modulated in CKO mice (Ppara −2.0-fold and Pparg 2.9-fold).
Carbohydrate metabolism gene expression changes unique to CKO mice could not be assigned to any one specific function. These changes include insulin signaling-associated genes—insulin-like growth factor-1 and phosphorylase kinase beta; glucose homeostasis regulator—arginine vasopressin receptor 1A and uridine diphosphate (UDP)-glucose pyrophosphorylase-2, which is involved in conversion of glucose-1P to UDP-glucose.
Biological function changes unique to genetic activation in CKO mice were also identified. Protein synthesis changes were identified due to modulation of eukaryotic translation initiation factors (Eifs). Eif2 alpha kinase-2 was downregulated (−2.3-fold), whereas Eif4E-binding protein-1 and Eif43 member-3 transcripts were increased 1.9- and 4.1-fold, respectively. Downregulation of complement system genes were identified in CKO mice, including complement component-6, complement component-8 alpha and complement component-8 beta. These complement components are associated with the membrane attack complex, a protein complex formed by complement components-6, -7, -8 and -9 that function to form a pore in a target cell membrane leading to cell lysis. Gene expression changes associated with downregulation of interferon signaling were also uniquely modulated in CKO mice, including interferon gamma-inducible 47, interferon-induced protein with tetratricopeptide repeats-3, interferon dependent positive acting transcription factor-3 gamma and signal transducer and activator of transcription-1.
CDDO-Im treatment results in unique gene expression changes
CDDO-Im treatment resulted in unique gene expression changes compared with genetic activation of Nrf2 signaling. Fewer biological functions were identified due to the small number of genes uniquely modulated by pharmacologic activation. Some of these changes were still associated with carbohydrate metabolism; however, many genes were involved in cell signaling and regulation of gene expression (Table III). Carbohydrate metabolism genes were associated with the pentose phosphate shunt, galactose metabolism and nucleotide–sugar biosynthesis. Many of the cell signaling genes are multifunctional, but include transcription factors, receptors and kinases that can be associated with several pathways. These changes can be associated with apoptosis (B-cell leukemia/lymphoma), nuclear factor kappa-B signaling (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha), transforming growth factor-β signaling (bone morphogenic protein-6 and Kruppel-like factor-10), insulin signaling (insulin-like growth factor binding protein-1) and cytokine signaling (interleukin-6 receptor alpha). In addition, CDDO-Im treatment results in upregulation of the multifunctional transcription factors CCAAT/enhancer-binding protein beta and BTB and CNC homology-1.
Table III.
Changes unique to CDDO-Im treatment
| Fold change | ||
| CDDO-Im | ||
| Carbohydrate metabolism | ||
| Ribose 5-phosphate isomerase A | Rpia | –3.22 |
| Galactose-4-epimerase, UDP | Gale | –2.78 |
| Phosphoglucomutase-3 | Pgm3 | –2.35 |
| Hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) | H6pd | 1.79 |
| Cell signaling | ||
| Kruppel-like factor-10 | Klf10 | –9.33 |
| Zinc finger protein 467 | Zfp467 | –3.28 |
| Basic helix-loop-helix domain containing, class B2 | Bhlhb2 | –2.69 |
| B-cell leukemia/lymphoma-3 | Bcl3 | –2.22 |
| Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia | 2.02 |
| CCAAT/enhancer-binding protein (C/EBP), beta | Cebpb | 2.06 |
| BTB and CNC homology 1 | Bach1 | 2.37 |
| Interleukin-6 receptor, alpha | Il6ra | 2.78 |
| Bone morphogenetic protein-6 | Bmp6 | 6.24 |
| Insulin-like growth factor-binding protein-1 | Igfbp1 | 8.82 |
All values reported are statistically significant (q-value < 10−10), as described in Materials and Methods.
A small group of genes were modulated in CKO mice treated with CDDO-Im. Only six genes in this group overlap with CDDO-Im modulated, Keap1–Nrf2-independent genes to provide secondary confirmation of Keap1–Nrf2 independence. As shown in supplementary Table 1 (available at Carcinogenesis Online), these overlapping genes are modulated in the same direction in both comparisons. In addition, it is interesting to note that the overlap of basal gene expression changes in CKO mice and CKO mice treated with CDDO-Im are modulated in opposite directions, suggesting a common regulatory mechanism. Additional genes modulated by CDDO-Im in CKO mice may reflect compensatory or secondary effects that remain to be addressed.
Genetic and pharmacologic modulation of lipid metabolism genes results in reduction in liver lipid levels
Liver lipid levels were measured to confirm the functional outcome of the downregulation of genes associated with lipid metabolism, which was the largest class of modulated genes. Liver triglyceride levels are reduced by genetic and pharmacologic activation of Nrf2 signaling (Figure 3). Compared with WT mice, CKO mice have a 69% reduction in liver triglycerides. Similarly, long-term treatment with CDDO-Im results in a 76% reduction in liver triglycerides. Short-term treatment with CDDO-Im does not significantly alter liver lipid levels. CKO mice also have a 41% reduction in free fatty acids compared with WT mice, but no significant modulation of liver cholesterol content. Chronic administration of CDDO-Im results in a 15% reduction in free fatty acids and 23% reduction in liver cholesterol levels. These results confirm that the observed downregulation of lipid metabolism genes caused by genetic and pharmacologic activation of Nrf2 signaling results in reduced liver lipid content.
Fig. 3.
Activation of Nrf2 signaling results in reduction in liver lipid levels. Triglyceride levels are reduced by genetic activation of Nrf2 signaling in CKO mice or following chronic CDDO-Im treatment. Short-term treatment with CDDO-Im does not significantly alter liver triglyceride levels. Bars indicate mean ± SEM, n = 4/group, *P < 0.05 versus WT, +P < 0.05 versus vehicle (V).
Discussion
Recent studies have identified inactivating mutations in Keap1, leading to an increase in Nrf2 function, in human cancers. Inducing the Nrf2-regulated cytoprotective response could provide a selective advantage to tumor cells, raising the question of whether it is hazardous to elicit these changes in the context of interventions for cancer chemoprevention. Our observation of the differing gene expression profiles, as well as magnitude of response in common genes modulated by genetic compared with pharmacologic activation of Nrf2, suggests that these are not equivalent means to modulate Nrf2 target genes. Less than half of the genes altered by deletion of Keap1 are also modulated by treatment with CDDO-Im. This suggests that Keap1 mutant tumor cells experience a much different signaling environment compared with cells in an at-risk individual given an Nrf2-inducing agent for chemoprevention. In addition, concerns regarding Keap1–Nrf2-mediated increases in resistance to chemotherapy are supported by induction of large groups of xenobiotic metabolism genes (including cytochrome P450s) and molecular transport genes. In addition, previous studies have shown that continuous induction of Nrf2 target genes is not necessary to affect chemoprevention (34). Intermittent, submaximal activation of Nrf2-regulated cytoprotective genes can be sufficient to inhibit carcinogen–DNA adduct formation resulting in reduced risk of tumorigenesis, while minimizing the chance of deleterious effects. Thus, the sustained activation of Nrf2 target genes in the genetic CKO model is very distinct from the modulation in gene expression achievable by chemoprevention.
It is important to note that induction of Nrf2-regulated genes such as those affecting xenobiotic metabolism is subject to sex differences (35). For example, greater induction of class Alpha, Mu and Pi glutathione S-transferase subunits is observed in the livers of female mice treated with butylated hydroxyanisole than in the livers of male mice treated with the same phenolic antioxidant. Gender differences have also been observed for the expression of genes affecting lipid metabolism (36). The comparisons described in our study were all conducted in male mice. While it is probable that the overall conclusions would remain the same, comparisons using female mice could result in different magnitudes of gene induction.
While Nrf2 is commonly described as a mediator of cytoprotective stress responses, our results suggest that the impact of activating this pathway must be considered more broadly. In fact, regulation of lipid metabolism appears to be a primary function of Nrf2 signaling in the mouse liver. Lipid metabolism genes are modulated by genetic activation and pharmacologic activation using CDDO-Im, hinting that Nrf2 signaling may also function to sense lipid levels or intermediates of lipid metabolism. Currently, it is unclear how Nrf2-regulated modulation of lipid metabolism may, if at all, impact the cytoprotective and detoxication changes that are desirable for cancer chemoprevention. However, it raises interesting possibilities for obesity-related cancers and suggests the possibility of targeting Nrf2 signaling for prevention of other disease states associated with lipid metabolism. Our results suggest that Nrf2-inducing agents such as CDDO-Im could be applied to prevent or reverse obesity and diabetes. In fact, recent studies conducted in our lab have shown that CDDO-Im protects against several aspects of high-fat diet-induced obesity and diabetes (S.Shin, J.Wakabayashi, M.S.Yates, N.Wakabayashi, P.M.Dolan, S.Aja, K.T.Liby, M.B.Sporn, M.Yamamoto, T.W.Kensler, in preparation). CDDO-Im prevents increases in total body weight and adipose tissue weight in mice fed a high-fat diet in an Nrf2-dependent manner. In addition, CDDO-Im improves glucose tolerance in mice fed a high-fat diet. Another known Nrf2 inducer, 3H-1,2-dithiole-3-thione, has also been shown to repress gene expression related to lipid metabolism (3,37), suggesting that this effect may be shared by other inducers. Epidemiologic evidence suggests that obesity is a risk factor for cancer incidence and mortality (38). A large cohort study conducted by the American Cancer Society estimates that 14% of all cancer deaths in men and 20% of all cancer deaths in women from several cancer types are attributable to overweight and obesity (39). The exact mechanisms responsible for the connection between obesity and carcinogenesis remain unclear; however, obesity-related increases in inflammation may play a role (40). CDDO-Im and other Nrf2 inducers could protect against both obesity and inflammation to prevent cancer.
Our current study provides unique insight into the pharmacodynamic action of CDDO-Im at low doses relevant to cancer chemoprevention. Our observation of Keap1–Nrf2 dependent and CDDO-Im-modulated changes in lipid metabolism may provide additional hints regarding CDDO-Im mechanism of action. The core triterpenoid structure is very similar to cholesterol, perhaps providing a mechanism for increased specificity or potency to activate Nrf2-signaling pathways by mimicking a lipid metabolism intermediate that could be sensed by Keap1. The cholesterol-like structure of this class of synthetic triterpenoids has been implicated in other molecular functions associated with apoptosis. While these latter experiments were conducted at concentrations much higher than is required for Nrf2 activation, the cholesterol-like action is still noteworthy. The analog CDDO-Me has been shown to act in a manner very similar to cholesterol to induce apoptosis by directly permeabilizing the inner mitochondrial membrane (41). Cholesterol deposition in the mitochondrial membrane alters membrane fluidity and can result in a loss of mitochondrial glutathione. Like cholesterol, CDDO-Me appears to target the inner mitochondrial membrane to induce apoptosis and inhibit mitochondrial electron transport.
While our study shows that Nrf2 signaling is the primary pathway modified by low-dose CDDO-Im treatment, molecular pathways modulated by triterpenoids vary depending on the dose. Low concentrations of triterpenoids (in picomolar range) have also been shown to activate an anti-inflammatory response. However, the anti-inflammatory activity of triterpenoids is closely correlated with Nrf2 activation, suggesting a common molecular mechanism (19). Additional gene expression changes were identified at the low dose used in our current study. These changes were not associated with any one predominant pathway, but suggest limited gene expression changes related to apoptosis, nuclear factor kappa-B, transforming growth factor-β, insulin and cytokine-signaling pathways at this low dose. While Nrf2 activation seems to be predominant and consistent in multiple tissue types and cell lines at low doses (10,16,20,42), higher doses of triterpenoids act on multiple signaling pathways. This suggests that the appropriate dose of triterpenoid could be fine tuned to target specific molecular pathways. For example, in vitro studies show that triterpenoids inhibit growth of many different cancer cell lines at nanomolar concentrations by modulation of cell cycle regulators such as Myc, p27, cyclin D1 and p21 (43). Triterpenoids also induce differentiation at nanomolar concentrations in a variety of cell lines, including leukemia cells and osteosarcoma cells (43). Micromolar concentrations of triterpenoids induce apoptosis in cancer cell lines including leukemia, breast cancer, multiple myeloma and lung cancer (43). Triterpenoids clearly modulate several different pathways depending on the dose and cell type. To date, four direct molecular targets have been identified for this class of triterpenoids. These direct targets include Keap1 (19), peroxisome proliferator-activated receptor gamma (44), I kappa-B kinase beta (45) and signal transducer and activator of transcription3 (46,47). These targets do not explain the many molecular pathways that are modulated by triterpenoids. Triterpenoids are known to reversibly react with nucleophiles, creating an adduct that converts back to the parent triterpenoid (48). This reversible adduct may be sufficient to initiate changes in signaling, but adduct instability makes identification of molecular targets especially challenging. It is also interesting to consider the possibility of a ‘cysteine code’. The differential reactivities of cysteines with triterpenoids, either within a target or between targets, may result in differing biological effects.
This study emphasizes the complex nature of biological responses that can be modulated by activation of Nrf2 signaling. While activation of Nrf2 signaling has been shown to mediate cytoprotective stress responses, our current results provide an important reminder that biological networks are rarely monofunctional. The ultimate biological outcome is dependent on many factors, including dose, duration, tissue or cell type and context (such as normal cell or tumor cell). This study also highlights the importance of characterizing pharmacodynamics in an unbiased manner to better understand the global effects of these molecular pathway changes. This can improve our success in applying pharmacologic agents to prevent or treat cancer. The current study suggests that CDDO-Im and other Nrf2 inducers may be effective chemopreventive agents against cancers associated with obesity or other lipid-related diseases, in addition to those directly mediated by electrophilic carcinogens. Although molecular pathway descriptions are often reduced to the most simplistic aspects and most commonly studied functions, we must move toward a systems biology view that embraces the reality of complex biological networks and their impact on health.
Supplementary material
Supplementary data and Table 1 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute (CA94076 and CA39416 to T.W.K.); National Institute of Environmental Health Sciences (ES03819); Reata Pharmaceuticals to M.B.S.; PhRMA Foundation to M.S.Y.; W.Harry Feinstone Center for Genomic Research to T.R.S.
Acknowledgments
We would like to thank Nadine Forbes in the Johns Hopkins School of Medicine Phenotyping Core for assistance with tissue lipid assays.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ATP
adenosine triphosphate
- CDDO-Im
1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole
- cDNA
complementary DNA
- CKO
conditional Keap1-null, Albumin-Cre:Keap1(flox/−)
- Eif
eukaryotic translation initiation factor
- Nrf2
NF-E2-related factor 2
- PCR
polymerase chain reaction
- UDP
uridine diphosphate
- WT
genetic control wild-type, Albumin-Cre:Keap1(flox/+)
References
- 1.Motohashi H, et al. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl Acad. Sci. USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwak MK, et al. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 4.Enomoto A, et al. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 5.Burton NC, et al. In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicology. 2006;27:1094–1100. doi: 10.1016/j.neuro.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 6.Osburn WO, et al. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int. J. Cancer. 2007;121:1883–1891. doi: 10.1002/ijc.22943. [DOI] [PubMed] [Google Scholar]
- 7.Ramos-Gomez M, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl Acad. Sci. USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahey JW, et al. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc. Natl Acad. Sci. USA. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C, et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 10.Yates MS, et al. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol. Cancer Ther. 2007;6:154–162. doi: 10.1158/1535-7163.MCT-06-0516. [DOI] [PubMed] [Google Scholar]
- 11.Yates MS, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-(2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66:2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 12.Liby K, et al. The synthetic triterpenoids CDDO-methyl ester and CDDO-ethyl amide prevent lung cancer induced by vinyl carbamate in A/J mice. Cancer Res. 2007;67:2414–2419. doi: 10.1158/0008-5472.CAN-06-4534. [DOI] [PubMed] [Google Scholar]
- 13.Sussan TE, et al. Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc. Natl Acad. Sci. USA. 2009;106:250–255. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyama T, et al. Cytoprotective role of Nrf2/Keap1 system in methylmercury toxicity. Biochem. Biophys. Res. Commun. 2007;363:645–650. doi: 10.1016/j.bbrc.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 15.Okawa H, et al. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 16.Osburn WO, et al. Genetic or pharmacologic amplification of nrf2 signaling inhibits acute inflammatory liver injury in mice. Toxicol. Sci. 2008;104:218–227. doi: 10.1093/toxsci/kfn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nioi P, et al. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem. Biophys. Res. Commun. 2007;362:816–821. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 19.Dinkova-Kostova AT, et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc. Natl Acad. Sci. USA. 2005;102:4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liby K, et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 21.Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 22.Honda T, et al. A novel dicyanotriterpenoid, 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-onitrile, active at picomolar concentrations for inhibition of nitric oxide production. Bioorg. Med. Chem. Lett. 2002;12:1027–1030. doi: 10.1016/s0960-894x(02)00105-1. [DOI] [PubMed] [Google Scholar]
- 23.Kwak MK, et al. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol. Med. 2001;7:135–145. [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, et al. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z, et al. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- 26.Gautier L, et al. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 27.Silkworth JB, et al. Toxicogenomic analysis of gender, chemical, and dose effects in livers of TCDD- or aroclor 1254-exposed rats using a multifactor linear model. Toxicol. Sci. 2008;102:291–309. doi: 10.1093/toxsci/kfm313. [DOI] [PubMed] [Google Scholar]
- 28.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 2003;31:2013–2035. [Google Scholar]
- 29.Nioi P, et al. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thimmulappa RK, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 31.Han ES, et al. The in vivo gene expression signature of oxidative stress. Physiol. Genomics. 2008;34:112–126. doi: 10.1152/physiolgenomics.00239.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu R, et al. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci. 2006;79:1944–1955. doi: 10.1016/j.lfs.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 33.Nair S, et al. Pharmacogenomics of phenolic antioxidant butylated hydroxyanisole (BHA) in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Pharm. Res. 2006;23:2621–2637. doi: 10.1007/s11095-006-9099-x. [DOI] [PubMed] [Google Scholar]
- 34.Primiano T, et al. Intermittent dosing with oltipraz: relationship between chemoprevention of aflatoxin-induced tumorigenesis and induction of glutathione S-transferases. Cancer Res. 1995;55:4319–4324. [PubMed] [Google Scholar]
- 35.Chanas SA, et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, et al. Gender disparity of hepatic lipid homeostasis regulated by the circadian clock. J. Biochem. 2009;145:609–623. doi: 10.1093/jb/mvp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, et al. Identification of novel transcriptional networks in response to treatment with the anticarcinogen 3H-1,2-dithiole-3-thione. Physiol. Genomics. 2006;24:144–153. doi: 10.1152/physiolgenomics.00258.2005. [DOI] [PubMed] [Google Scholar]
- 38.Pan SY, et al. Energy intake, physical activity, energy balance, and cancer: epidemiologic evidence. Methods Mol. Biol. 2009;472:191–215. doi: 10.1007/978-1-60327-492-0_8. [DOI] [PubMed] [Google Scholar]
- 39.Calle EE, et al. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 40.Hursting SD, et al. Energy balance and carcinogenesis: underlying pathways and targets for intervention. Curr. Cancer Drug Targets. 2007;7:484–491. doi: 10.2174/156800907781386623. [DOI] [PubMed] [Google Scholar]
- 41.Samudio I, et al. A novel mechanism of action of methyl-2-cyano-3,12 dioxoolean-1,9 diene-28-oate: direct permeabilization of the inner mitochondrial membrane to inhibit electron transport and induce apoptosis. Mol. Pharmacol. 2006;69:1182–1193. doi: 10.1124/mol.105.018051. [DOI] [PubMed] [Google Scholar]
- 42.Thimmulappa RK, et al. Preclinical evaluation of targeting the Nrf2 pathway by triterpenoids (CDDO-Im and CDDO-Me) for protection from LPS-induced inflammatory response and reactive oxygen species in human peripheral blood mononuclear cells and neutrophils. Antioxid. Redox Signal. 2007;9:1963–1970. doi: 10.1089/ars.2007.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liby KT, et al. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat. Rev. Cancer. 2007;7:357–369. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, et al. A synthetic triterpenoid, 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), is a ligand for the peroxisome proliferator-activated receptor gamma. Mol. Endocrinol. 2000;14:1550–1556. doi: 10.1210/mend.14.10.0545. [DOI] [PubMed] [Google Scholar]
- 45.Yore MM, et al. The synthetic triterpenoid 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole blocks nuclear factor-kappaB activation through direct inhibition of IkappaB kinase beta. Mol. Cancer Ther. 2006;5:3232–3239. doi: 10.1158/1535-7163.MCT-06-0444. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad R, et al. Triterpenoid CDDO-methyl ester inhibits the Janus-activated kinase-1 (JAK1)—>signal transducer and activator of transcription-3 (STAT3) pathway by direct inhibition of JAK1 and STAT3. Cancer Res. 2008;68:2920–2926. doi: 10.1158/0008-5472.CAN-07-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liby K, et al. Prevention and treatment of experimental estrogen receptor-negative mammary carcinogenesis by the synthetic triterpenoid CDDO-methyl Ester and the rexinoid LG100268. Clin. Cancer Res. 2008;14:4556–4563. doi: 10.1158/1078-0432.CCR-08-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Couch RD, et al. Studies on the reactivity of CDDO, a promising new chemopreventive and chemotherapeutic agent: implications for a molecular mechanism of action. Bioorg. Med. Chem. Lett. 2005;15:2215–2219. doi: 10.1016/j.bmcl.2005.03.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.