Abstract
O-GlcNAcylation is an essential, dynamic and inducible post-translational glycosylation of cytosolic proteins in metazoa and can show interplay with protein phosphorylation. Inhibition of OGA (O-GlcNAcase), the enzyme that removes O-GlcNAc from O-GlcNAcylated proteins, is a useful strategy to probe the role of this modification in a range of cellular processes. In the present study, we report the rational design and evaluation of GlcNAcstatins, a family of potent, competitive and selective inhibitors of human OGA. Kinetic experiments with recombinant human OGA reveal that the GlcNAcstatins are the most potent human OGA inhibitors reported to date, inhibiting the enzyme in the sub-nanomolar to nanomolar range. Modification of the GlcNAcstatin N-acetyl group leads to up to 160-fold selectivity against the human lysosomal hexosaminidases which employ a similar substrate-assisted catalytic mechanism. Mutagenesis studies in a bacterial OGA, guided by the structure of a GlcNAcstatin complex, provides insight into the role of conserved residues in the human OGA active site. GlcNAcstatins are cell-permeant and, at low nanomolar concentrations, effectively modulate intracellular O-GlcNAc levels through inhibition of OGA, in a range of human cell lines. Thus these compounds are potent selective tools to study the cell biology of O-GlcNAc.
Keywords: GlcNAcstatin, inhibition, O-GlcNAc, O-GlcNAcase
Abbreviations: GST, glutathione transferase; HEK, human embryonic kidney; Hex, hexosaminidase; 4MU-NAG, 4-methylumbelliferyl-N-acetyl-β-D-glucosaminide; OGA, O-GlcNAcase; CpOGA, Clostridium perfringens OGA; hOGA, human OGA; OGT, O-GlcNAc transferase; PUGNAc, O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenylcarbamate
INTRODUCTION
Reversible post-translational modification of many cytoplasmic and nuclear proteins in eukaryotic cells by glycosylation of serine and threonine residues with β-linked N-acetylglucosamine (O-GlcNAc) has been shown to play important roles in cellular processes as diverse as DNA transcription and translation, insulin sensitivity, protein trafficking and degradation [1–4]. Dysregulation of O-GlcNAc appears to play a role in human pathogenesis, such as cancer [5–7] and Alzheimer's disease [8–12]. O-GlcNAc is also implicated in Type 2 diabetes [13,14]; however, the precise mechanism is still controversial [15].
In higher eukaryotes only two enzymes are responsible for the dynamic cycling of O-GlcNAc, the OGT (O-GlcNAc transferase; CAZY family GT41 [16]) which transfers GlcNAc on to proteins from the UDP-GlcNAc donor, and the OGA (O-GlcNAcase; CAZY family GH84 [17]), which catalyses the removal of O-GlcNAc. The precise molecular mechanisms by which OGT and OGA recognize and act on hundreds of proteins remain to be discovered [18].
Inhibition of hOGA (human OGA) with PUGNAc [O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenylcarbamate] (Figure 1A) (Ki=50 nM [19,20]) has been used extensively to study the role of O-GlcNAc in a range of cellular processes [21–27]. Crystal structures of bacterial hOGA homologues have become available [28,29], and it has been shown that PUGNAc is a tight-binding inhibitor, with its imidolactone ring mimicking the half-chair/envelope conformation of the pyranose ring in the transition state by virtue of the stable oxime moiety [28,30]. However, PUGNAc also potently inhibits the human HexA/B (hexosaminidases A/B; CAZY family GH20), genetic inactivation of which has been associated with the Tay–Sachs and Sandhoff lysosomal storage disorders [31]. Structural analysis has revealed that the acetamido group of PUGNAc resides in a deep pocket that is significantly larger in OGAs than in the lysosomal HexA/HexB [28,29,32]. The feasibility of constructing more selective PUGNAc analogues, as well as other hOGA inhibitors that exploit the difference in the size of the N-acyl binding pocket, has previously been explored [20,33,34]. Increasing the size of the N-acyl substituents, however, also resulted in weaker (micromolar) inhibition of hOGA. Furthermore, PUGNAc is acid-labile [35]. Thiazoline, another inhibitor of GH20/84 enzymes [32,36,37], has also been similarly chemically modified to achieve more selective OGA inhibition, yielding derivatives that inhibited in the low micromolar range with three orders of magnitude selectivity towards hOGA [20,33], culminating in the recent report of the thiazoline derivative thiamet-G, with selectivity towards hOGA, inhibiting it with a Ki of 21 nM [38].
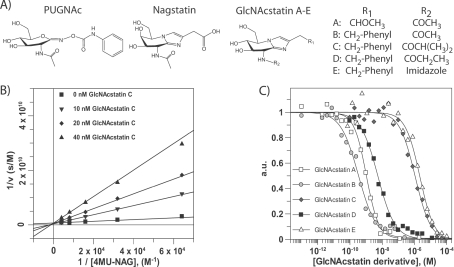
Figure 1. Kinetic characterization of GlcNAcstatin derivatives.
(A) Chemical structures of the glycosidase inhibitors PUGNAc, nagstatin and the GlcNAcstatin scaffold. (B) Lineweaver–Burk analysis of hOGA steady-state kinetics measured in the presence of 0–40 nM GlcNAcstatin C. Data were fitted using the standard equation for competitive inhibition in the GraFit program (Erithacus Software). (C) Dose–response curve of GlcNAcstatins A–E incubated with lysosomal HexA/HexB. Data were fitted using the standard IC50 equation in the GraphFit program (Erithacus Software).
We have recently reported a novel scaffold, GlcNAcstatin, a potent inhibitor of a bacterial OGA orthologue, exploiting the structural similarity of Z-PUGNAc and the naturally occurring potent Hex inhibitor nagstatin (Figure 1A) [39–41]. Inhibition of β-glycosidases with nagstatin-related sugar-imidazoles has been examined by Heightman and Vasella [42], who suggested that lateral protonation of the exo-cyclic nitrogen atom of the imidazole ring should account for the excellent inhibiting properties of these compounds. Indeed, GlcNAcstatin inhibited a bacterial OGA orthologue in the picomolar range, and structural analysis revealed a tight interaction between the catalytic acid and the (presumably protonated) imidazole [41].
In the present study, we report that GlcNAcstatin also potently inhibits hOGA, with a Ki in the low nanomolar range. We also show that this compound is able to induce hyper-O-GlcNAcylation in a range of human cell lines when used at low nanomolar concentrations. Furthermore, we report four new GlcNAcstatin derivatives that explore potency and selectivity of this scaffold, with one of these being the most potent hOGA inhibitor reported so far, inhibiting with a Ki of 420 pM. Guided by a crystal structure of one of these derivatives in complex with a bacterial OGA, we probe key interactions through mutagenesis. These novel molecules will be useful tools for the study of OGA in a range of cellular signal transduction pathways.
MATERIALS AND METHODS
Cloning, protein expression and purification
The previously described plasmid for expression of the OGA orthologue from CpOGA (Clostridium perfringens OGA) [28] was used as a template to carry out mutagenesis of Val331 to cysteine, using the QuikChange® kit (Stratagene) with the following primers: 5′-GGGAGATGTAAAACCATTAATAACATGCCCAACAGAGTATGATACTGGAGC-3′ and 5′-GCTCCAGTATCATACTCTGTTGGGCATGTTATTAATGGTTTTACATCTCCC-3′. W490A mutagenesis was performed using the same protocol and techniques with primers: 5′-GGACAATAAAACTGCGGCTAAATCAGGAAG-3′ and 5′-CTTCCTGATTTAGCCGCAGTT TTATTGTCC-3′.The constructs were verified by DNA-sequencing. V331C-CpOGA, W490A-CpOGA and wild-type protein were expressed and purified following the protocol described previously [28,41,43].
hOGA (amino acid residues 53–916) was cloned into a modified version of pGEX6P-1, lacking the BamHI site, and the hOGA sequence was inserted using EcoRI and NotI sites, after an internal EcoRI site was removed by introducing a silent mutation. The protein was expressed in Escherichia coli BL21 (DE3) cells overnight at 15 °C using 10 μM IPTG (isopropyl β-D-thiogalactoside; D600 of 0.4–0.6). The cells were harvested by centrifugation (3500 g for 30 min at 4 °C) and lysed with sonication in lysis buffer [50 mM Tris/HCl (pH 7.5), 250 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% 2-mercaptoethanol, 0.2 mM PMSF and 1 mM benzamidine]. The recombinant GST (glutathione transferase)-fusion protein was bound to glutathione–Sepharose beads that were pre-equilibrated in washing buffer [50 mM Tris/HCl (pH 7.5), 250 mM NaCl, 1 mM EGTA, 0.1% 2-mercaptoethanol, 0.2 mM PMSF and 1 mM benzamidine]. The fusion protein was eluted using the washing buffer supplemented with 20 mM glutathione and the pH was adjusted to 7.5. The eluted protein was dialysed into 50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA, 150 mM NaCl, 0.07% 2-mercaptoethanol, 0.1 mM PMSF and 1 mM benzamidine.
Determination of the CpOGA–GlcNAcstatin D complex structure
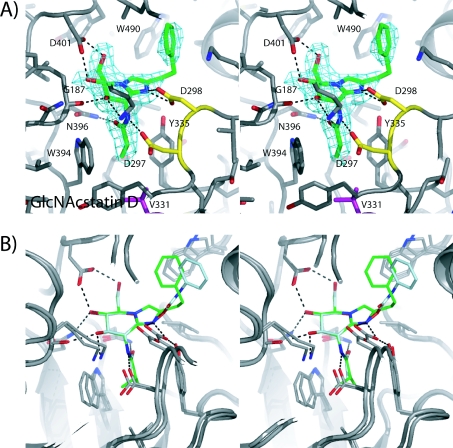
CpOGA was crystallized as described previously [28,41,43]. An aliquot of 1 μl of a suspension of GlcNAcstatin D in mother liquor was added to the crystallized protein in a 2.25 μl drop (1 μl of protein plus 1 μl of mother liquor plus 0.25 μl of 40% γ-butyrolactone). After 50 min at 20 °C (room temperature) the crystal was cryoprotected by 5 s immersion in 0.17 M ammonium sulfate, 0.085 M sodium cacodylate (pH 6.5), 25.5% PEG [poly(ethylene glycol)] 8000 and 15% glycerol, and frozen in a nitrogen cryostream. Data were collected at the European Synchrotron Radiation Facility on beamline ID14-1 to 2.3 Å (1 Å=0.1 nm), with an overall Rmerge of 0.074 and 98.2% completeness. Refinement was initiated from the native CpOGA–GlcNAcstatin C complex (PDB entry 2J62, [41]). Well-defined Fo−Fc electron density for the inhibitor was observed (Figure 2A), allowing placement of an inhibitor model, built with the help of a PRODRG [44] structure and topology. Iterative model building using COOT [45] and refinement with REFMAC [46] yielded the final model with good statistics (R, Rfree:18.5, 23.8).
Figure 2. Structural analysis of GlcNAcstatin derivatives and PUGNAc in complex with CpOGA.
(A) Stereo view of GlcNAcstatin D (sticks with green carbon, red oxygen and blue nitrogen atoms) in the active site of CpOGA (sticks with grey carbon). Hydrogen bonds are indicated by black broken lines. Unbiased |Fo|–|Fc|, ϕcalc electron density map (2.75 σ) is shown as a cyan chickenwire. (B) Stereo view of superimposed crystallographically determined complexes of CpOGA with GlcNAcstatin C (PDB number 2J62) (colour scheme as in A) and PUGNAc (PDB number 2CBJ) (sticks with light blue carbon), black broken lines showing hydrogen bonds for the CpOGA–GlcNAcstatin complex.
Inhibition measurements
Steady-state kinetics of wild-type hOGA and CpOGA mutants were determined using the fluorogenic substrate 4MU-NAG (4-methylumbelliferyl-N-acetyl-β-D-glucosaminide; Sigma). Standard reaction mixtures (50 μl) contained 2 pM CpOGA mutant in 50 mM citric acid, 125 mM Na2HPO4 (pH 5.5), 0.1 mg/ml BSA, and 1.5–25 μM of substrate in water. Steady-state kinetics of GlcNAcstatin C were performed in the presence of different concentrations of the inhibitor (0, 35, 70 and 140 pM). The reaction mixtures were incubated at 20 °C for 466 min. For hOGA, the 50 μl standard reaction volume contained 2 nM hOGA–GST (53–916), McIlvaine buffer-system (0.2 M Na2HPO4 mixed with 0.1 M citric acid, pH 5.7), 0.1 mg/ml BSA, 0–250 μM 4MU-NAG with various GlcNAcstatin C concentrations (0, 10, 20 and 40 nM). The reaction was run for 60 min.
All reactions were stopped (before more than 10% of the substrate was consumed) by the addition of 100 μl of 3 M glycine/NaOH (pH 10.3). The fluorescence of the released 4-methylumbelliferone was quantified using a FLX 800 Microplate Fluorescence Reader (Bio-Tek), with excitation and emission wavelengths of 360 and 460 nm respectively. The mode of inhibition was visually verified by the Lineweaver–Burk plot (Figure 1B), and the Ki was determined by fitting all fluorescence intensity data to the standard equation for competitive inhibition in GraFit (Erithacus Software) (Tables 1 and 2). IC50 measurements with a mixture of human Hex A/B activities (purchased from Sigma, catalogue number A6152) against GlcNAcstatin A–E and PUGNAc were performed using the fluorogenic 4MU-NAG substrate and standard reaction mixtures as described previously [41,43], with the following changes: 5 μ-units/ml enzyme mixture was used with a fixed substrate concentration at the Km (230 μM) in the presence of different concentrations of the inhibitors, 100 pM to 100 μM (GlcNAcstatins) and 10 pM to 1 mM (PUGNAc).
Table 1. Inhibition data of GlcNAcstatins A–E and PUGNAc against lysosomal HexA/HexB, hOGA and the compounds selectivity for hOGA.
Values are means±S.D. n.s., no selectivity for hOGA.
| Compound | Hex A/B, Ki (nM)* | hOGA, Ki (nM) | Selectivity (GH20/hOGA) |
|---|---|---|---|
| GlcNAcstatin A | 0.55±0.05 | 4.3±0.2 | n.s. |
| GlcNAcstatin B | 0.17±0.05 | 0.42±0.06 | n.s. |
| GlcNAcstatin C | 550±10 | 4.4±0.1 | 164 |
| GlcNAcstatin D | 2.7±0.4 | 0.74±0.09 | 4 |
| GlcNAcstatin E | 1100±100 | 8500±300 | n.s. |
| PUGNAc | 25±2.5 | 35±6* | n.s. |
*The Cheng–Prusoff equation {Ki=IC50/1+([S]/Km)} was used to convert the IC50 values into an absolute inhibition constant (Ki).
Table 2. Michaelis–Menten parameters of CpOGA wild-type and mutants and Ki values against GlcNAcstatin C.
Values are means±S.D.
| Enzyme | Ki (nM) | Km (μM) | kcat (s−1) |
|---|---|---|---|
| Wild-type | 4.6±0.1 | 2.9±0.2 | 10.5±0.2 |
| V331C | 98.1±6.4 | 6.8±0.4 | 17.3±0.7 |
| W490A | 74.0±5.6 | 100±10 | 65±3 |
Cell-based assays and Western blot analysis
Cell lines were maintained in DMEM [Dulbecco's modified Eagle's medium; containing 1 g/l glucose for HEK (human embryonic kidney)-293 cells, and 4.5 g/l glucose for HeLa, HT-1080, SH-SY5Y and U-2 OS cells] supplemented with 10% (v/v) foetal bovine serum. Subconfluent cells were treated with various concentrations of inhibitors in the presence of 0.1% DMSO. After 6 h, cells were washed once in ice-cold PBS and harvested in the following lysis buffer: 50 mM Tris (pH 7.4), 0.27 M sucrose, 1 mM sodium orthovanadate, 1 mM EDTA, 1 mM EGTA, 10 mM sodium β-glycerophosphate, 50 mM NaF, 1% Triton X-100, 0.1% 2-mercaptoethanol, 1 mM benzamidine, 0.1 mM PMSF and 5 μM leupeptin. Cell debris was removed by centrifugation (13000 g for 10 min at 4 °C), and supernatants were analysed by immunoblotting. For Western blot analysis, 15–30 μg of total cellular protein was separated by SDS/PAGE, and O-GlcNAcylation was detected with the anti-O-GlcNAc antibody CTD110.6. An anti-β-tubulin antiserum was used as a loading control. The O-GlcNAc signal in each lane was quantified using the AIDA (Advanaced Image Data Analyzer) software version 3.27, and normalized against the β-tubulin signal.
RESULTS AND DISCUSSION
GlcNAcstatins: a new family of potent hOGA inhibitors
Exploiting the available structural data of a complex of a C. perfringens OGA orthologue (CpOGA, [28,41]), GlcNAcstatins A–E were designed and synthesized (Figure 1A, details of synthesis to be reported elsewhere; for further details please contact the corresponding author). GlcNAcstatins A–E fall into two subfamilies, based on their modification on the C-2 position of the imidazole ring. GlcNAcstatin A carries the carboxymethyl group, whereas GlcNAcstatins B–E carry a phenylethyl moiety. GlcNAcstatin C carries an isobutyl group instead of the smaller N-acetyl group in GlcNAcstatin A and B, GlcNAcstatin D an N-propionyl group, and GlcNAcstatin E an N-imidazole group. The increased length of the N-acyl derivative was explored to determine the most useful substituent to specifically inhibit hOGA over the lysosomal Hexs.
GlcNAcstatins are (sub)nanomolar inhibitors of hOGA
A previous report on GlcNAcstatin C showed picomolar inhibiton of an apparent hOGA orthologue from the bacterium C. perfringens (CpOGA) [41], whereas inhibition of hOGA remained to be explored. GlcNAcstatins A–E were tested in dose–response experiments against recombinant hOGA, with subsequent Lineweaver–Burk analysis revealing competitive inhibition against the enzymes (Table 1 and Figure 1B). GlcNAcstatins A–D inhibit the human enzyme in the low nanomolar to subnanomolar range (Table 1 and Figure 1B). GlcNAcstatins B and D are the most potent hOGA inhibitors with Ki values of 0.4 and 0.7 nM respectively, being, to the best of our knowledge, the most potent hOGA inhibitors currently available. GlcNAcstatins A and C are equally potent with Ki values of 4.3 nM and 4.4 nM respectively. GlcNAcstatin E inhibits only in the micromolar range.
Tuning of hOGA/Hex selectivity
All GlcNAcstatin derivatives were evaluated against human lysosomal Hexs to investigate their potential selectivity towards hOGA (Table 1 and Figure 1C). GlcNAcstatin A is the smallest GlcNAcstatin family member. It carries the N-acetyl moiety and the carboxymethyl group, and inhibits hOGA in the nanomolar range. Assaying lysosomal HexA/B shows that the compound equally potently inhibits the GH20 enzymes (Ki of 550 pM) (Table 1). The first modification we addressed was to substitute the carboxymethyl group to obtain GlcNAcstatin B, similar to previous work showing that a β-glycosidase from C. saccharolyticum is more potently inhibited with a phenylethyl-substituted glucoimidazole [47]. Indeed, GlcNAcstatin B is approx. 10-fold more potent against hOGA (Ki of 420 pM) than GlcNAcstatin A. However, it also inhibits HexA/B more potently, giving a Ki of 170 pM (Table 1). The IC50 data show that GlcNAcstatin B is (to the best of our knowledge) also the most potent HexA/B inhibitor currently known, with a Ki of 170 pM, followed by GlcNAcstatin A (Ki of 550 pM). With these suitably potent inhibitors in hand we attempted to address selectivity by modifying the N-acetyl group. The addition of a single methyl group to obtain an N-propionyl sidechain (GlcNAcstatin D) already leads to different inhibition profiles between hOGA and human lysosomal HexA/HexB (Table 1), with a 15-fold reduction in inhibition of HexA/B. Further extension to an isobutyl group (GlcNAcstatin C) reduces HexA/B inhibition more than 200-fold, giving selectivities of 164-fold for hOGA, whereas incorporation of a larger imidazole group resulted in loss of both potency and selectivity (Table 1 and Figure 1C).
Probing key GlcNAcstatin binding residues
To further understand the structural basis for selectivity, we determined the crystal structure of CpOGA in complex with GlcNAcstatin D (Figure 2). These data show that GlcNAcstatin D binds deep into the active site of CpOGA, with the sugar moiety adopting a 4E conformation, and assuming the same conformation as observed for the GlcNAcstatin C complex [41]. Interestingly, the structural data point towards two active site residues that may play a key role in the surprisingly large difference in activity and selectivity of the GlcNAcstatins towards the hOGA/CpOGA and HexA/B. The only non-conserved residue near the N-acetyl binding pocket in CpOGA is Val331, corresponding to Cys215 in hOGA. Unbiased |Fo|−|Fc| density of the GlcNAcstatin D complex defines that the N-propionyl group points towards this non-conserved Val331 and occupies a single defined conformation, interacting with the Cβ/γ carbons (Figure 2A). Furthermore, the phenyl moiety from GlcNAcstatin D is seen to interact with a solvent-exposed tryptophan residue (Trp490), similar to the phenyl moiety from PUGNAc and GlcNAcstatin C in the respective CpOGA complexes [28,41] (Figure 2B). From a sequence alignment between CpOGA and hOGA, it is not clear whether an equivalently positioned aromatic residue exists in the human enzyme [28]. The relative contributions to binding of these residues were probed by mutagenesis in CpOGA. Mutation of Val331 to cysteine resulted in a mutant enzyme with unaltered steady-state kinetics compared with wild-type CpOGA (Table 2). However, Lineweaver–Burk analysis using GlcNAcstatin C as the inhibitor, gives a 25-fold reduced Ki of 98 pM (Table 2) compared with the wild-type enzyme. Thus although the cysteine, conserved in metazoan OGAs, is not involved in catalysis, it does contribute to defining the shape of the N-acetyl pocket.
Mutation of Trp490 to alanine resulted in a mutant enzyme that showed a 30-fold reduction in Km when assayed with the pseudo-substrate 4MU-NAG (Table 2). Inhibition by GlcNAcstatin C is similarly affected, with the Ki for W490A-CpOGA being almost 20-fold reduced (Ki=74 pM compared with Ki=4 pM for the wild-type enzyme; Table 2). In comparison, PUGNAc exhibits a 3-fold reduced binding affinity against W490A-CpOGA [28]. However, structural comparisons show that the PUGNAc phenylcarbamate moiety occupies a different position in the active site of CpOGA (Figure 2B) and lacks the stacking interactions with Trp490 that are observed in the GlcNAcstatin complexes. In the absence of an hOGA crystal structure, it remains unknown whether an equivalent of Trp490 (His433 in Bacillus thetaiotaomicron OGA, [29]) exists in the human enzyme.
GlcNAcstatins effectively induce cellular hyper-O-GlcNAcylation at low nanomolar concentrations
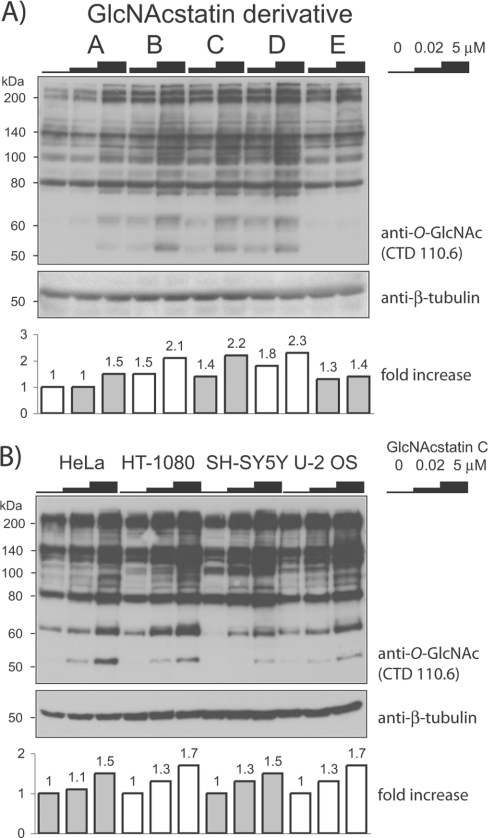
The intended application of the GlcNAcstatins is to inhibit hOGA in live human cells, resulting in hyper-O-GlcNAcylation by disrupting the balance between O-GlcNAc transfer and hydrolysis. Such modulation of O-GlcNAc levels would allow for the study of O-GlcNAc-dependent signal transduction processes. To evaluate the use of the GlcNAcstatins for cell biological studies, HEK-293 cells were exposed to various concentrations of GlcNAcstatins for 6 h, followed by investigation of O-GlcNAc levels on cellular proteins by Western blot analysis using an anti-O-GlcNAc antibody (CTD110.6) (Figure 3A). GlcNAcstatins B–D increase cellular O-GlcNAc levels of numerous intracellular proteins when used at concentrations as low as 20 nM. GlcNAcstatins A and E appear to be less potent as quantitatively assessed from the Western blots, requiring micromolar concentrations in the cell-based assay for a marked effect inside the cells. For GlcNAcstatin E, this is in agreement with the in vitro inhibition data that show that this compound is the weakest hOGA inhibitor (Table 1). The reduced cellular activity of GlcNAcstatin A could be due to differences in membrane permeability resulting from the less hydrophobic nature of the C-2 carboxymethyl substituent. Taken together, these results suggest that GlcNAcstatins are cell-permeant compounds that modulate O-GlcNAcylation levels within the cells by inhibiting hOGA.
Figure 3. Immunoblot detection of O-GlcNAc modifications on cellular proteins using an anti-O-GlcNAc antibody.
The increase in O-GlcNAc levels in comparison with untreated samples is shown in the histogram underneath the blot. (A) HEK-293 cells were treated with GlcNAcstatins A–E for 6 h with the concentrations indicated. (B) GlcNAcstatin C was added to HeLa, HT-1080, SH-SY5Y or U-2 OS cells for 6 h with the identical inhibitor concentrations as in (A). The molecular mass in kDa is indicated on the left-hand side of each blot.
We also have investigated the potency of the most selective GlcNAcstatin (GlcNAcstatin C) against a range of different cell lines (Figure 3B). HeLa (adenocarcinoma), HT-1080 (fibrosarcoma), SH-SY5Y (neuroblastoma) and U-2 OS (osteosarcoma) cells were treated for 6 h with 20 nM and 5 μM inhibitor concentrations. In comparison with the untreated cells, whole-cell lysate analysis with the anti-O-GlcNAc-antibody showed a concentration-dependent hyper-O-GlcNAcylation in all four cell lines, with an increase already detectable at a 20 nM concentration of the compound (Figure 3B).
Concluding remarks
We have explored a new family of potent and competitive OGA inhibitors, the GlcNAcstatins, based on the GlcNAc–imidazole scaffold. Based on structural analysis with GlcNAcstatin C [41] and GlcNAcstatin D in complex with CpOGA we can explain the potency of this inhibitor family. All GlcNAcstatins interact with the active site of OGAs forming at least eight conserved hydrogen bonds (Figures 2A and 2B), the pyranose ring adopts a favoured 4E conformation, and conserved interactions with Asp297 and Asn396 force the N2 substituent to adopt a conformation compatible with the proposed substrate-assisted catalytic mechanism [20,28,29], with the carbonyl oxygen approaching the sp2 hybridized carbon to within 3.4 Å.
GlcNAcstatins A–D inhibit hOGA in the low-nanomolar to sub-nanomolar range, while CpOGA is inhibited in the low picomolar range, implying that there are some (minor) differences in the GlcNAc-binding pocket between hOGA and CpOGA, even though the active sites of these enzymes are almost identical as assessed by sequence alignment [28]. We have identified two key active-site residues, Val331/Trp490, which are responsible for stronger interactions of GlcNAcstatins with CpOGA compared with hOGA. It still remains unknown to what extent the GlcNAcstatin aglycon can be modified in order to increase the binding affinity for the human active site, given the possible structural divergence between the human/bacterial enzymes in this area.
GlcNAcstatins are to the best of our knowledge the most potent competitive inhibitors of hOGA. They can be used in cell-based assays in nanomolar concentrations to increase cellular O-GlcNAc levels in vivo in a range of human cell lines. Thus GlcNAcstatins provide a valuable tool for functional investigations of this post-translational modification and its involvement in signalling pathways within the eukaryotic cell.
ACKNOWLEDGEMENTS
We thank the European Synchrotron Radiation Facility, Grenoble, for the time at beamline ID14-1. We thank Adel Ibrahim (University of Dundee, Cloning Service, DSTT) for cloning of the hOGA construct and Sharon Shepherd and Mark Dorward for protein expression and purification.
FUNDING
This work was supported by a Wellcome Trust Senior Fellowship and a Lister Institute for Preventive Medicine Research Prize. H.C.D. is supported by the College of Life Sciences Alumni Studentship.
References
- 1.Torres C. R, Hart G. W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes: evidence for O-linked GlcNAc. J. Biol. Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 2.Zachara N. E., Hart G. W. O-GlcNAc modification: a nutritional sensor that modulates proteasome function. Trends Cell Biol. 2004;14:218–221. doi: 10.1016/j.tcb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Love D. C., Hanover J. A. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Science STKE. 2005;312:1–14. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 4.Hart G. W., Housley M. P., Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 5.Chou T. Y., Hart G. W. O-linked N-acetylglucosamine and cancer: messages from the glycosylation of c-Myc. Adv. Exp. Med. Biol. 2001;491:413–418. doi: 10.1007/978-1-4615-1267-7_26. [DOI] [PubMed] [Google Scholar]
- 6.Liu K., Paterson A. J., Konrad R. J., Parlow A. F., Jimi S., Roh M., Chin E. J., Kudlow J. E. Streptozotocin, an O-GlcNAcase inhibitor, blunts insulin and growth hormone secretion. Mol. Cell. Endocrinol. 2002;194:135–146. doi: 10.1016/s0303-7207(02)00155-7. [DOI] [PubMed] [Google Scholar]
- 7.Donadio C., Lobo C., Tosina M., de la Rosa V., Martin-Rufian M., Campos-Sandoval J. A., Mates J. M., Marquez J., Alonso F. J., Segura J. A. Antisense glutaminase inhibition modifies the O-GlcNAc pattern and flux through the hexosaminepathway in breast cancer cells. J. Cell. Biochem. 2008;103:800–811. doi: 10.1002/jcb.21449. [DOI] [PubMed] [Google Scholar]
- 8.Griffith L. S., Schmitz B. O-linked N-acetylglucosamine is upregulated in Alzheimer brains. Biochem. Biophys. Res. Commun. 1995;213:424–431. doi: 10.1006/bbrc.1995.2149. [DOI] [PubMed] [Google Scholar]
- 9.Yao P., Coleman P. Reduction of O-linked N-acetylglucosamine-modified assembly protein-3 in Alzheimer's disease. J. Neurosci. 1998;18:2399–2411. doi: 10.1523/JNEUROSCI.18-07-02399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K., Paterson A., Zhang F., McAndrew J., Fukuchi K., Wyss J., Peng L., Hu Y., Kudlow J. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J. Neurochem. 2004;89:1044–1055. doi: 10.1111/j.1471-4159.2004.02389.x. [DOI] [PubMed] [Google Scholar]
- 11.Wells L., Hart G. W. O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 2003;546:154–158. doi: 10.1016/s0014-5793(03)00641-0. [DOI] [PubMed] [Google Scholar]
- 12.Dias W., Hart G. O-GlcNAc modification in diabetes and Alzheimer's disease. Mol. Biosyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 13.McClain D. A., Lubas W. A., Cooksey R. C., Hazel M., Parker G. J., Love D. C., Hanover J. A. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10695–10699. doi: 10.1073/pnas.152346899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Copeland R. J., Bullen J. W., Jr, Hart G. W. Crosstalk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 2008;295:17–28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macauley M. S., Bubb A. K., Martinez-Fleites C., Davies G. J., Vocadlo D. J. Elevation of global O-GlcNAc levels in 3T3-L1 adipocytes by selective inhibition of O-GlcNAcase does not induce insulin resistance. J. Biol. Chem. 2008;283:34687–34695. doi: 10.1074/jbc.M804525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coutinho P., Deleury E., Davies G., Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 17.Henrissat B., Davies G. Structural and sequence-based classification of glycosidehydrolases. Curr. Opin. Struct. Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 18.Hurtado-Guerrero R., Dorfmueller H. C., van Aalten D. M. F. Molecular mechanisms of O-GlcNAcylation. Curr. Opin. Struct. Biol. 2008;18:551–557. doi: 10.1016/j.sbi.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Dong L. Y., Hart G. W. Purification and characterization of an O-GlcNAc selective N-acetyl-β-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 20.Macauley M. S., Whitworth G. E., Debowski A. W., Chin D., Vocadlo D. J. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 2005;280:25313–25322. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- 21.Horsch M., Hoesch L., Vasella A., Rast D. M. N-acetylglucosaminono-1,5-lactoneoxime and the corresponding (phenylcarbamoyl)oxime: novel and potent inhibitors of β-N-acetylglucosaminidase. Eur. J. Biochem. 1991;197:815–818. doi: 10.1111/j.1432-1033.1991.tb15976.x. [DOI] [PubMed] [Google Scholar]
- 22.Haltiwanger R. S., Grove K., Philipsberg G. A. Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-β-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino-N-phenylcarbamate. J. Biol. Chem. 1998;273:3611–3617. doi: 10.1074/jbc.273.6.3611. [DOI] [PubMed] [Google Scholar]
- 23.Xing D., Feng W., Nöt L. G., Miller A. P., Zhang Y., Chen Y.-F., Majid-Hassan E., Chatham J. C., Oparil S. Increased protein O-GlcNAc modification inhibits inflammatory and neointimal responses to acute endoluminal arterial injury. Am. J. Physiol. Heart. Circ. Physiol. 2008;295:H335–H342. doi: 10.1152/ajpheart.01259.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lüdemann N., Clement A., Hans V. H., Leschik J., Behl C., Brandt R. O-Glycosylation of the tail domain of neurofilament protein M in human neurons and in spinal cord tissue of a rat model ofamyotrophic lateral sclerosis (ALS) J. Biol. Chem. 2005;280:31648–31658. doi: 10.1074/jbc.M504395200. [DOI] [PubMed] [Google Scholar]
- 25.Akimoto Y., Kawakami H., Yamamoto K., Munetomo E., Hida T., Hirano H. Elevated expression of O-GlcNAc-modified proteins and O-GlcNAc transferase in corneas of diabetic Goto–Kakizaki rats. Invest. Ophthalmol. Vis. Sci. 2003;44:3802–3809. doi: 10.1167/iovs.03-0227. [DOI] [PubMed] [Google Scholar]
- 26.Guinez C., Mir A.-M., Dehennaut V., Cacan R., Harduin-Lepers A., Michalski J.-C., Lefebvre T. Protein ubiquitination is modulated by O-GlcNAc glycosylation. FASEB J. 2008;22:2901–2911. doi: 10.1096/fj.07-102509. [DOI] [PubMed] [Google Scholar]
- 27.Kim J., Amorelli B., Abdo M., Thomas C. J., Love D. C., Knapp S., Hanover J. A. Distinctive inhibition of O-GlcNAcase isoforms by an α-GlcNAc thiolsulfonate. J. Am. Chem. Soc. 2007;129:14854–14855. doi: 10.1021/ja076038u. [DOI] [PubMed] [Google Scholar]
- 28.Rao V., Dorfmueller H. C., Villa F., Allwood M., Eggleston I. M., van Aalten D. M. F. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25:1569–1578. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dennis R. J., Taylor E. J., Macauley M. S., Stubbs K. A., Turkenburg J. P., Hart S. J., Black G. N., Vocadlo D. J., Davies G. J. Structure and mechanism of a bacterial β-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol. 2006;13:365–371. doi: 10.1038/nsmb1079. [DOI] [PubMed] [Google Scholar]
- 30.Whitworth G. E., Macauley M., Stubbs K., Dennis R., Taylor E., Davies G., Grieg I., Vocadlo D. Analysis of PUGNAc and NAG-thiazoline as transition state analogues for human O-GlcNAcase: mechanistic and structural insights into inhibitor selectivity and transition state poise. J. Am. Chem. Soc. 2007;129:635–644. doi: 10.1021/ja065697o. [DOI] [PubMed] [Google Scholar]
- 31.Mahuran D. J. Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim. Biophys. Acta. 1999;1455:105–138. doi: 10.1016/s0925-4439(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 32.Mark B. L., Vocadlo D. J., Zhao D. L., Knapp S., Withers S. G., James M. N. G. Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 β-N-acetylhexosaminidase inhibitor. J. Biol. Chem. 2001;276:42131–42137. doi: 10.1074/jbc.M107154200. [DOI] [PubMed] [Google Scholar]
- 33.Stubbs K. A., Zhang N., Vocadlo D. J. A divergent synthesis of 2-acyl derivatives of PUGNAc yields selective inhibitors of O-GlcNAcase. Org. Biomol. Chem. 2006;4:839–845. doi: 10.1039/b516273d. [DOI] [PubMed] [Google Scholar]
- 34.Kim E. J., Perreira M., Thomas C. J., Hanover J. A. An O-GlcNAcase-specific inhibitor and substrate engineered by the extension of the N-acetyl moiety. J. Am. Chem. Soc. 2006;128:4234–4235. doi: 10.1021/ja0582915. [DOI] [PubMed] [Google Scholar]
- 35.Shanmugasundaram B., Debowski A., Dennis R., Davies G., Vocadlo D., Vasella A. Inhibition of O-GlcNAcase by a gluco-configured nagstatin and a PUGNAc-imidazole hybrid inhibitor. Chem. Commun. (Camb.) 2006;42:4372–4374. doi: 10.1039/b612154c. [DOI] [PubMed] [Google Scholar]
- 36.Mark B. L., Mahuran D. J., Cherney M. M., Zhao D., Knapp S., James M. N. G. Crystal structure of human β-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay–Sachs disease. J. Mol. Biol. 2003;327:1093–1109. doi: 10.1016/s0022-2836(03)00216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemieux M. J., Mark B. L., Cherney M. M., Withers S. G., Mahuran D. J., James M. N. G. Crystallographic structure of human β-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J. Mol. Biol. 2006;359:913–929. doi: 10.1016/j.jmb.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuzwa S., Macauley M., Heinonen J., Shan X., Dennis R., He Y., Whitworth G., Stubbs K., McEachern E., Davies G., Vocadlo D. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 39.Aoyagi T., Suda H., Uotani K., Kojima F., Aoyama T., Horiguchi K., Hamada M., Takeuchi T. Nagstatin, a new inhibitor of N-acetyl-β-D-glucosaminidase, produced by Streptomyces amakusaensis MG846-fF3. Taxonomy, production, isolation, physico-chemical properties and biological activities. J. Antibiot. 1992;45:1404–1408. doi: 10.7164/antibiotics.45.1404. [DOI] [PubMed] [Google Scholar]
- 40.Aoyama T., Naganawa H., Suda H., Uotani K., Aoyagi T., Takeuchi T. The structure of nagstatin, a new inhibitor of N-acetyl-β-D-glucosaminidase. J. Antibiot. 1992;45:1557–1558. doi: 10.7164/antibiotics.45.1557. [DOI] [PubMed] [Google Scholar]
- 41.Dorfmueller H. C., Borodkin V. S., Schimpl M., Shepherd S. M., Shpiro N. A., van Aalten D. M. F. GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-GlcNAcylation levels. J. Am. Chem. Soc. 2006;128:16484–16485. doi: 10.1021/ja066743n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heightman T. D., Vasella A. T. Recent insights into inhibition, structure, and mechanism of configuration-retaining glycosidases. Angew. Chem.-Int. Edit. 1999;38:750–770. doi: 10.1002/(SICI)1521-3773(19990315)38:6<750::AID-ANIE750>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 43.Pathak S., Dorfmueller H. C., Borodkin V. S., van Aalten D. M. F. Chemical dissection of the link between streptozotocin, O-GlcNAc, and pancreatic cell death. Chem. Biol. 2008;15:799–807. doi: 10.1016/j.chembiol.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuettelkopf W., van Aalten D. M. F. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:D1355–D1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:D2126–D2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:D240–D255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Panday N., Canac Y., Vasella A. Very strong inhibition of glucosidases by C(2)-substituted tetrahydroimidazopyridines. Helv. Chim. Acta. 2000;83:58–79. [Google Scholar]