Abstract
Pharmacokinetic drug interactions can lead to serious adverse events, and the evaluation of a new molecular entity’s drug–drug interaction potential is an integral part of drug development and regulatory review prior to its market approval. Alteration of enzyme and/or transporter activities involved in the absorption, distribution, metabolism, or excretion of a new molecular entity by other concomitant drugs may lead to a change in exposure leading to altered response (safety or efficacy). Over the years, various in vitro methodologies have been developed to predict drug interaction potential in vivo. In vitro study has become a critical first step in the assessment of drug interactions. Well-executed in vitro studies can be used as a screening tool for the need for further in vivo assessment and can provide the basis for the design of subsequent in vivo drug interaction studies. Besides in vitro experiments, in silico modeling and simulation may also assist in the prediction of drug interactions. The recent FDA draft drug interaction guidance highlighted the in vitro models and criteria that may be used to guide further in vivo drug interaction studies and to construct informative labeling. This report summarizes critical elements in the in vitro evaluation of drug interaction potential during drug development and uses a case study to highlight the impact of in vitro information on drug labeling.
Key words: drug development, drug–drug interaction, new drug application, prediction, regulatory and guidance
INTRODUCTION
The desirable and undesirable effects of a drug are generally related to its concentration at the sites of action, which in turn is related to the amount administered (dose) and to the drug’s absorption, distribution, metabolism, and/or excretion (ADME). All these processes can be influenced by both intrinsic and extrinsic factors such as age, race, gender, disease states, concomitantly administered drugs, food, and juices (1). Observed changes arising from pharmacokinetic drug–drug interactions can be substantial such as an order of magnitude or more increase or decrease in the blood and tissue concentrations of a drug or its metabolites. Many of these interactions involved inhibition of metabolizing enzymes and transporters, resulting in increased systemic exposure and subsequent adverse drug reactions. In other cases, induction of metabolizing enzymes and transporters resulted in reduced systemic exposure leading to a risk of loss of efficacy of co-administered drugs. Therefore, drug interaction potential is recognized as an important consideration in the evaluation of a new molecular entity (NME) (2,3) and is an integral part of drug development and regulatory review prior to NME’s market approval.
Several FDA guidance documents developed since the mid-1990s and the most recent draft drug interaction guidance released in September 2006 reflect the Agency’s view that the metabolism of an NME and its potential on inhibition and induction of key metabolizing enzymes and transporters should be defined (4–6). Potential drug–drug interactions resulting from the effects of other drugs on NME and the effects of NME on other drugs should be explored during drug development to ensure an adequate assessment of an NME’s safety and effectiveness (6,7). An integrated approach (in vitro and in vivo) to the evaluation of an NME’s drug interaction potential may reduce the number of unnecessary studies and optimize knowledge. The recent FDA draft drug interaction guidance highlighted the in vitro models and criteria that may be used to guide further in vivo drug interaction studies (6). Besides in vitro experiments, in silico modeling and simulation may also assist in the prediction for drug interactions.
CURRENT STATUS AND RECOMMENDATION IN PREDICTING IN VIVO DRUG INTERACTIONS BASED ON IN VITRO EVALUATION
Pharmacokinetic drug interactions can occur via inhibition or induction of metabolic enzymes or transporters. Evaluation of an NME’s drug–drug interaction potential is an integrated part of drug development and regulatory review prior to its market approval. In general, three basic questions need to be addressed in the new drug application: (1) Will other drugs alter the exposure to an NME? (2) Will an NME alter the exposure to other drugs? (3) Are these alterations in exposure clinically relevant to warrant dose adjustment? While drug interactions can be evaluated via specific clinical studies in healthy subjects or patients, in vitro approaches are now becoming common as a critical first step in the assessment of drug interaction potential via specific pathways, and knowledge obtained from these studies may help reduce the number of unnecessary studies. The experiments are generally conducted during early phase of drug development process. Results from the in vitro studies can be used to predict in vivo interaction and guide the need for further in vivo study evaluation. The 2006 FDA draft drug interaction guidance has specific recommendation as to how to use in vitro models to address drug interaction potential and, for the first time, includes criteria for evaluating transporter-based drug interactions (6).
Prediction of Metabolism-Mediated Drug Interaction
The following cytochrome P450 (CYP) enzymes are recommended for routine assessment to identify potential P450-mediated drug interactions: CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A. Evaluation of phase II enzymes is highly encouraged if applicable (6).
Understanding Whether an NME is a Substrate for a Particular P450 Enzyme
Understanding which P450 enzyme is responsible for the metabolism of an NME is important in the evaluation of drug interaction potential. Drug interaction is likely to occur between such a drug and known inhibitors or inducers of that specific pathway if it contributes >25% to the total clearance of the NME. It is also important for selecting the interacting drugs to evaluate drug interaction in vivo, determining the impact of polymorphic enzyme activity on drug disposition, and deciding whether a multiple inhibitor study may be warranted. In general, the likelihood of drug interactions increases when a compound has a high affinity for a single metabolizing enzyme compared with a compound with affinity for a number of different enzymes.
A set of experiments (also known as reaction phenotyping) is conducted to identify the specific enzymes responsible for the metabolism of an NME. Oxidative and hydrolytic reactions involve cytochrome P450 (CYP) and non-CYP enzymes. For many drugs, transferase reactions (involving phase II enzymes) are preceded by oxidation or hydrolysis of the drug. However, direct transferase reactions may represent a major metabolic pathway for compounds containing the requisite functional groups. The guidance recommends that the metabolic profile of the NME be investigated using human liver tissues such as freshly isolated liver slices, freshly prepared or cryopreserved human hepatocytes, subcellular liver tissue fractions such as liver S9 fraction, liver microsomes, or recombinant complementary DNA (cDNA)-expressed microsomes for a particular CYP enzyme. If human in vivo data indicate that CYP enzymes contribute >25% of the total clearance of the NME, studies should be conducted using human liver microsomes or recombinant enzymes to determine the individual CYP enzymes responsible for the drug’s metabolism. If an NME is a substrate of a particular CYP, an in vivo interaction with a strong inhibitor or inducer for that CYP is needed to determine whether inhibition or induction of this particular pathway may lead to a change in the NME’s pharmacokinetics. Negative results would alleviate further in vivo studies with less strong inhibitors or inducers. If results are positive, further clinical studies with less potent inhibitors or inducers would generally be needed to provide guidance on dosage adjustment.
If an NME is metabolized by a polymorphic enzyme (such as CYP2D6, CYP2C9, or CYP2C19), the extent of drug interactions (inhibition or induction) may be different depending on the subjects’ genotype for the specific enzyme being evaluated. For example, subjects lacking the major, polymorphic clearance pathway will show reduced total metabolism. But alternative pathways may become quantitatively more important and need to be understood and studied appropriately. In general, the comparison of pharmacokinetic parameters of this NME in poor metabolizers versus extensive metabolizers may indicate the extent of interaction of this drug with strong inhibitors of these enzymes and make interaction studies with such inhibitors unnecessary. When the above study shows significant interaction, further evaluation with weaker inhibitors may be necessary.
Understanding Whether an NME is an Inhibitor for a Particular P450 Enzyme
If an NME is an inhibitor of a specific CYP enzyme, it may have the potential to inhibit the metabolism of a substrate drug of that CYP enzyme. The inhibition potential is usually evaluated using human liver microsomes or cDNA-expressed microsomes. An in vitro inhibition constant (Ki) that reflects the inhibitory effect of the NME is determined and its value is compared to clinically relevant concentrations. Because hepatocyte concentration is not easily measured, plasma concentrations are often used for this estimation. For an NME as a reversible inhibitor for a particular CYP enzyme, the guidance suggests that in vivo inhibition studies with representative substrates for that enzyme are needed if the calculated [I]/Ki is >0.1, where [I] is the estimated mean maximum total (bound and unbound) plasma concentration (Cmax) at steady state of the highest clinical dose and Ki is the inhibition constant for the NME measured in vitro. The total plasma concentration (instead of the free plasma concentration) is used as a conservative estimate to predict the expected higher hepatic concentration and to avoid false negative results when free plasma concentration is used in the I/Ki calculation. When evaluating the potential of the NME to inhibit CYP3A, at least two structurally different CYP3A substrates such as midazolam and testosterone should be used (8,9). If the [I]/Ki is >0.1 from either substrate, an in vivo interaction study is recommended.
In addition, time-dependent inhibition (TDI) potential for an NME should be evaluated. TDI is a collective term for a change (often an increase) in potency of CYP inhibitors during in vitro incubation or dosing period in vivo. Potential mechanisms include the formation of a more inhibitory metabolite or mechanism-based inhibition: the inactivation of enzymes by metabolic products that form haem or protein adducts. Over the past decade, time-dependent CYP inhibition has been recognized to be responsible for some important drug interactions in vivo (10). For example, the calcium channel blocker, mibefradil, is a potent mechanism-based CYP3A inhibitor and P-glycoprotein (P-gp) inhibitor (11). Mibefradil was withdrawn in 1998 shortly after its approval as a consequence of serious drug–drug interactions with substrates of CYP3A and/or P-gp (11,12). Therefore, TDI should be studied and its possible in vivo drug interaction potential needs to be projected.
Time-dependent inhibition is mainly assessed in vitro using microsomes or hepatocytes and has been incorporated increasingly in drug discovery process (13,14). Although inhibition parameters (i.e., kinact and KI) can be readily obtained in vitro, prediction of time-dependent inhibition in vivo remains challenging because of the complexity of the mechanism as compared to reversible inhibition. Thus, a decision tree with regard to the evaluation of mechanism-based inhibition in vivo based on in vitro parameters similar to the evaluation of reversible inhibition outlined in the FDA draft guidance needs to be developed.
Understanding Whether an NME is an Inducer for a Particular P450 Enzyme
An NME that induces a CYP enzyme can cause drug interactions with substrate drugs for that particular pathway leading to enhanced clearance. Human primary hepatocytes are the preferred experimental system for the evaluation of P450 induction. The results of a recent survey of the practice in pharmaceutical industries indicated general consensus that human hepatocyte culture induction studies are the best predictor of in vivo induction (15). However, there appeared to be no standard methods for conducting these studies and no consistent criteria for determining whether a clinical drug–drug interaction study should be carried out (15). The FDA guidance suggests that induction studies be carried out using freshly isolated or cryopreserved human hepatocytes or immortalized cell lines including a positive control. Hepatocytes need to be prepared from at least three individual donor livers because of the known inter-individual differences in induction potential. When using immortalized cell lines, the experiment needs to be conducted in triplicate. If the increase in enzyme activity for NME-treated cells is >40% of a positive control in any one batch of hepatocytes or immortalized cell lines, the NME is considered to be an enzyme inducer and in vivo induction studies are recommended. An alternative endpoint is the use of an EC50 (effective concentration at which 50% maximal induction occurs) value, an index that can be used to compare the potency of different compounds. Relative induction score approach has also been reported for prediction of induction potential (16).
Studies have indicated that activation of the nuclear receptor, pregnane X receptor, results in the co-induction of CYP3A and CYP2C. Thus, a negative in vitro result for CYP3A induction may eliminate the need for additional induction studies for both CYP3A and CYP2C enzymes. However, whether CYP2C and CYP3A are always co-induced may need further validation. Because CYP1A2 induction is mainly via aryl hydrocarbon receptor (AhR), CYP1A2 is not likely to be co-induced with CYP3A. For CYP2B6, although overlap exists between CYP2B6 and CYP3A inducers, there are data suggesting that certain CYP2B6 inducers selectively bind to the constitutive androstane receptor (CAR), and these inducers do not show significant induction for CYP3A (17). Therefore, the potential for induction of CYP1A2 and CYP2B6 should be evaluated separately regardless of the CYP3A induction result.
Phase II Enzymes
Phase II enzymes have been recognized to play important roles in the pharmacokinetics of drugs. Historically, these enzymes have attracted less attention than CYP enzymes in drug interaction potential evaluation, most likely due to the lack of tools to study them and/or a lower incidence of observed adverse drug–drug interactions. Exceptions include the polymorphisms of N-acetyltransferases (18) resulting in fast and slow acetylators and acyl glucuronidation by UDP glucuronosyl transferases (UGTs) (19); both can lead to the formation of toxic metabolites. Recently, there has been an increased interest in drug–drug interactions involving UGTs. For example, polymorphism of UGT1A1 was shown to affect exposure of SN-38, an active metabolite of irinotecan, which has efficacy and safety implications (20,21).
Similar to the CYP enzymes, UGTs are encoded by a UGT gene “superfamily” with 17 human UGT proteins identified to date (22). Unlike CYP enzymes, there is no consensus with respect to the tools, i.e., enzyme sources, selective substrates, inhibitors, and inducers for studying the UGT enzymes. Recombinant human UGTs, many are available from commercial sources, have been used to investigate the individual UGT enzymes responsible for the formation of a drug glucuronide metabolite. UGT1A1, 1A3, 1A4, 1A6, 1A9, 2B7, and 2B15 are considered to be the enzymes of the greatest importance in hepatic drug elimination (Zhang Y., et. al., book chapter submitted).
Establishing in vitro–in vivo correlation for drugs that are eliminated by glucuronidation has been challenging as compared to CYP enzymes. For example, the use of microsomes to determine the intrinsic clearance of drugs that are eliminated by glucuronidation is problematic because UGTs are integral proteins of the endoplasmic reticulum and are dependent on lipid for catalytic activity; both are variable parameters not controlled well in in vitro system.
Prediction of Transporter-Mediated Drug Interaction
In addition to the effects of drug metabolizing enzymes on the pharmacokinetics of drugs, increasing attention is being given to transporters where emerging evidence indicates their important role in modulating drug absorption, distribution, metabolism, and elimination as well as the historical importance of transporters in the development of drug resistant tumors. Transporters, acting alone or in concert with drug metabolizing enzymes, can affect the pharmacokinetics and/or pharmacodynamics of a drug. Of the various transporters, P-gp is the most well and extensively studied transporter.
Understanding Whether an NME is a Substrate for P-gp
To test whether the NME is a P-gp substrate, bidirectional experiments of cell transport are carried out with the NME to determine the net flux ratio for the basolateral to apical (B→ A) and apical to basolateral transport (A→B). If the efflux ratio, (B→A) to (A→B), is ≥2 and addition of P-gp inhibitors to the experiment decreases the net flux ratio by more than 50% or decreases the ratio to close to 1, then the NME is a potential P-gp substrate. A net flux ratio “cutoff” higher than 2 or a relative ratio to positive controls may be used to avoid false positives if a ratio of 2 is deemed non-discriminative as supported by prior experience with the cell system used. If in vitro experiments demonstrate that an NME is a P-gp substrate, additional drug-specific factors may be considered before determining whether an in vivo drug interaction study is warranted. For example, the bioavailability of the Biopharmaceutics Classification System Class 1 (23) or the Biopharmaceutics Drug Disposition Classification System Class 1 (24) NMEs that are highly soluble, highly permeable, and highly metabolized may not be significantly affected by a co-administered drug that is a P-gp inhibitor, and thus, an in vivo interaction study may not be needed. Nonetheless, it is recognized that the effects of P-gp inhibitors at the tissue levels (e.g., tumor or brain) cannot be easily assessed. If an NME is a substrate for both CYP enzyme and transporter, selection of inhibitors for studying inhibition needs to consider the significant overlap between enzymes and transporters (e.g., CYP3A and P-gp). A “dual” inhibitor for enzyme and transporter may be selected to study the maximal inhibition effect, although specific attribution of an AUC change to transporter or CYP enzyme may not be possible.
For NME that has low cell permeability due to lack of basolateral transporters in the cell lines for P-gp evaluation, the use of membrane vesicles may be an alternative method to understand the “intrinsic” affinity of NME to P-gp.
Understanding Whether an NME is an Inhibitor for P-gp
To test whether the NME is a P-gp inhibitor, bidirectional experiments of cell transport are carried out after adding varying concentrations of the NME to both sides of the monolayer followed by adding known P-gp substrates to the apical or basal side of the monolayer. The NME is a potential P-gp inhibitor if the net flux ratio of a P-gp probe substrate is decreased in the presence of the NME. To determine the potency of inhibition, an IC50 (concentration that inhibits 50%) or Ki value is determined.
The criteria for determining whether an in vivo drug interaction study is needed are evolving. The draft guidance published in Sept 2006 recommends [I]/IC50 of 0.1 as the “cutoff” for further in vivo evaluation, where [I] represents the total Cmax (bound plus unbound) at steady state at the highest clinical dose for NME (6). This ratio was adopted from criteria used to determine whether an NME is an inhibitor of P450 metabolizing enzymes. In contrast to P450 enzymes in the liver or transporters in the kidney where [I] reflects the systemic Cmax, [I] concentrations at the luminal side of GI may be more relevant when evaluating P-gp inhibition by the NME following oral administration.
To provide better criteria for recommending in vivo inhibition studies, in vitro IC50 (or Ki) values and in vivo inhibition data for marketed drugs and drugs under development, using the prototypic P-gp substrate digoxin, were collected and evaluated (25). Based on the evaluation results, the following alternative criteria are proposed: drugs that exhibit an [I]1/IC50 > 0.1 or [I]2/IC50 > 10 should be evaluated in vivo to determine whether there is clinically relevant P-gp inhibition with digoxin, a P-gp substrate with a narrow therapeutic index, where [I]1 is the mean NME steady-state total Cmax at the highest clinical dose and [I]2 is the theoretical maximal gastrointestinal NME concentration after oral administration estimated by the ratio of the highest clinical dose (mg) to a volume of 250 mL. If an NME meets either criterion, an in vivo drug interaction study with digoxin is recommended. Results from a recent publication from Fenner et al. (26) indicate that the proposed criteria for in vivo P-gp inhibition evaluation are reasonable.
Studying P-gp inhibition with digoxin is clinically relevant and useful because digoxin has a narrow therapeutic index and is one of the few known P-gp substrates that is not a CYP3A substrate. As more information about the interplay of P-gp and CYP3A emerges, the clinical relevance of P-gp may be better understood. Inhibition data obtained with digoxin may be applied to other “pure” P-gp substrates that have a narrow therapeutic index.
It is important to recognize the limitations of only using in vitro IC50 to predict in vivo interactions mediated by P-gp inhibition. The in vitro IC50 determination may be different across different laboratories. Appropriate controls are needed to compare results from different laboratories. Continued data collection is needed to further evaluate the adequacy of these criteria to predict possible in vivo interactions mediated by P-gp. A working group was formed following the October 2008 Transporter Workshop to continue further research in the area towards better predicting in vivo P-gp-mediated interaction based on in vitro data (Dr. Caroline Lee, personal communication).
Understanding Whether an NME is an Inducer for P-gp
Methods for in vitro evaluation for P-gp induction are not well understood. Thus, the P-gp induction potential of an investigational drug can only be evaluated in vivo. Because of similarities in the mechanism of CYP3A and P-gp induction, information from tests of CYP3A inducibility can inform decisions about the induction P-gp. If an NME is found not to induce CYP3A in vitro, no further tests of CYP3A and P-gp induction in vivo are necessary. If a study of the NME’s effect on CYP3A activity in vivo is indicated from a positive in vitro screen but the drug is shown not to induce CYP3A in vivo, then no further test of P-gp induction in vivo is necessary. However, if the in vivo CYP3A induction test is positive, then an additional study of the NME’s effect on a P-gp probe substrate is recommended (6).
Evaluation of Transporters Other Than P-gp
Reports of drug disposition mediated by membrane transporters other than P-gp continue to appear in the literature. For example, OATP1B1 and NTCP may play a major role in the disposition of the HMG-CoA reductase inhibitor rosuvastatin (27,28). A recent clinical study demonstrated that a genetic variation in the hepatic uptake transporter OCT1 is a determinant of metformin pharmacokinetics and may be associated with variation in response to this drug (29,30). Drug interaction potential exists if an NME is a substrate or inhibitor/inducer of transporters. However, routine in vitro studies cannot be recommended for transporters other than P-gp at this time because no consensus has been reached with regard to in vitro methods or probe substrates and inhibitors. Until additional knowledge and technologies are available, recommendations for evaluation of transporter-based drug interactions other than P-gp (e.g., OATP, BCRP, OATs, and OCTs) are on a case by case basis.
A recent DIA/FDA Critical Path Transporter Workshop has discussed the emerging science in transporters (7). An international working group, including members from academia, industry, and the FDA, are working on a whitepaper to highlight the recent progress in this field including in vitro tools and criteria for in vivo drug interaction evaluation for main transporters including P-gp, BCRP, OATP, OCT, and OAT.
In Silico Models
The current FDA guidance uses criteria that are based on in vitro Ki values in combination with in vivo total plasma/blood concentrations to predict the likelihood of drug interactions for NMEs as CYP inhibitors. The limitations of the I/Ki approach have been discussed elsewhere (31–36). For example, using total [I] may over-predict for drugs that are highly protein-bound in plasma. Conversely, using unbound plasma [I] may under-predict for drugs that are highly concentrated in the liver by uptake transporters (37). The use of single inhibitor concentration also poses a potential limitation because the in vivo drug interactions are expected to be dependent on the pharmacokinetic characteristics of both inhibitor and substrate.
Over the years, in vitro to in vivo prediction models have been developed to predict/simulate the magnitude of the interaction based on in vitro results (16,38–41). Commercial software products have been developed as well. Time–concentration profile and/or inter-individual variability of intrinsic factors influencing ADME processes of both substrate and inhibitor drugs based on physiological-based pharmacokinetic prediction approach have been integrated into the modeling algorithm (38,39,42–45). Modeling and simulation of drug interactions in vivo using the physiological-based pharmacokinetic approach appear to be valuable in evaluating the magnitude of drug interaction potential under different clinical scenarios, e.g., different dosing regimen (44,46,47). The tools are helpful not only in interaction prediction but also in clinical study design. Although progresses have been made in the in silico models to predict drug interactions, challenges remain because the lack of the true physiological representation in the models limits the ability to predict in vivo situations such as enzyme and transporter interplay at various tissues, e.g., different interplay of CYP3A4 and P-gp in the intestine vs. in the liver.
Recent NDA Examples
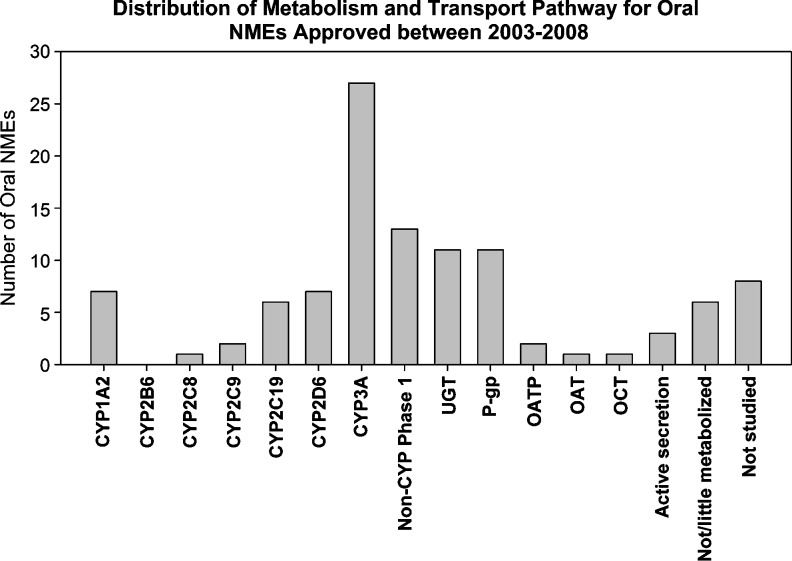
Recent IND and NDA reviews indicate that most pharmaceutical companies conduct recommended in vitro evaluation studies according to the guidance prior to drug approval. For example, a recent review of 121 new molecular entity drugs approved during 2003 and 2008 (up to Dec. 21, 2008) indicated that 88% (57 out of 65) of those intended for oral administration included in vitro study information with regard to which metabolic and/or transport pathways are involved in the ADME process of the drug (Fig. 1) (48). P450 3A is the main P450 enzyme involved in the metabolism of NMEs. In addition, most NMEs were studied for their inhibition or induction potential for major P450 enzymes. The information has greatly enhanced our ability to predict in vivo interaction potential to construct an informative labeling. Besides major CYPs and P-gp, in vitro evaluation studies are increasingly conducted with regard to whether an NME is a substrate or inhibitor for phase II enzymes (mostly UGTs) and transporters other than P-gp (e.g., BCRP, OATP1B1, OAT, and OCT). Although in vitro studies are being conducted, we found that positive in vitro findings are not necessarily always followed by in vivo drug interaction evaluation. In these situations, appropriate language is usually constructed in the labeling based on in vitro results. In some cases, post-marketing drug interaction studies are requested according to clinical need.
Fig. 1.
Distribution of metabolism and transport pathways for NMEs approved between 2003 and 2008 intended for oral administration
For example, ambrisentan, an endothelin receptor antagonist, was approved in 2007 for the treatment of pulmonary arterial hypertension (48). It was found to be a substrate of CYP3A, CYP2C19, UGT1A9, UGT2B7, UGT1A3, OATP, and P-gp. Because the relative contribution of each pathway for ambrisentan is not clear, specific interaction studies with inhibitors for these pathways were not conducted. The “HIGHLIGHT” section of the labeling states that “…based on in vitro data, interactions with P-glycoprotein (P-gp), the organic anion transport protein (OATP), CYP3A4,and CYP2C19 inhibitors, and uridine 5′-diphosphate glucuronosyltransferases (UGTs) would be expected.” The “WARNINGS AND PRECAUTIONS” section of the labeling states that caution should be exercised with ambrisentan when co-administered with cyclosporine A (a CYP3A, OATP, and P-gp inhibitor), strong CYP3A or CYP2C19 inhibitors. Post-marketing studies have been committed to explore the interactions between ambrisentan and a strong inhibitor of CYP2C19 (e.g., omeprazole), cyclosporine A (a strong inhibitor of OATP and P-gp), and rifampin (an inhibitor of OATP and inducer of P-gp, CYP3A, and CYP2C19).
Challenges in Predicting In Vivo Drug Interactions
Our understanding of the relationship between in vitro and in vivo drug–drug interactions and our ability to predict these interactions has improved over the years. The FDA drug interaction guidance (6) has included various decision trees for determining when clinical drug interaction studies are indicated. Depending on the study results, recommendations can then be made whether dosage adjustment is required including suitable language in the labeling. Even without in vivo evaluation, in vitro results are included in the labeling as the basis for cautionary language when appropriate.
In spite of these advances, unexpected drug–drug interactions do occur which could be due to several variables that we do not yet understand or cannot accurately measure. First, the interaction may be due to pharmacodynamic interactions or pharmacokinetic interaction involved with unknown mechanism (e.g., transporter or uncommon metabolic pathways). Second, prediction could be confounded when multiple enzymes or both metabolizing enzymes and transporters are involved in a drug’s disposition. The lack of in vitro models that represent the true physiological environment also limits our ability to predict in vivo situations where multiple drugs are co-administered and concomitant inhibition and induction of metabolic enzymes and transporter could occur. Third, although modeling and simulation approach using in silico models incorporating physiological parameter and inter-individual variability appears promising in comprehensively evaluating the drug interaction potential under different scenarios, an accurate prediction from in vitro data may continue to be hampered by unknown factors to be discovered as science evolves and as the knowledge of the drug accumulates. Therefore, the ability to quantitatively predict the magnitude of in vivo drug interaction from in vitro data is limited. Accurate predictions of the extent of in vivo drug interactions from in vitro studies will require continued efforts. Addressing these issues could improve our abilities to assess drug–drug interactions in vivo from in vitro data.
SUMMARY
Understanding of metabolism, transport, and drug–drug interactions is critical to the benefit/risk assessment of a drug during drug development and regulatory review. Using an integrated approach incorporating in vitro and in vivo metabolism and transport studies to elucidate the underlying mechanisms and to evaluate the potential for drug interactions can reduce the number of studies needed and optimize our knowledge to provide appropriate information in the labeling.
Abbreviations
- ADME
Absorption, distribution, metabolism or excretion
- AhR
Aryl hydrocarbon receptor
- BCRP
Breast cancer resistance protein
- CAR
Constitutive androstane receptor
- IND
Investigational new drug
- NDA
New drug application
- NME
New molecular entity
- OAT
Organic anion transporter
- OATP
Organic anion transporting polypeptide
- OCT
Organic cation transporter
- P-gp
P-glycoprotein
- UGT
UDP-glucuronosyltransferase
Footnotes
The opinions contained in this paper do not necessarily reflect the official views of the FDA.
References
- 1.Huang SM, Temple R. Is this the drug or dose for you? Impact and consideration of ethnic factors in global drug development, regulatory review, and clinical practice. Clin Pharmacol Ther. 2008;84(3):287–284. doi: 10.1038/clpt.2008.144. [DOI] [PubMed] [Google Scholar]
- 2.Huang SM, Temple R, Throckmorton DC, Lesko LJ. Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clin Pharmacol Ther. 2007;81(2):298–304. doi: 10.1038/sj.clpt.6100054. [DOI] [PubMed] [Google Scholar]
- 3.Huang SM, Strong JM, Zhang L, Reynolds KS, Nallani S, Temple R, Abraham S, Al HS, Baweja RK, Burckart GJ, Chung S, Colangelo P, Frucht D, Green MD, Hepp P, Karnaukhova E, Ko HS, Lee JI, Marroum PJ, Norden JM, Qiu W, Rahman A, Sobel S, Stifano T, Thummel K, Wei XX, Yasuda S, Zheng JH, Zhao H, Lesko LJ. New era in drug interaction evaluation: US food and drug administration update on CYP enzymes, transporters, and the guidance process. J Clin Pharmacol. 2008;48(6):662–670. doi: 10.1177/0091270007312153. [DOI] [PubMed] [Google Scholar]
- 4.Guidance for Industry: Drug metabolism/drug interactions in the drug development process: studies in vitro. http://www.fda.gov/cder/guidance.
- 5.In vivo drug metabolism/drug interaction studies—study design, data analysis, and recommendations for dosing and labeling. http://www.fda.gov/cder/guidance.
- 6.Draft guidance for industry: drug interaction studies—study design, data analysis, and implications for dosing and labeling. http://www.fda.gov/cder/guidance/6695dft.pdf. [DOI] [PubMed]
- 7.CDER Drug Development and Drug Interactions website. http://www.fda.gov/cder/drug/drugInteractions/default.htm.
- 8.Hosea NA, Miller GP, Guengerich FP. Elucidation of distinct ligand binding sites for cytochrome P450 3A4. Biochemistry. 2000;39(20):5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- 9.Kenworthy KE, Bloomer JC, Clarke SE, Houston JB. CYP3A4 drug interactions: correlation of 10 in vitro probe substrates. Br J Clin Pharmacol. 1999;48(5):716–727. doi: 10.1046/j.1365-2125.1999.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkatakrishnan K, Obach RS. Drug–drug interactions via mechanism-based cytochrome P450 inactivation: points to consider for risk assessment from in vitro data and clinical pharmacologic evaluation. Curr Drug Metab. 2007;8(5):449–462. doi: 10.2174/138920007780866861. [DOI] [PubMed] [Google Scholar]
- 11.Wandel C, Kim RB, Guengerich FP, Wood AJ. Mibefradil is a P-glycoprotein substrate and a potent inhibitor of both P-glycoprotein and CYP3A in vitro. Drug Metab Dispos. 2000;28(8):895–898. [PubMed] [Google Scholar]
- 12.FDA Talk Paper “ROCHE LABORATORIES ANNOUNCES WITHDRAWAL OF POSICOR FROM THE MARKET”. http://www.fda.gov/bbs/topics/ANSWERS/ANS00876.html (accessed 12-29-2008).
- 13.Zhao P. The use of hepatocytes in evaluating time-dependent inactivation of P450 in vivo. Expert Opin Drug Metab Toxicol. 2008;4(2):151–164. doi: 10.1517/17425255.4.2.151. [DOI] [PubMed] [Google Scholar]
- 14.Grime KH, Bird J, Ferguson D, Riley RJ. Mechanism-based inhibition of cytochrome P450 enzymes: an evaluation of early decision making in vitro approaches and drug–drug interaction prediction methods. Eur J Pharm Sci. 2009;36:175–191. doi: 10.1016/j.ejps.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Hewitt NJ, de Kanter R, LeCluyse E. Induction of drug metabolizing enzymes: a survey of in vitro methodologies and interpretations used in the pharmaceutical industry—do they comply with FDA recommendations? Chem Biol Interact. 2007;168(1):51–65. doi: 10.1016/j.cbi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Fahmi OA, Boldt S, Kish M, Obach RS, Tremaine LM. Prediction of drug–drug interactions from in vitro induction data: application of the relative induction score approach using cryopreserved human hepatocytes. Drug Metab Dispos. 2008;36(9):1971–1974. doi: 10.1124/dmd.108.021907. [DOI] [PubMed] [Google Scholar]
- 17.Faucette SR, Zhang TC, Moore R, Sueyoshi T, Omiecinski CJ, LeCluyse EL, Negishi M, Wang H. Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J Pharmacol Exp Ther. 2007;320(1):72–80. doi: 10.1124/jpet.106.112136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, Devanaboyina US, Nangju NA, Feng Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000;9(1):29–42. [PubMed] [Google Scholar]
- 19.Aithal GP, Day CP. Nonsteroidal anti-inflammatory drug-induced hepatotoxicity. Clin Liver Dis. 2007;11(3):563–575. doi: 10.1016/j.cld.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Hahn KK, Wolff JJ, Kolesar JM. Pharmacogenetics and irinotecan therapy. Am J Health Syst Pharm. 2006;63(22):2211–2217. doi: 10.2146/ajhp060155. [DOI] [PubMed] [Google Scholar]
- 21.CAMPTOSAR Labeling. http://www.fda.gov/cder/foi/label/2006/020571s030lbl.pdf.
- 22.Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15(10):677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- 23.Guidance for Industry: waiver of in vivo bioavailability and bioequivalence studies for immediate-release solid oral dosage forms based on a Biopharmaceutics Classification System. http://www.fda.gov/cder/guidance.
- 24.Wu CY, Benet LZ. Predicting drug disposition via application of BCS: transport/absorption/ elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm Res. 2005;22(1):11–23. doi: 10.1007/s11095-004-9004-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Zhang YD, Strong JM, Reynolds KS, Huang SM. A regulatory viewpoint on transporter-based drug interactions. Xenobiotica. 2008;38(7–8):709–724. doi: 10.1080/00498250802017715. [DOI] [PubMed] [Google Scholar]
- 26.Fenner K, Troutman M, Kempshall S, Cook J, Ware J, Smith D, Lee C. Drug–drug interactions mediated through P-glycoprotein: clinical relevance and in vitro–in vivo correlation using digoxin as a probe drug. Clin Pharmacol Ther. 2009;85:173–181. doi: 10.1038/clpt.2008.195. [DOI] [PubMed] [Google Scholar]
- 27.Simonson SG, Raza A, Martin PD, Mitchell PD, Jarcho JA, Brown CD, Windass AS, Schneck DW. Rosuvastatin pharmacokinetics in heart transplant recipients administered an antirejection regimen including cyclosporine. Clin Pharmacol Ther. 2004;76(2):167–177. doi: 10.1016/j.clpt.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Ho RH, Tirona RG, Leake BF, Glaeser H, Lee W, Lemke CJ, Wang Y, Kim RB. Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology. 2006;130(6):1793–1806. doi: 10.1053/j.gastro.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 29.Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, Sheardown SA, Yue L, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther. 2008;83(2):273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117(5):1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito K, Brown HS, Houston JB. Database analyses for the prediction of in vivo drug–drug interactions from in vitro data. Br J Clin Pharmacol. 2004;57(4):473–486. doi: 10.1111/j.1365-2125.2003.02041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito K, Iwatsubo T, Kanamitsu S, Nakajima Y, Sugiyama Y. Quantitative prediction of in vivo drug clearance and drug interactions from in vitro data on metabolism, together with binding and transport. Annu Rev Pharmacol Toxicol. 1998;38:461–499. doi: 10.1146/annurev.pharmtox.38.1.461. [DOI] [PubMed] [Google Scholar]
- 33.Ito K, Chiba K, Horikawa M, Ishigami M, Mizuno N, Aoki J, Gotoh Y, Iwatsubo T, Kanamitsu S, Kato M, Kawahara I, Niinuma K, Nishino A, Sato N, Tsukamoto Y, Ueda K, Itoh T, Sugiyama Y. Which concentration of the inhibitor should be used to predict in vivo drug interactions from in vitro data? AAPS PharmSci. 2002;4(4):E25. doi: 10.1208/ps040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanamitsu S, Ito K, Sugiyama Y. Quantitative prediction of in vivo drug–drug interactions from in vitro data based on physiological pharmacokinetics: use of maximum unbound concentration of inhibitor at the inlet to the liver. Pharm Res. 2000;17(3):336–343. doi: 10.1023/A:1007509324428. [DOI] [PubMed] [Google Scholar]
- 35.Houston JB, Galetin A. Methods for predicting in vivo pharmacokinetics using data from in vitro assays. Curr Drug Metab. 2008;9(9):940–951. doi: 10.2174/138920008786485164. [DOI] [PubMed] [Google Scholar]
- 36.Kato M, Shitara Y, Sato H, Yoshisue K, Hirano M, Ikeda T, Sugiyama Y. The quantitative prediction of CYP-mediated drug interaction by physiologically based pharmacokinetic modeling. Pharm Res. 2008;25(8):1891–1901. doi: 10.1007/s11095-008-9607-2. [DOI] [PubMed] [Google Scholar]
- 37.Yao C, Kunze KL, Kharasch ED, Wang Y, Trager WF, Ragueneau I, Levy RH. Fluvoxamine-theophylline interaction: gap between in vitro and in vivo inhibition constants toward cytochrome P4501A2. Clin Pharmacol Ther. 2001;70(5):415–424. doi: 10.1067/mcp.2001.119724. [DOI] [PubMed] [Google Scholar]
- 38.Dickins M, van de Waterbeemd H. Simulation models for drug disposition and drug interactions. Drug Discovery Today: BIOSILICO. 2004;2(1):38–45. doi: 10.1016/S1741-8364(04)02388-1. [DOI] [Google Scholar]
- 39.McGinnity DF, Waters NJ, Tucker J, Riley RJ. Integrated in vitro analysis for the in vivo prediction of cytochrome P450-mediated drug–drug interactions. Drug Metab Dispos. 2008;36(6):1126–1134. doi: 10.1124/dmd.108.020446. [DOI] [PubMed] [Google Scholar]
- 40.Obach RS, Walsky RL, Venkatakrishnan K, Gaman EA, Houston JB, Tremaine LM. The utility of in vitro cytochrome P450 inhibition data in the prediction of drug–drug interactions. J Pharmacol Exp Ther. 2006;316(1):336–348. doi: 10.1124/jpet.105.093229. [DOI] [PubMed] [Google Scholar]
- 41.Venkatakrishnan K, von Moltke LL, Obach RS, Greenblatt DJ. Drug metabolism and drug interactions: application and clinical value of in vitro models. Curr Drug Metab. 2003;4(5):423–459. doi: 10.2174/1389200033489361. [DOI] [PubMed] [Google Scholar]
- 42.Rostami-Hodjegan A, Tucker GT. Simulation and prediction of in vivo drug metabolism in human populations from in vitro data. Nat Rev Drug Discov. 2007;6(2):140–148. doi: 10.1038/nrd2173. [DOI] [PubMed] [Google Scholar]
- 43.Youdim KA, Zayed A, Dickins M, Phipps A, Griffiths M, Darekar A, Hyland R, Fahmi O, Hurst S, Plowchalk DR, Cook J, Guo F, Obach RS. Application of CYP3A4 in vitro data to predict clinical drug–drug interactions; predictions of compounds as objects of interaction. Br J Clin Pharmacol. 2008;65(5):680–692. doi: 10.1111/j.1365-2125.2007.03070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chien JY, Lucksiri A, Ernest CS, 2nd, Gorski JC, Wrighton SA, Hall SD. Stochastic prediction of CYP3A-mediated inhibition of midazolam clearance by ketoconazole. Drug Metab Dispos. 2006;34(7):1208–1219. doi: 10.1124/dmd.105.008730. [DOI] [PubMed] [Google Scholar]
- 45.Vossen M, Sevestre M, Niederalt C, Jang IJ, Willmann S, Edginton AN. Dynamically simulating the interaction of midazolam and the CYP3A4 inhibitor itraconazole using individual coupled whole-body physiologically-based pharmacokinetic (WB-PBPK) models. Theor Biol Med Model. 2007;4:13. doi: 10.1186/1742-4682-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Kjellsson M, Rostami-Hodjegan A, Tucker GT. The effects of dose staggering on metabolic drug–drug interactions. Eur J Pharm Sci. 2003;20(2):223–232. doi: 10.1016/S0928-0987(03)00200-8. [DOI] [PubMed] [Google Scholar]
- 47.Zhao P, Ragueneau-Majlessi I, Zhang L, Strong JM, Reynolds KS, Levy RH, Thummel KE, Huang SM. Quantitative evaluation of pharmacokinetic inhibition of CYP3A substrates by ketoconazole: a simulation study. J Clin Pharmacol. 2009;49(3):351–359. doi: 10.1177/0091270008331196. [DOI] [PubMed] [Google Scholar]
- 48.Drug Information in Drugs@FDA (http://www.accessdata.fda.gov/scripts/cder/drugsatfda/index.cfm).