Abstract
The Arabidopsis thaliana NPR1 has been shown to be a key regulator of gene expression during the onset of a plant disease-resistance response known as systemic acquired resistance. The npr1 mutant plants fail to respond to systemic acquired resistance-inducing signals such as salicylic acid (SA), or express SA-induced pathogenesis-related (PR) genes. Using NPR1 as bait in a yeast two-hybrid screen, we identified a subclass of transcription factors in the basic leucine zipper protein family (AHBP-1b and TGA6) and showed that they interact specifically in yeast and in vitro with NPR1. Point mutations that abolish the NPR1 function in A. thaliana also impair the interactions between NPR1 and the transcription factors in the yeast two-hybrid assay. Furthermore, a gel mobility shift assay showed that the purified transcription factor protein, AHBP-1b, binds specifically to an SA-responsive promoter element of the A. thaliana PR-1 gene. These data suggest that NPR1 may regulate PR-1 gene expression by interacting with a subclass of basic leucine zipper protein transcription factors.
Systemic acquired resistance (SAR) is a general plant-resistance response that can be induced during a local infection by an avirulent pathogen. Although early studies of SAR were conducted by using tobacco mosaic virus and its Solanaceous hosts (1), SAR has been demonstrated in many plant species and shown to be effective against not only viruses but also bacterial and fungal pathogens (2, 3). A necessary signal for SAR induction is salicylic acid (SA); plants that fail to accumulate SA because of the expression of an SA-oxidizing enzyme, salicylate hydroxylase, are impaired in SAR (4). Conversely, an elevation in the endogenous level of SA or exogenous application of SA or its synthetic analogs, such as 2,6-dichloroisonicotinic acid (INA), not only results in an enhanced, broad-spectrum resistance but also stimulates concerted expression of a battery of genes known as pathogenesis-related (PR) genes (5–12). PR genes may play direct roles in conferring resistance because their expression coincides with the onset of SAR and some of the PR genes encode enzymes with antimicrobial activities (11, 12). Therefore, understanding the regulation of PR gene expression has been a focal point of research in plant disease resistance.
Using PR genes as reporters, two classes of Arabidopsis thaliana mutants have been identified. One class constitutively expresses PR genes whereas the other class is impaired in the SA- or INA-induced PR gene expression (13–20). Interestingly, from the second class of mutants only one genetic locus, NPR1 (also known as NIM1), has been identified. NPR1 has been shown to be a key component of the SA-regulated PR gene expression and disease resistance because npr1 mutants fail to express PR1, PR2, and PR5 and display enhanced susceptibility to infection even after treatment with SA or INA (17–20). Furthermore, transgenic plants overexpressing NPR1 display a more dramatic induction of PR genes during an infection and show complete resistance to Pseudomonas syringae pv. maculicola 4326 and Peronospora parasitica Noco, two very different pathogens that are virulent on wild-type A. thaliana plants (21).
How does NPR1 transduce the SA signal and up-regulate the PR genes? Sequence analysis of NPR1 does not reveal any obvious homology to known transcription factors (22, 23). Therefore, it is unlikely that NPR1 is involved directly in transactivating the promoters of PR genes. However, NPR1 contains at least four ankyrin repeats, which are found in proteins with very diverse biological functions and are involved in protein–protein interactions (24, 25). The functional importance of the ankyrin repeat domain has been demonstrated by mutations found in the npr1–1 and the nim1–2 alleles, where the highly conserved histidine residues in the third and the second ankyrin repeats, respectively, are changed to a tyrosine. Because these conserved histidine residues are involved in the formation of hydrogen bonds, which are crucial in stabilizing the three-dimensional structure of the ankyrin-repeat domain (26), npr1–1 and nim1–2 mutations may cause disruption in the local structure within the ankyrin-repeat domain and abolish its ability to interact with other proteins. These data suggest that NPR1 probably exerts its regulatory function by interacting with other proteins.
To understand further the function of NPR1, we performed a yeast two-hybrid screen to search for genes encoding NPR1-interacting proteins. We found that NPR1 interacts specifically with a subclass of basic leucine zipper protein (bZIP) transcription factors.
MATERIALS AND METHODS
Strains and Plasmids.
The yeast two-hybrid system that contains the yeast strain EGY48 and plasmids pEG202, pSH18–34, pJK101, pRFHM1, pSH17–4, and pJG4–5 was obtained from R. Brent (27). The full-length tomato NPR1 homolog was cloned into pEG202 after amplification from the cDNA clone (pTomNPR1) by using PCR. The full-length A. thaliana NPR1 cDNA was amplified from pKExNPR1 (22) and cloned into pEG202 similarly. PCR also was used to construct the truncated NPR1 baits. pEGNPR11–177 encodes the amino-terminal 177 aa of NPR1, pEGNPR11–432 contains the amino-terminal portion and the ankyrin repeats, and pEGNPR1178–593 encodes the ankyrin repeats and the carboxyl end of NPR1. The A. thaliana bZIP transcription factor genes AHBP-1b, TGA6, and OBF5 were obtained by PCR from a cDNA preparation and cloned into pJG4–5. AHBP-1b and OBF5 also were cloned into pET24C(+) (Novagen) to add a (His)6-tag at the carboxyl end of the protein. The resulting plasmids were designated pET-AHBP-1b and pET-OBF5. The bait constructs containing the npr1–1 and npr1–2 mutations (22) were generated by site-directed mutagenesis by using a PCR-based “link-scanning” method (28).
Isolation of an NPR1 Homolog from Tomato.
Approximately 1 million plaques of a tomato leaf cDNA library (29) were screened by using both an A. thaliana NPR1 cDNA and an NPR1 homolog from Nicotiana glutinosa (M.K. and X.D., unpublished data). Colony/plaque screen nylon filters were hybridized at 37°C in 40% formamide/5×SSC/5×Denhardt’s solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA)/1% SDS/10% dextran sulfate. The final wash was for 20 min at 37°C in 2× SSC and 1% SDS. Three independent clones were sequenced and found to be identical in their overlapping regions. The clone that contains the full-length cDNA was designated pTomNPR1.
Yeast Two-Hybrid Screen and Assay.
The yeast two-hybrid screen was performed as described (30). The prey library was constructed in plasmid pJG4–5 in B. Baker’s laboratory (U.S. Department of Agriculture) by using cDNA (average size of 1.8 kb) extracted from tobacco mosaic virus-infected tomato VF36 leaves and contains 107 independent clones.
Overexpression and Purification of the Transcription Factor Proteins.
The E. coli strain BL21(DE3) carrying pET-AHBP-1b or pET-OBF5 was grown (OD600 = 1.0) in LB (1 liter), and the expression of AHBP-1b or OBF5 was induced by isopropyl β-d-thiogalactoside (0.1 mM). After 2 hr, the bacteria were harvested, ground in alumina powder (two times the weight of the cell pellet), and then resuspended in 50 ml buffer A [50 mM Tris⋅HCl, pH 7.5/50 mM KCl/1× proteinase inhibitor mixture (21)/6 mM 2-mercaptoethanol/10% glycerol]. The cell extract was spun twice, and Ni-NTA resin (Qiagen) was added to the supernatant and incubated for 1 hr. The mix was loaded into a column and washed with buffer B (1 M KCl/50 mM Tris⋅HCl, pH 7.5/10% glycerol/6 mM 2-mercaptoethanol/10 mM imidazole). The proteins then were eluted in buffer C (50 mM Tris⋅HCl, pH 7.5/50 mM KCl/150 mM imidazole/6 mM 2-mercaptoethanol/10% glycerol). The eluted protein solution was dialyzed against 10 mM Tris⋅HCl, pH 7.5/50 mM KCl/6 mM 2-mercaptoethanol/10% glycerol.
Overexpression of NPR1 by Using the Baculovirus System.
The NPR1 cDNA was amplified by PCR and cloned into pVL1392 by using the restriction sites NotI and BglII and then recombined in vivo into BaculoGold (PharMingen). The amplified virus preparation was used to infect Sf9 insect cells (2 × 106 cells/ml). The cells were harvested and the total protein extract was prepared as described (PharMingen). The presence of NPR1 in the protein extract was confirmed by using an antiserum prepared against the protein (21).
In Vitro Analysis of Protein–Protein Interactions.
Partially purified, His-tagged transcription factors (10 μg) were mixed in buffer A with the insect cell extract (50 μl) expressing A. thaliana NPR1. The protein mix was incubated on ice for 2 hr. Ni-NTA resin then was added to the protein mix and incubated for another hour. The Ni-NTA resin was pelleted and washed five times with buffer B. Proteins were eluted from the Ni-NTA in buffer C, and a quarter of the proteins were run on an SDS/PAGE gel and subsequently transferred to a Protran nitrocellulose membrane (Schleicher & Schuell). Immunoblot analysis was performed by using antibodies raised against NPR1 (21) to check for copurification of NPR1 with the His-tagged transcription factors.
Gel Mobility Shift Assay.
Oligonucleotide probes used in the gel mobility shift assay were designed according to the sequence of the INA- and SA-responsive promoter element identified in the A. thaliana PR-1 gene (31). The wild-type oligonucleotide probe used in the assay was 5′-CTCTACGTCACTATTTTACTTACGTCATAGATG-3′, and the mutant used was 5′-CTCTAttctACTATTTTACTTAttctATAGATG-3′. Each strand of the probes was end-labeled by incubating 10 pmol of oligonucleotide in a 20-μl reaction with 10 units of polynucleotide kinase (NEB) and 50 μCi of [γ-32P]ATP. The two complementary strands then were mixed, annealed, and purified by using a Nucleotide Removal Kit (Qiagen). For each binding reaction, 1 μg of the partially purified transcription factor protein (10% purity) was added together with 100 ng of poly[dI-dC] and 20 μl binding buffer (12 mM Hepes, pH 7.9/60 mM KCl/2 mM MgCl2/10% glycerol/1 mM DTT/1× protease inhibitor cocktail). The mixture was incubated at room temperature for 10 min before addition of the labeled probe (0.02 pmol, 40,000 cpm per reaction). The reaction was incubated for another 30 min and then run on a 4% (wt/vol) native polyacrylamide gel in 0.5× TBE buffer (90 mM Tris·borate/2 mM EDTA, pH 8.3). After electrophoresis, the gel was dried and autoradiographed.
RESULTS
Tomato NPR1 Homolog Interacts with a bZIP Transcription Factor in the Yeast Two-Hybrid Screen.
To identify genes that encode NPR1 interactors, we performed a yeast two-hybrid screen by using a full-length tomato NPR1 homolog as the bait (pEGTomNPR1) and a tomato cDNA library. We chose to perform the screen in tomato for two reasons. First, the quality of a cDNA library is critical to the success of a yeast two-hybrid screen, and in tomato, this could be determined by using the well studied PTO–PTI interaction as a positive control (32). Second, by performing the yeast two-hybrid assay in tomato and, later, in A. thaliana we can evaluate more effectively the general significance of the candidate genes. We believe that the tomato cDNA clone used in our yeast two-hybrid screen is a true homolog of the A. thaliana NPR1 because significant homology (54% identity and 73% similarity) is detected throughout the protein and the functionally important residues as defined by various npr1 mutant alleles are conserved in this clone.
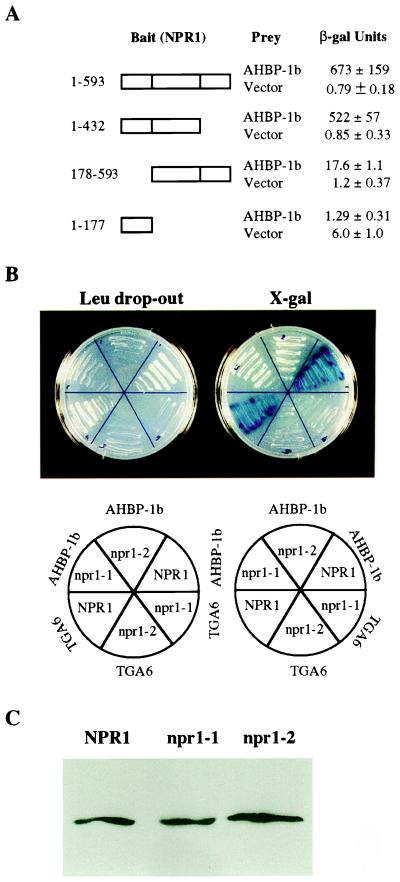
Before performing the yeast two-hybrid screen, the bait constructs containing TomNPR1 or A. thaliana NPR1 first were tested for self-activation. We found that the bait alone did not activate the expression of the reporter genes. The cDNA library and the bait plasmid pEGTomNPR1 were cotransformed into yeast strain EGY48, and 2.5 × 106 colonies were obtained. From these primary transformants, 2.5 × 107 cells were plated onto leucine drop-out plates. Seven distinct classes of tomato genes were found to interact with TomNPR1 in the yeast two-hybrid system. One class, NIF1 (NPR1-interacting factor 1), which gave the highest reporter activity, was characterized in more detail. The NIF1 plasmid (pJGNIF1) was isolated and retransformed into EYG48 to confirm the interaction. Colonies carrying both pJGNIF1 and pEGTomNPR1 grew on plates lacking leucine and turned dark blue on 5-bromo-4-chloro-3-indolyl β-d-galactoside plates within 24 hr (Fig. 1). The restoration of leucine prototrophy and the expression of β-galactosidase activity were dependent on the presence of galactose, indicating that the expression of NIF1 driven by the promoter of the yeast GAL1 gene was required for the expression of both reporter genes. This clone also was transformed into EGY48 together with vector pEG202 to test whether NIF1 by itself activates the transcription of the reporter genes. Expression of NIF1 alone did not restore leucine prototrophy or result in detectable β-galactosidase activity.
Figure 1.
Yeast two-hybrid assay of interactions between NPR1 and bZIP transcription factors. (A) Yeast cells (EGY48) containing TomNPR1 and NIF1, NPR1 and AHBP-1b, NPR1 and TGA6, or NPR1 and OBF5 were grown for 2 days at 30°C on galactose medium lacking leucine for detection of the LEU2 reporter expression. (B) β-Galactosidase activity was calculated as described previously (33). Three independent colonies were taken for each combination, and four measurements were performed on a culture grown from each colony. The mean ± SD is presented.
NIF1 was sequenced, and a search of the GenBank database identified three independent, closely related A. thaliana genes encoding the bZIP transcription factors AHBP-1b (34), TAG6 (35), and OBF5 (36). Sequence comparisons of NIF1, AHBP-1b, TAG6, and OBF5 reveal that the NIF1 clone identified in the yeast two-hybrid screen encodes the carboxyl two-thirds of a bZIP transcription factor, which does not include the DNA-binding or leucine zipper domains. The NIF1 clone shares 69–75% identity and 83–87% similarity with these A. thaliana homologs at the amino acid level (Fig. 2).
Figure 2.
Sequence alignment of tomato NIF1 and A. thaliana AHBP-1b, TGA6, and OBF5. The tomato NIF1 sequence was aligned with AHBP-1b (residues 96–330) (34), TGA6 (residues 90–325) (35), and OBF5 (residues 90–324) (36) by using multalin (37). Red letters represent the residues of high consensus, blue letters indicate the amino acids with low consensus, and black letters highlight the residues with no consensus.
A. thaliana NPR1 Interacts Strongly with AHBP-1b and TGA6 but Weakly with OBF5 in the Yeast Two-Hybrid System.
To determine whether A. thaliana NPR1 interacts with the A. thaliana homologs of NIF1, DNA fragments containing the full-length AHBP-1b, TGA6, and OBF5 genes were amplified and cloned into pJG4–5. Interactions between the A. thaliana NPR1 and all three transcription factors then were tested in the yeast two-hybrid system. As observed for TomNPR1 and NIF1, where 1,286 units of β-galactosidase activity were detected, A. thaliana NPR1 interacts strongly with AHBP-1b and TGA6, resulting in 673 and 367 units of β-galactosidase activity, respectively (Fig. 1). Intriguingly, only 7.6 units of β-galactosidase activity was detected in yeast carrying NPR1 and OBF5 (Fig. 1). This may indicate that OBF5 interacts with NPR1 with a much lower affinity. But it is also possible that the protein is not expressed as well as AHBP-1b and TGA6 or does not fold properly in yeast.
NPR1 Interacts with AHBP-1B and OBF5 in Vitro.
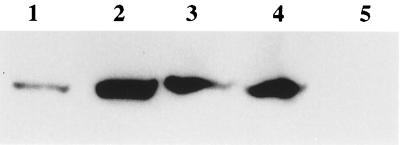
To test whether NPR1 interacts with AHBP-1b and OBF5 in vitro, His-tagged AHBP-1b and OBF5 proteins were expressed in E. coli and purified by using a Ni-NTA column. Purified AHBP-1b or OBF5 protein was mixed with extracts containing baculovirus-expressed NPR1 protein. The AHBP-1b and OBF5 proteins then were “pulled down” by using Ni-NTA resin. Immunoblot analysis using an antiserum against NPR1 showed that the NPR1 protein copurified with AHBP-1b (Fig. 3, lanes 2 and 4), demonstrating that AHBP-1b physically associates with NPR1 in vitro. As a negative control, Ni-NTA resin alone was mixed with the NPR1 protein extract. The results showed that NPR1 does not bind to Ni-NTA resin by itself (Fig. 3, lane 5). When the partially purified OBF5 protein preparation was used in the experiments, copurification of NPR1 also was detected (Fig. 3, lane 3). This indicates that OBF5 can interact with NPR1 in vitro.
Figure 3.
In vitro analysis of interactions between NPR1 and AHBP-1b or OBF5. Immunoblot analysis was performed on protein preparations purified by using Ni-NTA resin to detect copurification of NPR1 with His-tagged AHBP-1b or OBF5. The blot was probed with an antiserum generated against the carboxyl-terminal 16 aa of NPR1. Lanes: 1, crude extract (0.2 μl) of Sf9 insect cells expressing NPR1 by baculovirus; 2, NPR1 copurified with AHBP-1b; 3, NPR1 copurified with OBF5; 4, NPR1 copurified with AHBP-1b from a separate purification; 5, NPR1 purified with Ni-NTA resin alone.
NPR1 Interacts with AHBP-1b Through the Ankyrin-Repeat Domain.
To define the region in NPR1 that interacts directly with AHBP-1b, NPR1 gene fragments encoding different domains of the protein were cloned into the bait vector. The truncations were made at the exon–intron boundaries because these boundaries are conserved between NPR1 and its homologs and, therefore, may define distinct functional domains of the protein. pEGNPR11–177 carries the first exon of the NPR1 gene and encodes the amino-terminal 177 residues; pEGNPR11–432 includes both the amino-terminal and the ankyrin-repeat domain of NPR1 (exons 1 and 2); pEGNPR1178–593 contains the ankyrin-repeat domain and the carboxyl end of NPR1 (exons 2, 3, and 4). These truncated NPR1 proteins were coexpressed with the transcription factor AHBP-1b in yeast, and β-galactosidase reporter gene activity was measured. In yeast expressing NPR11–432 and AHBP-1b, the β-galactosidase reporter gene activity (522 units) was similar to that observed in the cells expressing the full-length NPR1 (673 units) (Fig. 4A). This shows that NPR1 interacts with AHBP-1b through the amino-terminal and/or the ankyrin-repeat domain. In cells expressing NPR1178–593, which lacks the amino-terminal 177 residues, the NPR1-AHBP-1b interaction still occurred but at a much lower level (17.6 units). This suggests that the amino-terminal region of NPR1 contributes to the NPR1-AHBP-1b interaction and the ankyrin-repeat domain interacts with AHBP-1b. Because the amino-terminal region alone does not interact with AHBP-1b (Fig. 4A), this region probably serves to stabilize the ankyrin-repeat domain. Surprisingly, the amino-terminal region of NPR1 alone seems to have a low level of intrinsic transactivation activity (6 units) detected when it is expressed in yeast without the prey (Fig. 4A).
Figure 4.
Interactions between mutant npr1 proteins and the bZIP transcription factors. (A) Truncated versions of the NPR1 protein were tested for their interactions with AHBP-1b by measuring the β-galactosidase activity resulting from the interactions in yeast. Three independent colonies were taken for each combination, and four measurements were performed on a culture grown from each colony. The mean ± SD is presented. (B) Yeast two-hybrid assay performed by using npr1–1 and npr1–2 point mutants as bait. Yeast cells carrying mutant npr1 and either AHBP-1b or TGA6 were grown for 2 days at 30°C on galactose medium lacking leucine for detection of the LEU2 reporter expression (Left). The same yeast cells also were grown for 2 days at 30°C in medium containing 5-bromo-4-chloro-3-indolyl β-d-galactoside for detection of the β-galactosidase reporter expression (Right). (C) Immunoblot analysis of the expression of npr1 mutant proteins in yeast. Ten micrograms of total yeast protein was loaded in each lane, and immunoblot analysis was carried out by using the NPR1 antiserum.
The NPR1-AHBP-1b and NPR1-TGA6 Interactions Are Abolished by the npr1 Mutations.
To further determine the specificity of the NPR1-AHBP-1b and NPR1-TGA6 interactions, we generated bait constructs containing either the npr1–1 or npr1–2 point mutation (22). In npr1–1, the highly conserved histidine-334 in the ankyrin-repeat domain is changed to a tyrosine whereas in npr1–2, cysteine-150 in the amino-terminal region of NPR1 is converted to a tyrosine. These mutant constructs were cotransformed into yeast with either AHBP-1b or TGA6 clone, and β-galactosidase activity was measured in the resulting transformants. In both transformants, only background levels of β-galactosidase activity (<1 unit, data not shown) were detected, indicating that the npr1–1 and npr1–2 mutations abolish the ability of NPR1 to interact with AHBP-1b or TGA6 (Fig. 4B). Immunoblot analysis of the total protein preparations from these yeast strains showed that npr1–1 and npr1–2 were expressed at levels similar to the wild-type NPR1 protein (Fig. 4C). Therefore, the lack of reporter gene expression was not a result of poor expression of the mutant bait proteins but a consequence of impaired interaction caused by the point mutations. Because the npr1–1 mutation is in the ankyrin-repeat domain and the npr1–2 mutation is in the amino-terminal region, this indicates further that both these regions are required for the interaction with AHBP-1b.
AHBP-1b Binds to a Promoter Element in the PR-1 Gene.
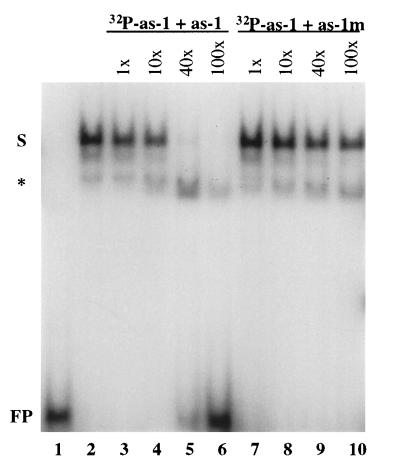
To analyze the role of AHBP-1b in regulating SA-responsive gene expression, we performed a gel mobility shift assay to determine whether AHBP-1b could bind to a promoter fragment of the PR-1 gene. The A. thaliana PR-1 gene promoter fragment used contains an as-1-like element, which has been identified previously as a binding motif of bZIP transcription factors and has been shown by linker-scanning mutagenesis to be essential for both INA- and SA-induced PR-1 gene expression in planta (31). With the partially purified transcription factor, a mobility shift was observed for the oligonucleotide probe (Fig. 5, lane 2). To demonstrate that this mobility shift was due to the binding of AHBP-1b and not other nonspecific proteins in the preparation, a control reaction was carried out by using an unrelated protein purified under identical conditions. As shown in Fig. 5 (lane 1), the control protein preparation did not bind to the probe. To examine further the specificity of the binding, a competition experiment was performed by using an excess amount of unlabeled probe containing the bZIP transcription factor-binding site. When a 40-fold excess of unlabeled oligonucleotides was included in the reaction, binding of AHBP-1b to the labeled probe was abolished completely (Fig. 5, lane 5). As a negative control, an oligonucleotide containing point mutations in the bZIP-binding motif also was used in the competition experiment. Binding of AHBP-1b to the labeled probe was unaffected by the presence of the mutant fragment even when its concentration was 100 times higher than the labeled probe (Fig. 5, lane 10).
Figure 5.
AHBP-1b interacts specifically with the A. thaliana PR-1 promoter sequence containing an as-1-like element. All binding reactions contain 4 × 104 cpm 32P-labeled probe, incubated with 1 μg of either a control protein preparation (lane 1) or AHBP-1b (lanes 2–10), in the absence (lanes 1 and 2) or presence of specific (lanes 3–6) or nonspecific (lanes 7–10) competitor probes. In lanes 3–6, 1×, 10×, 40×, and 100× molar excess of the unlabeled as-1-like probe were added, respectively. In lanes 7–10, 1×, 10×, 40×, and 100× molar excess of the unlabeled probe containing point mutations in the as-1-like element were added, respectively. S, specific band shifting; ∗, nonspecific banding; FP, free probe.
DISCUSSION
To understand better how NPR1 functions to regulate the expression of genes associated with disease resistance, we used the yeast two-hybrid system to identify proteins that physically interact with NPR1. The tomato NPR1 homolog originally was found to interact strongly with a protein (NIF1) that is highly homologous to a class of plant bZIP transcription factors. We then tested the interactions between A. thaliana NPR1 and three A. thaliana transcription factors (AHBP-1b, TGA6, and OBF5) that are closely related to the tomato NIF1. The amino acid sequences of these three A. thaliana proteins share more than 80% identity and 90% similarity with each other, with AHBP-1b and TGA6 being slightly more similar to NIF1 than OBF5. In the yeast two-hybrid assay, both AHBP-1b and TGA6 strongly interact with A. thaliana NPR1. The interaction between NPR1 and AHBP-1b was confirmed further by an in vitro binding assay. Interestingly, the interaction between OBF5 and NPR1 is much weaker as measured in the yeast two-hybrid assay. This result suggests that the interactions between A. thaliana NPR1 and its transcription factor partners may be highly specific. The specificity of the interaction probably is defined in the transcription factors by the residues downstream of the DNA-binding domain and the leucine zipper domain (Fig. 2) because only the carboxyl two-thirds of the protein is present in the NIF1 clone identified in our initial screen. In this region, only a few amino acids are different between AHBP-1b, TGA6, and OBF5, with OBF5 displaying the most divergence. In NPR1, the ankyrin-repeat domain is probably the region involved in the interaction because truncated NPR1 proteins containing the ankyrin-repeat domain still interact with AHBP-1b in the yeast two-hybrid assay.
The specificity of the interaction between A. thaliana NPR1 and the transcription factors was demonstrated further by using two point mutations that are shown to abolish NPR1 function. A single amino acid change corresponding to the npr1–1 or npr1–2 mutation completely abolished the ability of NPR1 to interact with both AHBP-1b and TGA6 even though they had little effect on the accumulation of the mutant proteins in yeast. This suggests further that the interactions between NPR1 and its interactors have a high degree of specificity. The inability of npr1–1 and npr1–2 to interact with AHBP-1b and TAG6 may contribute directly to these mutants’ lack of PR gene expression in response to SAR induction.
Our finding that NPR1 interacts specifically with two bZIP transcription factors brings together the genetic data on the function of NPR1 in SAR with the previous molecular studies of SA-regulated gene expression. SA-responsive promoter elements such as the as-1 element in the 35S promoter of cauliflower mosaic virus and the ocs and nos elements in opine synthase promoters of Agrobacterium have been identified and characterized previously (38–40). The as-1 element has been shown to bind to a tobacco transcription factor, SARP (SA response protein), which is related immunologically to the tobacco protein TAG1a, a bZIP transcription factor (41). In A. thaliana, there are at least six bZIP genes identified that have homology to the tobacco TAG transcription factor (34–36, 42–44). These TAG transcription factors have been shown to have different affinities for the as-1 element in in vitro binding assays (44). Whereas strong binding of AHBP-1b requires two tandem copies of the TGACG motif present in the as-1 element, binding of TGA6 appears to be unaffected by the number of motifs because a single copy seems to be sufficient. The in vivo regulation of the activities of the transcription factors in the bZIP family may be much more complex. They may exert a broad range of promoter specificities in regulating genes in diverse signal-transduction pathways by forming homodimers and heterodimers between family members and by possible interactions with other proteins. Our results suggest that AHBP-1b and TGA6 may regulate PR gene expression through their interactions with NPR1.
Recently, the promoter of the A. thaliana PR-1 gene has been analyzed thoroughly by using deletion and linker-scanning mutagenesis performed in transgenic plants as well as in vivo footprinting analysis (31). Through these analyses, two INA-responsive elements have been defined. One element at −610 is similar to a recognition sequence for the transcription factor NF-κB, whereas the other promoter element around residue −640 contains a CGTCA motif (the complementary sequence is TGACG), which is present in the as-1 element. The CGTCA motif was shown by linker-scanning mutagenesis to be essential for both SA and INA induction of PR-1 gene expression. In this paper, we show that purified AHBP-1b protein specifically binds to this as-1-like element found in the PR-1 promoter. This suggests that the SA-responsive PR-1 gene expression may be regulated by the transcription factor AHBP-1b, and the activity of AHBP-1b probably is affected by its association with NPR1. Because TGA6 is highly homologous to AHBP-1b, it is possible that AHBP-1b and TGA6 have redundant functions with respect to their regulation of PR gene expression. This may explain why plants with mutations in these transcription factors were not discovered in the previous screens for npr1-like mutants whereas 12 npr1 alleles were isolated.
Based on both genetic and molecular data, the function of NPR1 is postulated. In support of our finding that NPR1 interacts with a subclass of bZIP transcription factors is evidence that shows NPR1 is nuclear-localized upon SAR induction (M.K. and X.D., unpublished data). In the nucleus, AHBP-1b and TGA6 may be sequestered by an unidentified repressor protein(s) under uninduced conditions. Upon induction, NPR1 is nuclear-localized and binds to AHBP-1b or TGA6. Binding of NPR1 to AHBP-1b or TGA6 then leads to the release of the repressor protein and the expression of the PR genes. The presence of a repressor has been implicated by both molecular and genetic evidence. In a gel mobility shift assay, the addition of dissociation agents to a tobacco protein extract has been shown to enhance significantly the SARP binding to as-1 (41). Furthermore, an A. thaliana mutant recently has been found to suppress the npr1 phenotype and fully restore the responsiveness of PR gene expression to SA induction. Because the mutation is recessive, it probably abolishes the function of a repressor (X.L. and X.D., unpublished data). An alternative mode of action for NPR1 is that it may serve directly as a transcriptional activator of PR genes when bound to the bZIP proteins. The detection of intrinsic transactivation activity in the amino-terminal region of NPR1 is in support of this hypothesis.
In summary, we have shown that NPR1 strongly interacts with a subclass of the bZIP transcription factors in yeast and in vitro and that these transcription factors bind to the as-1-like element of the PR-1 promoter, which is required for SA-induced gene expression. In future studies, the NPR1-bZIP interaction will be examined in vivo, and the role of the bZIP transcription factors in PR gene regulation will be characterized by using both knockout and dominant-negative mutants as well as transgenic plants overexpressing the transcription factors. Because there are multiple bZIP transcription factors in A. thaliana that can interact with NPR1, in the mutant analysis all of these factors may have to be affected before a biological effect is observed. These experiments will provide further genetic evidence for the roles of NPR1 and the bZIP transcription factors in regulating SAR.
Acknowledgments
We thank G. Martin and B. Baker for sharing their tomato libraries with us. We also thank J. Moomaw and P. Casey for helping with the expression of NPR1 in baculovirus system, H.-S. Jung for assisting with experiments, and J. Siedow, J. Clarke, and L. Anderson for helpful comments on the manuscript. This work was supported in part by National Science Foundation Grant MCB-9728111 and by a grant from Monsanto Company awarded to X.D.
ABBREVIATIONS
- bZIP
basic leucine zipper protein
- INA
2,6-dichloroisonicotinic acid
- PR
pathogenesis related
- SA
salicylic acid
- SAR
systemic acquired resistance
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF143442).
References
- 1.Ross A F. Virology. 1961;14:340–358. doi: 10.1016/0042-6822(61)90319-1. [DOI] [PubMed] [Google Scholar]
- 2.Kuc J. Bioscience. 1982;32:854–860. [Google Scholar]
- 3.Ryals J A, Neuenschwander U H, Willits M G, Molina A, Steiner H-Y, Hunt M D. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 5.Malamy J, Carr J P, Klessig D F, Raskin I. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- 6.Métraux J-P, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B. Science. 1990;250:1004–1006. doi: 10.1126/science.250.4983.1004. [DOI] [PubMed] [Google Scholar]
- 7.Rasmussen J B, Hammerschmidt R, Zook M N. Plant Physiol. 1991;97:1342–1347. doi: 10.1104/pp.97.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yalpani N, Silverman P, Wilson T M A, Kleier D A, Raskin I. Plant Cell. 1991;3:809–818. doi: 10.1105/tpc.3.8.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White R F. Virology. 1979;99:410–412. doi: 10.1016/0042-6822(79)90019-9. [DOI] [PubMed] [Google Scholar]
- 10.Métraux J-P, Ahl-Goy P, Staub T, Speich J, Steinemann A, Ryals J, Ward E. In: Advances in Molecular Genetics of Plant-Microbe Interactions. Hennecke H, Verma D P S, editors. Vol. 1. Dordrecht, The Netherlands: Kluwer; 1991. pp. 432–439. [Google Scholar]
- 11.Ward E R, Uknes S J, Williams S C, Dincher S S, Wiederhold D L, Alexander D C, Ahl-Goy P, Metraux J-P, Ryals J A. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawton K, Uknes S, Friedrich L, Gaffney T, Alexander D, Goodman R, Métraux J-P, Kessmann H, Ahl-Goy P, Gut-Rella M, Ward E, Ryals J. In: Mechanisms of Defense Responses in Plants. Fritig B, Legrand M, editors. Dordrecht, The Netherlands: Kluwer; 1993. pp. 422–432. [Google Scholar]
- 14.Bowling S A, Guo A, Cao H, Gordon A S, Klessig D F, Dong X. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowling S A, Clarke J D, Liu Y, Klessig D F, Dong X. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke J D, Liu Y, Klessig D F, Dong X. Plant Cell. 1998;10:557–569. doi: 10.1105/tpc.10.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao H, Bowling S A, Gordon A S, Dong X. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaney T P, Friedrich L, Ryals J A. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glazebrook J, Rogers E E, Ausubel F M. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah J, Tsui F, Klessig D F. Mol Plant–Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 21.Cao H, Li X, Dong X. Proc Natl Acad Sci USA. 1998;95:6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao H, Glazebrook J, Clarke J D, Volko S, Dong X. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 23.Ryals J A, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner H Y, Johnson J, Delaney T P, Jesse T, Vos P, Uknes S. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bork P. Proteins Struct Funct Genet. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 25.Michaely P, Bennet V. Trends Cell Biol. 1992;2:127–129. doi: 10.1016/0962-8924(92)90084-z. [DOI] [PubMed] [Google Scholar]
- 26.Gorina S, Pavletich N P. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 27.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Shapiro L. Nucleic Acids Res. 1993;21:3745–3748. doi: 10.1093/nar/21.16.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin G B, Brommonschenkel S H, Chungwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 30.Finley R L, Jr, Brent R. In: DNA Cloning—Expression Systems: A Practical Approach. Glover D, Hames B D, editors. Oxford: Oxford Univ. Press; 1996. pp. 169–203. [Google Scholar]
- 31.Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Plant J. 1998;16:223–234. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, Loh Y T, Bressan R A, Martin G B. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds A, Lundblad V. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Greene & Wiley; 1989. pp. 13.6.1–13.6.4. [Google Scholar]
- 34.Kawata T, Imada T, Shiraishi H, Okada K, Shimura Y, Iwabuchi M. Nucleic Acids Res. 1992;20:1141. doi: 10.1093/nar/20.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang C, Miao Z, Lam E. Plant Mol Biol. 1997;34:403–415. doi: 10.1023/a:1005873500238. [DOI] [PubMed] [Google Scholar]
- 36.Zhang B, Foley R C, Singh K B. Plant J. 1993;4:711–716. doi: 10.1046/j.1365-313x.1993.04040711.x. [DOI] [PubMed] [Google Scholar]
- 37.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam E, Benfey P N, Gilmartin P M, Fang R-X, Chua N-H. Proc Natl Acad Sci USA. 1989;86:7890–7894. doi: 10.1073/pnas.86.20.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin X-F, Holuigue L, Horvath D M, Chua N-H. Plant Cell. 1994;6:863–874. doi: 10.1105/tpc.6.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellis J G, Tokuhisa J G, Llewellyn D J, Bouchez D, Singh K, Dennis E S, Peacock W S. Plant J. 1993;4:433–443. doi: 10.1046/j.1365-313x.1993.04030433.x. [DOI] [PubMed] [Google Scholar]
- 41.Jupin I, Chua N-H. EMBO J. 1996;15:5679–5689. [PMC free article] [PubMed] [Google Scholar]
- 42.Schindler U, Beckmann H, Cashmore A R. Plant Cell. 1992;4:1309–1319. doi: 10.1105/tpc.4.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miao Z H, Liu X, Lam E. Plant Mol Biol. 1994;25:1–11. doi: 10.1007/BF00024193. [DOI] [PubMed] [Google Scholar]
- 44.Lam E, Lam Y K-P. Nucleic Acids Res. 1995;23:3778–3785. doi: 10.1093/nar/23.18.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]