Abstract
Treating spontaneously hypertensive rats (SHR) with l-arginine, taurine, and vitamins C and E (ATCE) during nephrogenesis (2 weeks before to 4 weeks after birth) persistently lowers blood pressure. Hypothetically, differential gene expression in kidney of SHR vs. normotensive Wistar–Kyoto rats (WKY) is partially corrected by maternal ATCE in SHR. Differential gene expression in 2-days, 2-weeks, and 48-week-old rats was studied using oligonucleotide chips. Transcription factor binding sites (TFBS) of differentially expressed genes were analyzed in silico. Differential gene expression varied between SHR+ATCE and SHR, suggesting both direct and indirect effects; but, few genes were modulated toward WKY level and there was little overlap between ages. TFBS analysis suggests less Elk-1-driven gene transcription in both WKY and SHR+ATCE vs. SHR at 2 days and 2 weeks. Concluding, in SHR, persistent antihypertensive effects of maternal ATCE are not primarily due to persistent corrective transcription. Less Elk-1-driven transcription at 2 days and 2 weeks may be involved.
Keywords: Development, Gene expression, Hypertensive rats, Kidney, Transcription factor binding sites
Introduction
Transplantation studies indicate that blood pressure “follows” the kidney [16, 25, 43] and all monogenic forms of hypertension have a renal origin [30]. In studies reporting on the renal transcriptome in SHR [19, 33, 35, 41], the earliest time point studied was 3–4 weeks of age [19, 41]. This is commonly termed the prehypertensive stage, a disputable term since small differences in blood pressure may not be detectable at this age [26]. At 2 days, the rat kidney contains at least 26 terminally differentiated cell types, recognizable by morphology, location, and function [2]. At 2 weeks, renal vascular and nephron development is complete [17, 31]. Even by 1 year, there is little renal injury in male SHR [10] and practically none in female SHR [9].
We previously reported persistent reduction of hypertension up to 48 weeks of age in offspring of SHR dams that were perinatally treated with a combination of l-arginine and antioxidants (taurine, vitamins C and E; ATCE) in the last 2 weeks of pregnancy and up to 4 weeks of lactation [39]. This alleviation of hypertension could be related to differences between the renal transcriptome of perinatally treated and untreated SHR. Hence, we hypothesized that differential gene expression in suckling 2 days and 2 weeks SHR vs. age-matched normotensive Wistar–Kyoto rats (WKY) would be partially corrected by maternal treatment with ATCE in SHR.
Primary objectives were to identify differential gene expression in suckling newborn (2 days) and 2-week-old SHR vs. WKY and associated transcription factor bindings sites (TFBS) to ascertain whether partial correction of this differential expression occurred with maternal treatment of SHR with ATCE and, if so, whether this partial correction involved the same associated TFBS. The TFBS were identified using in silico techniques. A secondary objective was to determine whether consistent differential gene expression and associated TFBS occurring in very young (prehypertensive) SHR vs. WKY persisted at 48 weeks in SHR vs. WKY and to ascertain whether these persistent variables were also persistently affected in SHR by maternal treatment with ATCE.
Materials and methods
Breeding and supplement protocol
Untreated SHR and normotensive Wistar–Kyoto rats (WKY; Harlan, Horst, The Netherlands) were kept under standard conditions and were fed non-synthetic rodent chow (Special Diets Services, Witham, Essex, England). SHR dams were perinatally treated with l-arginine, taurine, and vitamins C and E in the last 2 weeks of pregnancy and the first 4 weeks of lactation in order to increase NO and reduce oxidative stress during nephrogenesis. Female offspring of these treated SHR dams were used in the present study (SHR+ATCE). l-arginine, taurine, and vitamin C were dissolved in drinking water at doses of 20 g/L, 25 g/L, and 594 mg/L, respectively. Vitamin E was mixed in finely grounded chow (9 g/kg) that contained 2 and 82 mg/kg vitamins C and E, respectively. No difference was found in intake of food and water between treated and untreated rats (data not shown). Sentinel animals, housed under the same conditions, were regularly monitored for infections. All rats were housed in the same room under automated temperature control and a 12/12 h light/dark cycle. The Utrecht University Board for studies in experimental animals approved the protocol. Adult SHR and WKY females were mated with adult males from the same strain and supplier. At age of 2 days, the female pups were killed by decapitation, and at age of 2 and 48 weeks, the female rats were exsanguinated under pentobarbital anesthesia.
Total RNA isolation, microarray procedures, and analysis
Although kidneys are highly inhomogeneous, no differences in morphology and structure were found between rats of the same age (data not shown). To achieve homogeneity, the renal cortex of the adult kidney was collected from the poles of the kidney and snap frozen. However, in the pups, separation of cortex and medulla was not clear. Thus, the whole and half of the kidney was taken from 2-day and 2-week-old rats, respectively. Details of the procedures are available in Web Appendix (http://www.nephrogenomics.net/data/appendices/SHR-Development/). In short, total RNA was extracted (TRIzol™) from frozen samples. The RNA was stored at −80°C in distilled H2O. For microarray analysis, total RNA was pooled per group at each age in equal amounts per subject. At 2 days, 2 weeks, and 48 weeks of age, we studied WKY (n = 6, n = 6, and n = 8, respectively), SHR (n = 6, n = 5, and n = 12), and SHR+ATCE (n = 6, n = 5, and n = 6). Total RNA was reverse-transcribed (RT) with allyl-dUTP incorporation and labeled with Cy3 or Cy5 dyes (Amersham Biosciences, Piscataway, NJ, USA). Samples were hybridized to rat 7.5k Oligo Chips manufactured in the Genomics Laboratory, University Medical Center Utrecht, containing Rat Genome Array-Ready Oligo Set Version 1.1 of Qiagen-Operon [46], spotted in duplicate. Samples of WKY or SHR+ATCE were compared to age-matched untreated SHR and a dye switch procedure was applied [29]. SHR Cy3- and Cy5-labeled cDNA were also compared to allow elimination of unreliable spots. After washing, slides were stored in the dark until scanning using Agilent Scanner (BioDiscovery, El Segundo, CA, USA). Images were quantified using ImaGene Software (BioDiscovery) and data were normalized as described previously [29]. Duplicates of the genes were averaged. A ratio of a spot is defined as log2(Cy5/Cy3). Microarray data of SHR vs. SHR consisted of six arrays (control arrays). We used the average ratios and standard deviation (SD = 0.189) of the six control arrays to determine which genes were considered for the analyses.
To define the significance of ratio, we set the cutoff at three times SD. Considering the fact that statistics cannot be performed on treated vs. untreated because they were hybridized together on a single slide, the cutoff was set at a higher stringency level, 0.7. Thus, a ratio of >0.7 or <−0.7 were considered significant. The z test for two proportions indicated that a spot may have two out of six ratios >0.7 or <−0.7 before being indicated as unreliable. Out of 5,889 spots, 5,614 were considered reliable. Only the reliable spots, determined as described above, were used in data analysis. Considering the fact that homogeneity of kidneys may vary between ages, we compared the SHR samples to age-matched WKY samples. Thus, the reliable genes in WKY and SHR+ATCE compared to age-matched untreated SHR with a ratio of >0.7 or <−0.7 were considered differentially expressed. Microarray data were submitted to MIAME (EMBL-EBI) with accession numbers for array (A-MEXP-562) and experiment (E-MEXP-1550).
Clustering analysis
Hierarchical clustering of microarray data was performed using the Expression Profile data CLUSTering and analysis (EPCLUST, http://www.bioinf.ebc.ee/EP/EP/). Average linkage (average distance, UPGMA) clustering based on correlation measure-based distance was performed on data for ratios >0.7 or <−0.7.
Identification of genes with consistent differential expression
Observing genes at each age in WKY as compared to age-matched untreated SHR revealed more than 700 differentially expressed genes. We evaluated whether and for how many of these genes this was consistent at 2 days and 2 weeks. The rationale to do this was as follows. If a gene in the WKY vs. SHR comparison was consistently expressed differentially at 2 days and 2 weeks (but not at 48 weeks), this would support a potential role in early development. We also evaluated whether differential expression between WKY vs. SHR and SHR+ATCE vs. SHR overlapped, reasoning that this would reflect “correction” of the SHR transcriptome. Genes differentially expressed only in 48-week-old animals are most likely related to age and/or evolving damage.
Analysis of frequency of binding sites for transcription factors
To assess whether the genes expressed at different ages and responsive to perinatal ATCE treatment had different frequencies of transcription factor (TF) binding sites (TFBS) in their promoter regions, the 1,000-bp upstream regions of these genes were analyzed as previously described [4]. First, the upstream sequence of 1,000 bp of each gene was obtained via BIOMART (http://www.biomart.org). This sequence was subjected to TFBS analysis with a library of mononucleotide weight matrices from TRANSFAC® 6.0 using MATCH™ [23] (http://www.gene-regulation.com), and matrix and core similarity cutoffs were set at 0.95. Two sets of genes were subjected to this analysis: The differentially expressed genes and a set of 200 genes that was centered on a log2 ratio of zero. Thus, for each experimental group, two sets of frequencies of potential binding sites for TFs were obtained. A size test was used to determine the significance in difference of frequency of a TFBS between differentially expressed and non-differentially expressed gene set at each age. The same method for in silico analysis was applied to genes that were consistently and consecutively differentially expressed in at least two ages in WKY/SHR. This analysis was also performed on genes in SHR+ATCE that were shifted toward WKY level.
Quantitative PCR
For real-time PCR, total RNA from individual samples (five to six per group) was used. Laboratory details on semi-quantitative RT-PCR and the primer conditions are available as Web Appendix (http://www.nephrogenomics.net/data/appendices/SHR-Development/). TaqMan® Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) were used for real-time quantitative PCR, and the protocol was performed as recommended by the supplier. In short, cDNA (50 ng of starting material total RNA) was mixed with Taqman Universal PCR Master Mix (with AmpErase UNG) and TaqMan® Gene Expression Assay in end-volume of 25 μL. Quantitative PCR was performed with the ABI 7900HT Fast Real-Time PCR System. Gene threshold was determined by the SDS 2.2.2 software utility (Applied Biosystems). Genes studied with quantitative PCR were Cd36 (Rn00580728), connective tissue growth factor (CTGF; Rn00573960), Ephx2 (Rn00576023), Gstm1 (Rn00755117), and Ptger2 (Rn00579419). Calculation of the ratios is explained in the Web Appendix (http://www.nephrogenomics.net/data/appendices/SHR-Development/). All individual samples were compared to 2-day WKY as the calibrator group.
Statistical analyses
Results are expressed as mean ± SEM. Group and age-dependent effects were analyzed using two-way ANOVA where appropriate followed by a Student–Newman–Keuls post hoc test. The frequency of TFBS linked to differentially expressed genes was compared to that of the same TFBS linked to non-differentially expressed genes using the size test. P < 0.05 was considered significant.
Results
Animal data
The kidney/body weight (BW) ratio in SHR was higher than in WKY at 2 days and 2 weeks but not at 48 weeks (Table 1). Some of the kidneys at 48 weeks were from our previous study [39]. At 48 weeks, systolic blood pressure (measured every 4 to 6 weeks from age of 4 weeks) was 65–70 mmHg higher in SHR than in WKY (P < 0.001). BW was slightly lower in SHR+ATCE than in SHR at 2 and 48 weeks, but kidney/BW ratio was unaffected. At 2 days and 2 weeks, 24-h urine collection was not yet possible. At 48 weeks, there were no difference in urine NOx and TBARS between groups. In our previous study [39], we found no difference in NOx and TBARS excretion from 20 weeks onward.
Table 1.
Animal data
| WKY | SHR | SHR+ATCE | |
|---|---|---|---|
| 2 days old | |||
| n | 6 | 6 | 6 |
| Body weight (g) | 4.3 ± 0.3 | 4.8 ± 0.1 | 4.4 ± 0.2 |
| Right kidney weight (mg) | 21.3 ± 3.0 | 28.5 ± 1.4 | 27.0 ± 2.1 |
| RKW/BW (mg/g) | 4.8 ± 0.3 | 5.9 ± 0.2a | 6.1 ± 0.2 |
| 2 weeks old | |||
| n | 6 | 5 | 5 |
| Body weight (g) | 29.9 ± 0.5 | 20.4 ± 0.2b | 17.0 ± 1.8c |
| Right kidney weight (mg) | 150 ± 2.5 | 121 ± 1.9b | 98.2 ± 8.7c |
| RKW/BW (mg/g) | 5.0 ± 0.1 | 5.9 ± 0.1b | 5.8 ± 0.2 |
| 48 weeks old | |||
| n | 8 | 12 | 6 |
| Systolic blood pressure (mmHg) | 122 ± 2 | 187 ± 6b | 176 ± 5c |
| Body weight (g) | 267 ± 9 | 254 ± 8 | 228 ± 4c |
| Right kidney weight (mg) | 968 ± 53 | 882 ± 27 | 784 ± 32 |
| RKW/BW (mg/g) | 3.6 ± 0.2 | 3.4 ± 0.1 | 3.4 ± 0.2 |
aP < 0.05 vs. WKY
bP < 0.01 vs. WKY
cP < 0.05 vs. SHR
Global analysis of differential gene expression data
The number of differentially expressed genes at 2 days was higher than at 2 weeks, both in WKY vs. SHR and in SHR+ATCE vs. SHR. The old rats showed the highest number of differentially expressed genes (Table 2). As perinatal treatment with ATCE may partially correct the renal transcriptome in SHR, i.e., bring it closer to that of WKY, WKY (vs. SHR), and SHR+ATCE (vs. SHR) were compared using Venn diagrams. At 2 days and 2 weeks, 18 and nine of the differentially expressed genes (about 11%) were in the same direction in WKY and SHR+ATCE respectively, while at 48 weeks, this applied to 28 genes (about 7%). Hierarchical clustering was performed on the whole dataset for visualization of renal transcriptomes and how the groups related to each other (Fig. 1 in Web Appendix).
Table 2.
Number of differentially expressed genes
| Number of differentially expressed genes | |||||||
|---|---|---|---|---|---|---|---|
| 2 days | 2 weeks | 48 weeks | Total | ||||
| Criteria | WKY/SHR | SHR+ATCE/SHR | WKY/SHR | SHR+ATCE/SHR | WKY/SHR | SHR+ATCE/SHR | |
| Induced | 76 | 117 | 38 | 53 | 222 | 197 | 703 |
| Reduced | 45 | 46 | 56 | 35 | 272 | 143 | 597 |
| Total | 121 | 163 | 94a | 88a | 494a | 340a | 1,300 |
Induced indicates a ratio >0.7; Reduced indicates a ratio <−0.7 Differences in the number of differentially expressed genes between ages were determined using a z test. Venn diagrams compare differential expression of genes in WKY/SHR and SHR+ATCE/SHR. Upward and downward arrows indicate up- and down-regulation, respectively. Symbiosis of upward and downward arrows indicates counter-regulation
aP < 0.001 vs. all other ages with same treatment
This global analysis (both Venn diagram and clustering) revealed that, vs. WKY, young SHR displayed less differential renal gene expression than aged SHR. There was little consistent differential expression between consecutive ages. Third, perinatal ATCE treatment resulted in differential expression of the renal SHR transcriptome; however, differential expression did not become similar to WKY vs. SHR. Although at 48 weeks, perinatal ATCE in SHR still had a profound impact on differential gene expression (vs. control SHR); there was very little overlap with differential gene expression in WKY vs. SHR.
(Non)consecutive differential expression of genes at different ages
We first considered genes that were consistently differentially expressed in WKY/SHR (Table 3; n = 26). SHR was chosen as the denominator, so that in Tables 3 and 4, differential expression induced by perinatal treatment would be “towards” WKY/SHR. Twenty-one genes were consistently differentially expressed at 2 days and 2 weeks, and six genes were consistently differentially expressed at all three ages. Five other genes were consistently differentially expressed at 2 weeks and 48 weeks. At 2 days and 2 weeks, two vascular receptors, prostaglandin E2 receptor (Ptger2) and GABA receptor rho3 (Gabrr3), were higher in WKY/SHR. At all ages, Ephx2 (soluble epoxide hydrolase), Grin3a (glutamate receptor 3A), and Gstm1 (glutathione-S-transferase) were consistently lower in WKY/SHR. At 2 and 48 weeks, CTGF and CD74 are lower in WKY/SHR. Among the consistently differentially expressed genes in WKY/SHR, 15 were also among the most strongly differentially expressed genes (Web Appendix; Table 2).
Table 3.
Genes consistently and consecutively differentially expressed at more than one age in WKY/SHR (n = 26)
| Symbol | Description | Control arrays | 2 days | 2 weeks | 48 weeks | |||
|---|---|---|---|---|---|---|---|---|
| WKY | SHR+ATCE | WKY | SHR+ATCE | WKY | SHR+ATCE | |||
| LOC171573 | Spleen protein 1 precursor | 0.0 | −1.8 | −0.1 | −1.1 | 0.0 | 0.6 | 0.6 |
| Cdk5rap1 | CDK5 regulatory subunit associated protein 1 | −0.5 | −1.0 | 0.5 | −0.9 | 1.3 | 0.1 | 0.2 |
| Grin3a | Glutamate receptor, ionotropic, N-methyl-d-aspartate 3A | 0.0 | −0.9 | −1.0 | −0.8 | 1.5 | −0.9 | −0.7 |
| Ephx2 | Epoxide hydrolase 2, cytoplasmic | 0.0 | −0.8 | −0.1 | −1.9 | 0.0 | −1.4 | 0.1 |
| Skiv2l | Superkiller viralicidic activity 2-like | −0.1 | −0.7 | 0.1 | −0.7 | 0.0 | −0.2 | 0.3 |
| Ctsl | Cathepsin L | 0.0 | 0.7 | 0.0 | 0.7 | 0.0 | 0.3 | 0.0 |
| Gstm1 | Glutathione-S-transferase | −0.1 | 0.8 | 0.0 | 0.7 | −0.1 | −0.2 | 0.2 |
| Psg19 | Heart and neural crest derivatives expressed transcript 1 | −0.1 | 0.8 | 0.0 | 0.8 | −0.1 | 0.2 | 0.0 |
| Ptprj | Protein tyrosine phosphatase, receptor type, J | 0.0 | 0.8 | 0.2 | 1.1 | −0.1 | 1.0 | 0.2 |
| Sval2 | Seminal vesicle antigen-like 2 | −0.1 | 0.9 | 1.1 | 0.9 | 0.0 | 0.2 | 0.0 |
| V1rb5 | Vomeronasal V1r-type receptor V1rb5 | 0.0 | 0.9 | −1.1 | 1.0 | 1.5 | 0.0 | −0.3 |
| RT1-N1 | RT1 class Ib gene, H2-TL-like, grc region (N1) | −0.1 | 1.0 | 0.0 | 1.1 | 0.1 | 0.3 | 0.1 |
| Uts2r | Urotensin 2 receptor | 0.4 | 1.1 | −1.7 | 1.9 | 0.2 | 0.1 | 0.3 |
| Retsat | All-trans-13,14-dihydroretinol saturase | 0.0 | 1.1 | 0.0 | 0.9 | −0.1 | 1.3 | 0.0 |
| Gabrr3 | Gamma-aminobutyric acid (GABA) receptor, rho 3 | −0.1 | 1.4 | 0.2 | 1.3 | 0.4 | −0.3 | 0.1 |
| Nmi | N-myc (and STAT) interactor | −0.2 | 1.5 | 1.3 | 1.0 | −0.4 | 0.9 | 0.4 |
| Ptger2 | Prostaglandin E receptor 2, subtype EP2 | −0.1 | 1.5 | −0.7 | 0.8 | −1.7 | −0.3 | 0.1 |
| Cfi | Complement factor I | 0.0 | 1.5 | −0.6 | 1.2 | 0.1 | −0.1 | −0.4 |
| Il2 | Interleukin 2 | −0.1 | 1.6 | −0.2 | 1.0 | 0.2 | −0.3 | −0.2 |
| Spp2 | Secreted phosphoprotein 2 | 0.0 | 1.7 | −0.4 | 0.9 | 0.1 | 0.7 | 0.3 |
| Cd36 | Cd36 antigen | −0.1 | 2.8 | 0.9 | 1.5 | −0.1 | 0.5 | −0.2 |
| RT1-Bb | RT1 class II, locus Bb | 0.0 | −0.2 | −0.1 | −1.2 | 0.0 | −3.4 | −0.1 |
| Adam6 | A disintegrin and metallopeptidase domain 6 | 0.0 | 0.1 | 0.1 | −0.8 | −0.1 | −1.4 | 0.1 |
| Ctgf | Connective tissue growth factor | 0.0 | −0.4 | 0.5 | −0.9 | −0.3 | −1.3 | −0.1 |
| Cd74 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) | 0.0 | −0.5 | −0.1 | −0.8 | −0.1 | −0.9 | 0.0 |
| Adam18 | A disintegrin and metallopeptidase domain 18 | 0.2 | −0.1 | 0.1 | 0.8 | Below threshold | 1.8 | −0.1 |
Ratios in bold indicate absolute ratio >0.7
Table 4.
Genes differentially expressed in SHR+ATCE/SHR toward age-matched WKY/SHR level at 2 days (n = 15) and 2 weeks (n = 9)
| Symbol | Description | Control arrays | 2 days | 2 weeks | 48 weeks | |||
|---|---|---|---|---|---|---|---|---|
| WKY | SHR+ATCE | WKY | SHR+ATCE | WKY | SHR+ATCE | |||
| Cyp2c13 | Cytochrome P450 2c13 | −0.2 | −2.5 | −2.4 | 1.2 | −1.5 | 0.5 | 0.1 |
| Cyp11b2 | Cytochrome P450, family 11, subfamily B, poly peptide 2 | 0.1 | −2.3 | −1.8 | 0.0 | 0.8 | 0.0 | 0.0 |
| Cyp21a1 | Cytochrome P450, subfamily 21A, poly peptide 1 | 0.2 | −1.8 | −1.4 | −0.1 | −0.5 | 0.0 | 0.1 |
| Cyp11b1 | Cytochrome P450, subfamily 11B, poly peptide 1 | −0.1 | −2.6 | −1.4 | −0.1 | −0.3 | 0.1 | −0.1 |
| Grin3a | Glutamate receptor, ionotropic, N-methy l-d-aspartate 3A | 0.0 | −0.9 | −1.0 | −0.8 | 1.5 | −0.9 | −0.7 |
| Atf3 | Activating transcription factor 3 | −0.1 | −0.9 | −1.0 | 0.3 | 0.1 | 0.1 | −0.1 |
| Akr1b7 | Aldo-keto reductase family 1, member B7 | 0.2 | −1.2 | −0.9 | 0.7 | −0.1 | 0.6 | 0.2 |
| Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | 0.0 | −0.7 | −0.9 | 0.0 | 0.3 | 1.1 | −0.1 |
| Cyp3a62 | Cytochrome P450, subfamily 3A, poly peptide 62 | −0.2 | 1.0 | 0.7 | −0.1 | 0.8 | 0.1 | −0.3 |
| Fut9 | Fucosy ltransferase 9 | 0.2 | 1.7 | 0.7 | 0.5 | −0.5 | 0.2 | 0.1 |
| Ptgfr | Prostaglandin F receptor | 0.2 | 0.9 | 0.9 | −1.3 | 0.3 | 0.0 | −0.1 |
| Cd36 | Cd36 antigen | −0.1 | 2.8 | 0.9 | 1.5 | −0.1 | 0.5 | −0.2 |
| Sval2 | Seminal vesicle antigen-like 2 | −0.1 | 0.9 | 1.1 | 0.9 | 0.0 | 0.2 | 0.0 |
| Cars | Cysteinyl-tRNA synthetase | −0.2 | 1.0 | 1.2 | 0.4 | 0.3 | −0.1 | 0.3 |
| Nmi | N-my c (and STAT) interactor | −0.2 | 1.5 | 1.3 | 1.0 | −0.4 | 0.9 | 0.4 |
| Ndufa12 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 | −0.5 | 1.3 | −0.5 | −0.8 | −1.7 | −0.2 | −0.4 |
| Hist1h4b | Histone cluster 1, H4b | 0.2 | 0.1 | −0.2 | −1.0 | −1.3 | 0.0 | −0.9 |
| Prss1 | Protease, serine, 1 (trypsin 1) | −0.1 | −0.3 | −1.2 | −0.8 | −1.1 | 0.0 | 0.0 |
| Slco1a1 | Solute carrier organic anion transporter family, member 1a1 | 0.1 | −0.1 | −2.5 | −2.2 | −1.1 | 0.9 | 0.1 |
| Prl8a3 | Prolactin family 8, subfamily a, member 3 | 0.1 | −0.4 | 0.0 | −1.3 | −0.8 | 0.0 | 0.4 |
| Krt1 | Keratin 1 | 0.1 | −0.2 | −0.3 | 1.2 | 0.8 | −0.1 | 0.3 |
| Slco1a5 | Solute carrier organic anion transporter family, member 1a6 | 0.2 | 0.1 | 1.6 | 1.1 | 1.0 | 0.1 | 0.1 |
| Ryr2 | Ryanodine receptor 2, cardiac | 0.2 | −0.5 | −0.1 | 1.6 | 1.3 | −0.1 | 0.5 |
| V1rb5 | Vomeronasal V1r-type receptor V1rb5 | 0.0 | 0.9 | −1.1 | 1.0 | 1.5 | 0.0 | −0.3 |
Ratios in bold indicate absolute ratio >0.7
We then focused on differential expression of genes in SHR+ATCE/SHR that shifted toward WKY/SHR levels. The Venn diagram showed that 55 genes had congruent differential expression in both WKY and SHR+ATCE as compared to SHR (Table 2; all ages together). Only six of these 55 genes were shifted in the same direction in at least two ages (consistent change). At both 2 days and 2 weeks, this was the case for five genes, namely two organic anion solute carriers (Slco1a1 and Slco1a5), two cytochrome P450 oxidases (Cyp2c13 and Cyp3a62), and a trypsin (Prss1; Table 4). At 48 weeks, only one of the 28 genes, where differential expression of SHR+ATCE/SHR had shifted toward WKY/SHR, coincided with one of the six genes consistent changed by ATCE treatment at age of 2 days, namely the glutamate receptor Grin3a.
Analysis of frequency of binding sites for transcription factors
In an attempt to identify transcription factors involved in the regulation of gene expression in the renal cortex of the SHR at different ages, we performed an in silico analysis of the 1,000 bp upstream regions of the differentially expressed genes for the presence of TFBS and compared this to the upstream regions of genes that were not differentially expressed.
The first analysis describes the frequency, for each age, with which a TFBS can be detected in a group of differentially expressed genes vs. the non-differentially expressed control set. Comparing upstream regions in differentially expressed genes vs. non-differentially expressed genes in WKY vs. SHR revealed 12 TFBS to be significantly affected (Table 5). Only Elk-1 and S8 were identified at both 2 days and 2 weeks, and Elk-1 was the only TF with a reduced frequency of TFBS. Whether reduced or induced frequency correlated with increased or reduced gene differential expression in WKY vs. SHR is undetermined because all genes were taken into the calculation, and TF activity at TFBS can be both stimulatory and inhibitory.
Table 5.
Analysis of frequency of transcription factor binding sites (TFBS) in WKY/SHR and SHR+ATCE/SHR
| WKY/SHR | SHR+ATCE/SHR | |||||
|---|---|---|---|---|---|---|
| z test | z test | |||||
| TF | 2 days | 2 weeks | 48 weeks | 2 days | 2 weeks | 48 weeks |
| Elk-1 | ↓ | ↓ | – | ↓↓↓ | ↓ | – |
| S8 | ↑ | ↑ | – | – | – | ↑↑ |
| HFH-8 | ↑ | – | – | – | – | – |
| Tal-1alpha/E47 | ↑ | – | – | – | – | – |
| Tal-1beta/E47 | ↑↑ | – | – | – | – | – |
| GATA-1 | – | ↑ | – | – | – | – |
| GATA-2 | – | ↑ | – | – | – | – |
| GATA-X | – | ↑↑↑ | – | – | – | – |
| Lmo2 | – | ↑ | – | – | – | – |
| Poly | – | ↑ | – | – | – | – |
| HFH-1 | – | – | ↑ | – | – | – |
| HFH-3 | – | – | ↑↑ | – | – | – |
| c-Myc/Max | – | – | – | ↓ | – | – |
| c-Rel | – | – | – | ↓ | – | – |
| NRF-2 | – | – | – | ↓ | – | – |
| ER | – | – | – | – | – | ↑ |
| HNF-3beta | – | – | – | – | – | ↑↑ |
| HNF-4 | – | – | – | – | – | ↑ |
For each experimental group, two sets of frequencies of potential TFBS were obtained (differentially expressed and non-differentially expressed genes). Note that frequency was either up or down. A size test was applied to determine the significance in difference of frequency of a TFBS between the differentially expressed and non-differentially expressed genes (one, two, or three arrows indicate P < 0.05, P < 0.01, and P < 0.001, respectively). Reduced/increased frequency indicates that the binding site is less/more frequently activated or is inhibited/stimulated, respectively
↓ reduced frequency, ↑ increased frequency
In silico analysis of SHR+ATCE vs. SHR datasets revealed eight TFBS to be involved. Interestingly, both SHR+ATCE/SHR and WKY/SHR had reduced Elk-1 frequency at 2 days and 2 weeks, indicating that ATCE treatment shifted Elk-1 frequency in SHR toward that of WKY. The observation that the frequency of Elk-1 TFBS was reduced in both WKY and SHR+ATCE vs. SHR suggests that SHR have enhanced Elk-1 driven gene transcription as compared to WKY at 2 days and 2 weeks. Elk-1 was not identified by this approach at 48 weeks, suggesting a specific role in the prehypertensive stage.
In the second analysis, we determined whether the genes consistently and consecutively altered at different ages (at all ages and at 2 days and 2 weeks in WKY/SHR) were related to each other with respect to their TFBS. Four (CDP, GATA-X, HFH-8, and XFD-2) had significantly altered frequency of their TFBS (Table 6). Note that the reduction of Elk-1 binding sites was not associated with consistently differentially expressed genes. For genes differentially expressed at all ages, TFBS for CDP were more frequent at day 2. Both GATA-X and XFD-2 had TFBS with a higher frequency at 2 days and 2 weeks for the genes with consecutive differential expression in the 2 days to 2 weeks gene set of WKY/SHR. For HFH-8, an increased frequency of TFBS in differentially expressed genes was detected at day 2 in the gene set of 2 days to 2 weeks of WKY/SHR. Note that HFH-8 also appears in whole WKY/SHR data set at 2 days (Table 5).
Table 6.
Analysis of frequency of transcription factor binding sites (TFBS) in WKY/SHR that are linked to consistently and consecutively differentially expressed genes and in SHR+ATCE/SHR that are linked to genes differentially expressed either up or down
| All ages in WKY/SHR | 2 days and 2 weeks in WKY/SHR | SHR+ATCE/SHR shifted toward age-matched WKY/SHR | ||||||
|---|---|---|---|---|---|---|---|---|
| z test | z test | z test | ||||||
| TF | 2 days | 2 weeks | 48 weeks | 2 days | 2 weeks | 2 days | 2 weeks | 48 weeks |
| CDP | ↑ | – | – | – | – | – | – | – |
| GATA-X | – | – | – | ↑ | ↑↑ | – | – | – |
| HFH-8 | – | – | – | ↑ | – | – | – | – |
| XFD-2 | – | – | – | ↑↑ | ↑↑ | – | – | – |
| ER | – | – | – | – | – | ↑↑ | – | – |
| GATA-3 | – | – | – | – | – | ↑ | – | – |
| Gfi-1 | – | – | – | – | – | ↑ | – | – |
| RFX1 | – | – | – | – | – | ↑ | – | – |
| TGIF | – | – | – | – | – | ↑ | – | – |
| GR | – | – | – | – | – | – | ↑↑ | – |
| Oct-1 | – | – | – | – | – | – | – | ↑ |
| XFD-1 | – | – | – | – | – | – | – | ↑↑ |
A size test was applied to determine the difference of frequency of a TFBS between the differentially expressed and non-differentially expressed genes (one or two arrows indicate P < 0.05 and P < 0.01, respectively). Increased frequency indicates that the binding site is more frequently activated or is stimulated
↑ increased frequency
For each age, the TFBS level of differentially expressed genes in SHR+ATCE/SHR that had shifted toward WKY/SHR was determined (Table 6). Increased frequencies at eight TFBS were found: five at 2 days, one at 2 weeks, and two at 48 weeks. The TFBS for ER appeared in both analyses (cf., Tables 5 and 6), but not at the same age.
Gene expression by PCR
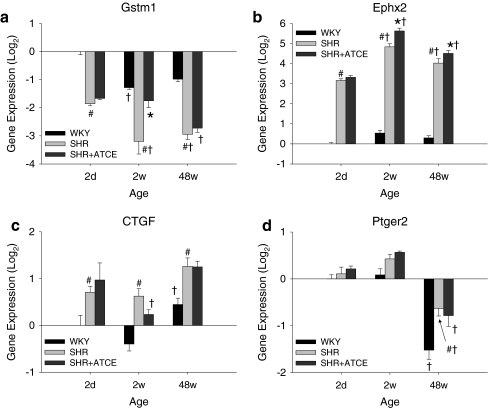
Semi-quantitative PCR was performed on ten genes in 2-days and 2-week-old SHR (Table 5; Web Appendix). Quantitative PCR was performed on Cd36, Gstm1, Ephx2, CTGF, and Ptger2 in WKY, SHR, and SHR+ATCE at all ages. Note that, in contrast to differential expression of the microarray data in Table 3, the data in Fig. 1 are normalized to expression in WKY at 2 days. Due to deletion in Cd36 caused by unequal recombination [14], gene expression of Cd36 cannot be directly compared in WKY vs. SHR. Gstm1 gene expression was reduced in SHR compared to WKY at all ages (Fig. 1a). ATCE treatment in SHR temporarily induced Gstm1 to the level of WKY at 2 weeks. Ephx2 gene expression was strongly increased in SHR as compared to WKY at all ages, confirming that the microarray data and ATCE treatment induced the expression slightly more at 2 and 48 weeks (Fig. 1b). CTGF and Ptger2 were increased in SHR vs. WKY (all ages for CTGF and 48 weeks for Ptger2) but were not consistently affected by perinatal ATCE treatment (Figs. 1c and d).
Fig. 1.
Real-time quantitative PCR of Gstm1 (a), Ephx2 (b), CTGF (c), and Ptger2 (d). There were five to six rats per group. All data are expressed relative to WKY at 2 days. Data were analyzed by two-way ANOVA. Different degrees of post hoc significances are all collected in P < 0.05 in order to decrease complexity of presentation. #P < 0.05 vs. WKY (same age), *P < 0.05 vs. SHR (same age), †P < 0.05 vs. younger age (same strain)
Discussion
Extensive differential expression involving hundreds of genes was present at a very early age, namely 2 days and 2 weeks, in SHR vs. WKY. However, a relatively small group of these displayed consistent and consecutive differential expression at 2 days and 2 weeks. The renal transcriptome also varied substantially at different ages for SHR+ATCE vs. SHR, suggesting both direct and indirect effects of early treatment. However, perinatal treatment of SHR with ATCE induced practically no congruent direct change in differential expression for the two comparisons. In silico analysis of TFBS for both sets of differentially expressed genes revealed that only ELK-1 was consistently related to differentially expressed genes in both comparisons and at both 2 days and 2 weeks. At 48 weeks, there were very few genes with congruent differential expression in the two comparisons and no shared TFBS.
Differential renal gene expression was quite extensive at 2 days and 2 weeks. Because of our interest in early development of hypertension, this was the focus of our attention. In the light of the perinatal intervention with ATCE, we first investigated the transporters of l-arginine and taurine, as reported elsewhere [24]. The expression of Slc7a7 (an important basolateral arginine transporter) was decreased in 2 days SHR vs. WKY, and RT-PCR revealed slc7a7 also to be decreased in 2 weeks SHR. This suggests defective renal arginine transport, which may be related to the reduced renal NO production that we recently documented in 2-week-old female SHR [24]. The present study shows that ATCE supplements did not affect slc7a7 expression in SHR (Web Appendix, Table 5). Hence, perinatal ATCE may not directly be able to improve renal nitric oxide (NO) production, and the increased renal NO observed previously [39] may have resulted from decreased oxidative stress. Another gene of interest was soluble (cytoplasmic) epoxide hydrolase 2 (Ephx2), an enzyme that metabolizes epoxyeicosatrienoic acids, potent vasodilator agents, to less active dihydroxyeicosatrienoic acids [21]. Inhibition of Ephx2 has been shown to reduce blood pressure in various models of hypertension [20]. Our finding that Ephx2 was consistently and strongly upregulated from 2 days of age makes it an obvious target for inhibition at an early age, and these experiments are indeed underway. However, Ephx2 was not inhibited by ATCE, on the contrary.
Well-known candidate genes with reduced expression in SHR include fatty acid translocase (Cd36) and glutathione-S-transferase (Gstm1). Cd36 was identified as an insulin-resistance gene causing defective fatty acid and glucose metabolism in SHR [1]. This is due to deletion in Cd36 caused by unequal recombination [14]. Our amplification plot of qPCR clearly confirms the existence of different gene products of Cd36 in WKY and SHR. Transgenic rescue of defective Cd36 prevents insulin resistance but not high blood pressure [38]. Moreover, the Cd36 mutation was absent in the original SHR [15, 35]. However, a recent report from the same group does implicate Cd36 in the pathogenesis of hypertension in SHR [37]. Prevention of oxidative stress may ameliorate development of hypertension in SHR [40, 48, 49]. Oxidative stress is increased in the kidney of aging SHR [6, 12]. This has been ascribed to the well-documented reduced expression of Gstm1 [19, 32, 33]. However, similarly to Cd36, earlier work has shown that down-regulation of Gstm1 is not common to all SHR substrains [35]. Moreover, recent data document that GSTM variants are not associated with hypertension in humans [8]. Thus, for Gstm1, convincing evidence suggests relations with the metabolic but not the hypertensive aspects of the SHR phenotype. Neither Cd36 nor Gstm1 was substantially directly or indirectly affected by perinatal ATCE in SHR, despite persistently reduced blood pressure [39].
TFs importantly regulate transcription [27]. Conversely, a gene expression profile may indirectly reflect the actions of TFs. Therefore, TFs can be related to differentially expressed genes in WKY vs. SHR and in SHR+ATCE vs. SHR. We previously developed an approach to derive potential TFBS from a differential gene expression set [4]. While we recognize the limitations of such a screening approach, it can lead us to TFs that are involved in both the development of hypertension and in perinatal programming phenomena in the SHR. The generated inventory of documented TFBS, recognized in the upstream 1,000 bp of the differentially expressed vs. non-differentially expressed genes, revealed that only binding sites of Elk-1 are significantly less associated with differential gene expression between WKY and SHR and between SHR+ATCE and SHR. This was observed at 2 days and at 2 weeks. The observation that the frequency of Elk-1 TFBS was reduced in both WKY and SHR+ATCE vs. SHR suggests that untreated SHR have enhanced Elk-1 driven gene transcription at 2 days and 2 weeks. Elk-1 is reportedly down-stream of ERK 1/2, MAPK, and JNK (Fig. 2; Web Appendix) [13, 28]. Down-stream signaling from Elk-1 is via c-Fos and other pathways [44]. With respect to the treatment with arginine and antioxidants, it could be noted that ERK1/2 is influenced by NO through cAMP [5, 18] and JNK and Elk-1 by NO and ROS [22]. Very little information is available on the role of epigenetic modification of TFs in early development. However, recently, it was reported that intrauterine growth retardation is associated with progressive epigenetic silencing of Pdx1, a pancreatic and duodenal homeobox 1 TF that regulates pancreas development and β cell differentiation [36]. Ultimately, this appears to lead to the development of type 2 diabetes in adults [42]. However, in our model, Elk-1 binding sites are not significantly linked to individual genes that were consistently differentially expressed at more than one age, underlining the complexity of transcriptional regulation down-stream from Elk-1.
In SHR vs. WKY, the number of differentially expressed genes increased fivefold from 2 to 48 weeks. Changes in our study were less consistent than previously reported [19]. This is not unexpected because the age span in that study was only from 4 to 18 weeks [19], which falls within the 2- to 48-week step in our study. This supports the notion that in the face of persistent hypertension, secondary changes occur in the renal cortex and in other target organs [11, 47]. However, with respect to our secondary objective, we could identify very few persistent consistent changes in differential gene expression in the WKY/SHR gene set, and none of these were affected by perinatal ATCE. The latter is possibly due to the fact that perinatal ATCE only reduced systolic blood pressure by about 35 mmHg between 14 and 26 weeks and that this difference fell to about 10 mmHg by 48 weeks. This is similar to the reduction obtained by continuous antioxidant supplements administered from the first week of pregnancy up to 24 weeks of age [48, 49]. Moreover, there were no persistent TFBS. Had we observed multiple persistent and consistent changes, then we would sorely have missed a group of WKY perinatally treated with ATCE. However, for the current data set, this does not appear to be a major drawback. An interesting gene in the light of aging, hypertension, and target organ damage is CTGF. Although CTGF is widely studied, little is known on renal CTGF in SHR. CTGF is induced in adult SHR and is reduced by antihypertensive treatments [7], suggesting that CTGF expression is dependent on blood pressure. However, we already found increased CTGF expression at 2 days in SHR, a stage when presumably no hypertension, let alone fibrosis, is present.
Some limitations of the present study have to be discussed. First, the kidney is highly inhomogeneous. However, there was no a priori reason to choose a single structure (e.g., only glomeruli), so no differential sieving was applied. Moreover, histology confirmed that there were no differences in renal morphology between WKY and SHR. Thus, cortex was sampled from all the adult rats by sampling the renal poles. However, in the small pups, separation of cortex and medulla was not clear. Thus, all or half of the kidney was taken from 2-day and 2-week-old rats, respectively. This ensured that the degree of homogeneity remained constant between age-matched SHR and WKY. Second, an absolute stringent cutoff of 0.7, much higher than three times the standard deviation of the six control-vs.-control arrays, was applied to identify differential gene expression. Applying the stringent cutoff lowers the chance of false positives, but subtle changes in differential gene expression may be missed. Lastly, microarray technology is extremely powerful but is not a technology that is easily applicable to test mechanisms, in particular not when considering the complex structure of the kidney. Nevertheless, there are numerous other microarray studies, which have pointed at potential mechanisms, and then have been followed by studies specifically directed at these potential pathways, including one from our group [24].
In conclusion, analysis of the renal transcriptome over the whole life span of WKY and SHR+ATCE vs. SHR revealed considerable differential expression from a very early age, but with very little overlap. The number of differentially expressed genes increases with age, suggesting secondary changes. At an early age, a common factor to both sets of differentially expressed genes was a reduction in TFBS of Elk-1. Our genomics approach suggests increased activity of the TF ELK-1 in SHR in the prehypertensive stage. This deserves further study.
Perspective
There is accumulating evidence that poor conditions in early life have long-term deleterious consequences, including hypertension [34]. The present study shows that perinatal treatment of hypertensive rats with micronutrients such as amino acids and vitamins that persistently lowered blood pressure in their offspring directly affects the renal cortical transcriptome. As such, improved conditions in early life may have long-term advantages, especially in people with a predisposition to hypertension such as indigenous communities exposed to a Western life style [3, 45].
Acknowledgments
The Dutch Kidney Foundation (Grants NS6013 and C03.2039) and the European Union Sixth Framework Program for Research and Technical Development of the European Community—The Early Nutrition Programming Project (FOOD-CT-2005-007036) supported this study. A Research Fellowship of the Royal Dutch Academy of Arts and Sciences supported Branko Braam. The technical assistance of Adèle Dijk, Nel Willekes-Koolschijn, and Paula Martens (Univ. Med. Center Utrecht—Nephrology) was greatly appreciated.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St Lezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J (1999) Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet 21:76–83 [DOI] [PubMed]
- 2.Al-Awqati Q, Oliver JA (2002) Stem cells in the kidney. Kidney Int 61:387–395 [DOI] [PubMed]
- 3.Al Salmi I, Hoy WE, Kondalsamy-Chennakesavan S, Wang Z, Gobe GC, Barr EL, Shaw JE (2008) Disorders of glucose regulation in adults and birth weight: results from the Australian Diabetes, Obesity and Lifestyle (AUSDIAB) Study. Diabetes Care 31:159–164 [DOI] [PubMed]
- 4.Braam B, de Roos R, Bluyssen H, Kemmeren P, Holstege F, Joles JA, Koomans H (2005) Nitric oxide-dependent and nitric oxide-independent transcriptional responses to high shear stress in endothelial cells. Hypertension 45:672–680 [DOI] [PubMed]
- 5.Cavanaugh JE, Ham J, Hetman M, Poser S, Yan C, Xia Z (2001) Differential regulation of mitogen-activated protein kinases ERK1/2 and ERK5 by neurotrophins, neuronal activity, and cAMP in neurons. J Neurosci 21:434–443 [DOI] [PMC free article] [PubMed]
- 6.de Cavanagh EM, Toblli JE, Ferder L, Piotrkowski B, Stella I, Inserra F (2006) Renal mitochondrial dysfunction in spontaneously hypertensive rats is attenuated by losartan but not by amlodipine. Am J Physiol Regul Integr Comp Physiol 290:R1616–1625 [DOI] [PubMed]
- 7.de las Heras N, Ruiz-Ortega M, Miana M, Ruperez M, Sanz-Rosa D, Aragoncillo P, Mezzano S, Cachofeiro V, Egido J, Lahera V (2007) Interactions between aldosterone and connective tissue growth factor in vascular and renal damage in spontaneously hypertensive rats. J Hypertens 25:629–638 [DOI] [PubMed]
- 8.Delles C, Padmanabhan S, Lee WK, Miller WH, McBride MW, McClure JD, Brain NJ, Wallace C, Marcano AC, Schmieder RE, Brown MJ, Caulfield MJ, Munroe PB, Farrall M, Webster J, Connell JM, Dominiczak AF (2008) Glutathione S-transferase variants and hypertension. J Hypertens 26:1343–1352 [DOI] [PubMed]
- 9.Feld LG, Brentjens JR, Van Liew JB (1981) Renal injury and proteinuria in female spontaneously hypertensive rats. Ren Physiol 4:46–56 [DOI] [PubMed]
- 10.Feld LG, Van Liew JB, Brentjens JR, Boylan JW (1981) Renal lesions and proteinuria in the spontaneously hypertensive rat made normotensive by treatment. Kidney Int 20:606–614 [DOI] [PubMed]
- 11.Fornage M, Swank MW, Boerwinkle E, Doris PA (2003) Gene expression profiling and functional proteomic analysis reveal perturbed kinase-mediated signaling in genetic stroke susceptibility. Physiol Genomics 15:75–83 [DOI] [PubMed]
- 12.Fortepiani LA, Reckelhoff JF (2005) Increasing oxidative stress with molsidomine increases blood pressure in genetically hypertensive rats but not normotensive controls. Am J Physiol Regul Integr Comp Physiol 289:R763–R770 [DOI] [PubMed]
- 13.Fuchs SY, Xie B, Adler V, Fried VA, Davis RJ, Ronai Z (1997) c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J Biol Chem 272:32163–32168 [DOI] [PubMed]
- 14.Glazier AM, Scott J, Aitman TJ (2002) Molecular basis of the Cd36 chromosomal deletion underlying SHR defects in insulin action and fatty acid metabolism. Mamm Genome 13:108–113 [DOI] [PubMed]
- 15.Gotoda T, Iizuka Y, Kato N, Osuga J, Bihoreau MT, Murakami T, Yamori Y, Shimano H, Ishibashi S, Yamada N (1999) Absence of Cd36 mutation in the original spontaneously hypertensive rats with insulin resistance. Nat Genet 22:226–228 [DOI] [PubMed]
- 16.Grisk O, Kloting I, Exner J, Spiess S, Schmidt R, Junghans D, Lorenz G, Rettig R (2002) Long-term arterial pressure in spontaneously hypertensive rats is set by the kidney. J Hypertens 20:131–138 [DOI] [PubMed]
- 17.Guron G, Friberg P (2000) An intact renin–angiotensin system is a prerequisite for normal renal development. J Hypertens 18:123–137 [DOI] [PubMed]
- 18.Hayashi T, Juliet PA, Matsui-Hirai H, Miyazaki A, Fukatsu A, Funami J, Iguchi A, Ignarro LJ (2005) l-Citrulline and l-arginine supplementation retards the progression of high-cholesterol-diet-induced atherosclerosis in rabbits. Proc Natl Acad Sci U S A 102:13681–13686 [DOI] [PMC free article] [PubMed]
- 19.Hinojos CA, Boerwinkle E, Fornage M, Doris PA (2005) Combined genealogical, mapping, and expression approaches to identify spontaneously hypertensive rat hypertension candidate genes. Hypertension 45:698–704 [DOI] [PubMed]
- 20.Imig JD (2005) Epoxide hydrolase and epoxygenase metabolites as therapeutic targets for renal diseases. Am J Physiol Renal Physiol 289:F496–F503 [DOI] [PubMed]
- 21.Imig JD, Navar LG, Roman RJ, Reddy KK, Falck JR (1996) Actions of epoxygenase metabolites on the preglomerular vasculature. J Am Soc Nephrol 7:2364–2370 [DOI] [PubMed]
- 22.Ishrath A, Kumar N, Dey CS (2002) Differential activation of ERK and JNK by arsenite in mouse muscle cells. Comp Biochem Physiol C Toxicol Pharmacol 132:375–384 [DOI] [PubMed]
- 23.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E (2003) MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 31:3576–3579 [DOI] [PMC free article] [PubMed]
- 24.Koeners MP, van Faassen EE, Wesseling S, de Sain-van der Velden M, Koomans HA, Braam B, Joles JA (2007) Maternal supplementation with citrulline increases renal nitric oxide in young spontaneously hypertensive rats and has long-term antihypertensive effects. Hypertension 50:1077–1084 [DOI] [PubMed]
- 25.Kurtz TW, Al-Bander HA, Morris RC Jr (1987) “Salt-sensitive” essential hypertension in men. Is the sodium ion alone important? N Engl J Med 317:1043–1048 [DOI] [PubMed]
- 26.Lais LT, Rios LL, Boutelle S, DiBona GF, Brody MJ (1977) Arterial pressure development in neonatal and young spontaneously hypertensive rats. Blood Vessels 14:277–284 [DOI] [PubMed]
- 27.Latchman DS (1997) Transcription factors: an overview. Int J Biochem Cell Biol 29:1305–1312 [DOI] [PubMed]
- 28.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG (2004) Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell 14:43–55 [DOI] [PubMed]
- 29.Liang M, Briggs AG, Rute E, Greene AS, Cowley AW Jr (2003) Quantitative assessment of the importance of dye switching and biological replication in cDNA microarray studies. Physiol Genomics 14:199–207 [DOI] [PubMed]
- 30.Lifton RP, Gharavi AG, Geller DS (2001) Molecular mechanisms of human hypertension. Cell 104:545–556 [DOI] [PubMed]
- 31.Marquez MG, Cabrera I, Serrano DJ, Sterin-Speziale N (2002) Cell proliferation and morphometric changes in the rat kidney during postnatal development. Anat Embryol (Berl) 205:431–440 [DOI] [PubMed]
- 32.McBride MW, Brosnan MJ, Mathers J, McLellan LI, Miller WH, Graham D, Hanlon N, Hamilton CA, Polke JM, Lee WK, Dominiczak AF (2005) Reduction of Gstm1 expression in the stroke-prone spontaneously hypertension rat contributes to increased oxidative stress. Hypertension 45:786–792 [DOI] [PubMed]
- 33.McBride MW, Carr FJ, Graham D, Anderson NH, Clark JS, Lee WK, Charchar FJ, Brosnan MJ, Dominiczak AF (2003) Microarray analysis of rat chromosome 2 congenic strains. Hypertension 41:847–853 [DOI] [PubMed]
- 34.McMillen IC, Robinson JS (2005) Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev 85:571–633 [DOI] [PubMed]
- 35.Okuda T, Sumiya T, Iwai N, Miyata T (2002) Difference of gene expression profiles in spontaneous hypertensive rats and Wistar–Kyoto rats from two sources. Biochem Biophys Res Commun 296:537–543 [DOI] [PubMed]
- 36.Park JH, Stoffers DA, Nicholls RD, Simmons RA (2008) Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest 118:2316–2324 [DOI] [PMC free article] [PubMed]
- 37.Pravenec M, Churchill PC, Churchill MC, Viklicky O, Kazdova L, Aitman TJ, Petretto E, Hubner N, Wallace CA, Zimdahl H, Zidek V, Landa V, Dunbar J, Bidani A, Griffin K, Qi N, Maxova M, Kren V, Mlejnek P, Wang J, Kurtz TW (2008) Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat Genet 40:952–954 [DOI] [PubMed]
- 38.Pravenec M, Landa V, Zidek V, Musilova A, Kren V, Kazdova L, Aitman TJ, Glazier AM, Ibrahimi A, Abumrad NA, Qi N, Wang JM, St Lezin EM, Kurtz TW (2001) Transgenic rescue of defective Cd36 ameliorates insulin resistance in spontaneously hypertensive rats. Nat Genet 27:156–158 [DOI] [PubMed]
- 39.Racasan S, Braam B, van der Giezen DM, Goldschmeding R, Boer P, Koomans HA, Joles JA (2004) Perinatal L-arginine and antioxidant supplements reduce adult blood pressure in spontaneously hypertensive rats. Hypertension 44:83–88 [DOI] [PubMed]
- 40.Rodriguez-Iturbe B, Ferrebuz A, Vanegas V, Quiroz Y, Mezzano S, Vaziri ND (2005) Early and sustained inhibition of nuclear factor-kappaB prevents hypertension in spontaneously hypertensive rats. J Pharmacol Exp Ther 315:51–57 [DOI] [PubMed]
- 41.Seubert JM, Xu F, Graves JP, Collins JB, Sieber SO, Paules RS, Kroetz DL, Zeldin DC (2005) Differential renal gene expression in prehypertensive and hypertensive spontaneously hypertensive rats. Am J Physiol Renal Physiol 289:F552–F561 [DOI] [PubMed]
- 42.Simmons RA, Templeton LJ, Gertz SJ (2001) Intrauterine growth retardation leads to the development of type 2 diabetes in the rat. Diabetes 50:2279–2286 [DOI] [PubMed]
- 43.Smallegange C, Hale TM, Bushfield TL, Adams MA (2004) Persistent lowering of pressure by transplanting kidneys from adult spontaneously hypertensive rats treated with brief antihypertensive therapy. Hypertension 44:89–94 [DOI] [PubMed]
- 44.Tanimura S, Nomura K, Ozaki K, Tsujimoto M, Kondo T, Kohno M (2002) Prolonged nuclear retention of activated extracellular signal-regulated kinase 1/2 is required for hepatocyte growth factor-induced cell motility. J Biol Chem 277:28256–28264 [DOI] [PubMed]
- 45.Thame M, Osmond C, Wilks RJ, Bennett FI, McFarlane-Anderson N, Forrester TE (2000) Blood pressure is related to placental volume and birth weight. Hypertension 35:662–667 [DOI] [PubMed]
- 46.van de Peppel J, Kemmeren P, van Bakel H, Radonjic M, van Leenen D, Holstege FC (2003) Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep 4:387–393 [DOI] [PMC free article] [PubMed]
- 47.Wesseling S, Joles JA, van Goor H, Bluyssen HA, Kemmeren P, Holstege FC, Koomans HA, Braam B (2007) Transcriptome-based identification of pro- and antioxidative gene expression in kidney cortex of nitric oxide-depleted rats. Physiol Genomics 28:158–167 [DOI] [PubMed]
- 48.Zhan CD, Sindhu RK, Pang J, Ehdaie A, Vaziri ND (2004) Superoxide dismutase, catalase and glutathione peroxidase in the spontaneously hypertensive rat kidney: effect of antioxidant-rich diet. J Hypertens 22:2025–2033 [DOI] [PubMed]
- 49.Zhan CD, Sindhu RK, Vaziri ND (2004) Up-regulation of kidney NAD(P)H oxidase and calcineurin in SHR: reversal by lifelong antioxidant supplementation. Kidney Int 65:219–227 [DOI] [PubMed]