Abstract
Campylobacter is a leading foodborne bacterial pathogen, which causes gastroenteritis in humans. This pathogenic organism is increasingly resistant to antibiotics, especially fluoroquinolones and macrolides, which are the most frequently used antimicrobials for the treatment of campylobacteriosis when clinical therapy is warranted. As a zoonotic pathogen, Campylobacter has a broad animal reservoir and infects humans via contaminated food, water or milk. Antibiotic usage in both animal agriculture and human medicine, can influence the development of antibiotic-resistant Campylobacter. This review will describe the trend in fluoroquinolone and macrolide resistance in Campylobacter, summarize the mechanisms underlying the resistance to various antibiotics and discuss the unique features associated with the emergence, transmission and persistence of antibiotic-resistant Campylobacter. Special attention will be given to recent findings and emphasis will be placed on Campylobacter resistance to fluoroquinolones and macrolides. A future perspective on antibiotic resistance and potential approaches for the control of antibiotic-resistant Campylobacter, will also be discussed.
Keywords: antibiotic resistance, Campylobacter, ecological fitness, fluoroquinolone, food safety, macrolide, public health
Thermophilic Campylobacter species, particularly Campylobacter jejuni, have been recognized as a major cause of acute bacterial gastroenteritis in humans since the late 1970s and it is estimated that Campylobacter sp. are responsible for 400–500 million cases of diarrhea each year, worldwide [1]. As an enteric organism, Campylobacter is carried in the intestinal tracts of a wide range of domestic animals and poultry as well as wild animals and birds [2,3]. Transmission of Campylobacter to humans occurs mainly through the consumption of contaminated foods of animal origin, especially undercooked poultry meat, unpasteurized milk and dairy products, as well as by the ingestion of other foods that are cross-contaminated by raw poultry meat during food preparation [3,4]. Although most Campylobacter infections are mild, self-limiting and usually resolve within a few days without antibiotic treatment, severe or prolonged infections can occur, particularly in the young, elderly and in individuals with compromised immunity. In these circumstances, therapeutic intervention is usually warranted [4,5]. For clinical therapy of campylobacteriosis, erythromycin (a macrolide) is considered the drug of choice, but fluoroquinolone (FQ) antimicrobials (e.g., ciprofloxacin) are also frequently used owing to their broad spectrum of activity against enteric pathogens [4–6]. Alternative drugs include tetracyclines and gentamicin, which are used in cases of systemic infection with Campylobacter [5]. However, Campylobacter is increasingly resistant to the clinically important antibiotics and this rising resistance is a concern for public health. Development and transmission of antibiotic-resistant Campylobacter is complicated by the fact that Campylobacter is a zoonotic pathogen and is therefore exposed to antibiotics used in both animal production and human medicine. Thus, an ecological approach is required to understand the emergence, transmission and persistence of antibiotic-resistant Campylobacter.
Prevalence of antibiotic resistance in Campylobacter
A rapid increase in the proportion of Campylobacter strains resistant to antimicrobial agents, particularly to FQs, has been reported in many countries worldwide [6–8]. Prior to 1992, FQ-resistant Campylobacter was rarely observed in the USA and Canada, but several recent reports have indicated that approximately 19–47% of Campylobacter strains isolated from humans were resistant to ciprofloxacin [9–11]. A steady increase in FQ resistance among Campylobacter isolates has also been observed in many European countries and 17–99% of Campylobacter strains isolated from humans and animals in this region were resistant to FQs, with the highest resistance levels reported in Spain [5,8,12–18]. FQ-resistant Campylobacter has also become prevalent in Africa and Asia. In both continents, FQ resistance among clinical Campylobacter isolates was not detected before 1991, however, since 1993 the frequency of FQ-resistant Campylobacter strains has increased remarkably and the FQ-resistance rates have reached more than 80% in Thailand and Hong Kong [19–22]. Although FQ resistance in Campylobacter isolates was also observed in Australia and New Zealand, the rate of FQ-resistant Campylobacter isolates in this region is significantly lower than that in other regions [8,23, 24].
A trend for increased macrolide resistance in Campylobacter has been observed in some countries [25]. Generally, the prevalence of erythromycin resistance among Campylobacter strains (including both C. jejuni and Campylobacter coli) isolated from humans, broilers and cattle in the USA and Canada has been reported at 10% or lower [25–30]. In contrast, more than 40% of C. coli, isolated from turkeys and swine in the USA, were resistant to this antimicrobial agent [26,29,31]. Likewise, macrolide resistance among Campylobacter isolates from humans and C. jejuni isolates from chickens and cattle has been low and stable in most European countries, especially in Scandinavia, but a high prevalence of macrolide resistance, ranging from 15–80%, was observed in C. coli, isolated from chickens and swine [12,25,32–36]. Interestingly, high erythromycin resistance levels were observed among human clinical Campylobacter isolates from Africa, but low resistance levels to this antibiotic were noticed in C. jejuni and C. coli isolated from food-producing animals [25,37,38]. In Asia, less than 5% of C. jejuni isolated from humans, broilers, swine and cattle were resistant to macrolides, while 14–62% of C. coli isolated from humans, broilers and swine were resistant to this class of antimicrobial agents [39–43]. Similar to findings from other continents, macrolide resistance was mainly observed among C. coli isolates from swine in Australia [44]. These surveillance data clearly indicate that C. coli isolates harbor more macrolide resistance than C. jejuni isolates. The exact reasons for this difference are unknown and warrant further investigation.
Resistance mechanisms
In Campylobacter, the resistance to FQs is mainly mediated by point mutations in the quinolone resistance-determining region (QRDR) of DNA gyrase A (GyrA) [45,46]. No mutations in DNA gyrase B have been associated with FQ resistance in Campylobacter [47–49]. The genes encoding topoisomeraseIV (parC/parE) are also involved in FQ resistance in Gram-negative bacteria, however, these genes are absent in Campylobacter. Thus, it is not surprising that parC/parE mutations are not implicated in Campylobacter resistance to FQ antimicrobials [47–52]. Unlike FQ resistance in other enteric organisms (e.g., Salmonella and Escherichia coli), in which acquisition of high-level FQ resistance requires stepwise accumulation of point mutations in gyrA and parC, a single point mutation in the QRDR of gyrA is sufficient to substantially reduce the susceptibility of Campylobacter to FQ antimicrobials [45,50,53]. The most frequently observed mutation in FQ-resistant isolates of Campylobacter is the C257T change in the gyrA gene, which leads to the T86I substitution in the gyrase and confers high-level resistance to FQs [45]. Other reported resistance-associated mutations include T86K, A70T and D90N, which are less common and do not confer FQ resistance as high as that observed for the T86I mutation [6,45]. In addition to the mutations in GyrA, the multidrug efflux pump, CmeABC, also contributes to FQ resistance by reducing the accumulation of the agents in Campylobacter cells [50,53,54]. Thus, CmeABC functions synergistically with the gyrA mutations in mediating FQ resistance. All of the known FQ resistance determinants in Campylobacter are chromosomally encoded and plasmid-mediated quinolone-resistance determinants, such as qnr, aac(6′)-Ib-cr and qepA, have not been reported in Campylobacter.
Macrolide resistance in Campylobacter is mainly associated with target modification and active efflux [55–59]. Modification of the ribosomal target, leading to macrolide resistance in Campylobacter, can occur either by enzyme-mediated methylation or by point mutation in the 23S rRNA and/or ribosomal proteins L4 and L22 [25,45]. To date, macrolide resistance mediated by rRNA methylation has been reported only in Campylobacter rectus [60]. Point mutations in domain V of the 23S rRNA, on the other hand, have been recognized as the most common mechanism for macrolide resistance in C. jejuni and C. coli [25,45,61]. These point mutations occur at positions 2074 and 2075 of the 23S rRNA, corresponding to positions 2058 and 2059, respectively, in E. coli. Among the reported resistance-associated mutations, the A2074C, A2074G and A2075G mutations are found to confer a high-level resistance to macrolide antibiotics (erythromycin MIC >128 μg/ml) in C. jejuni and C. coli [55,57,58,61,62]. In clinical and field isolates, the A2075G mutation is observed most frequently [25,45,63]. C. coli and C. jejuni have three copies of the rrn operon [64]. A mutation associated with macrolide resistance in Campylobacter is usually present in all three copies of the 23S rRNA gene; however, some mutations, such as A2074T, which confers a low level of erythromycin resistance, may not be present in all copies of the 23S rRNA gene [55,58,65].
In addition to the target modification, active efflux also contributes to macrolide resistance in Campylobacter [57–59,62,63,66,67]. In isolates with intermediate- or low-level macrolide resistance, inactivation of the CmeABC efflux pump completely restored the susceptibility of the isolates [57,58,68]. Even in the highly resistant Campylobacter strains with the A2074G or A2075G mutation, inactivation of CmeABC also significantly reduced the resistance level to macrolide antibiotics, suggesting that this efflux system functions synergistically with target mutations [58,59,66,68]. Additionally, the synergy between the CmeABC efflux pump and mutations in the ribosomal proteins L4 (G74D) and L22 (insertions at position 86 or 98), was also shown to confer macrolide resistance in C. jejuni and C. coli [59,62]. The target mutations and active efflux confer resistance in Campylobacter not only to macrolides (e.g., erythromycin, clarithromycin, azithromycin and tylosin), but also to ketolides (e.g., telithromycin) [45,68].
Resistance to tetracycline in Campylobacter is conferred by tet(O), which is widely present in Campylobacter isolates recovered from various animal species [7]. To date, no other tet resistance genes have been found in Campylobacter. tet(O) encodes a ribosomal protection protein [69]. Recent work demonstrates that this protein recognizes an open A site on the bacterial ribosome and binds it in such a manner that it induces a conformational change that results in the release of the bound tetracycline molecule [70]. Furthermore, the conformational change persists for an extended period of time, thus allowing for continued protein elongation in an efficient manner [70,71]. Based on G–C content, sequence homology, codon usage and hybridization studies, it appears that Campylobacter tet(O) was probably acquired by horizontal gene transfer (HGT) from either Streptomyces, Streptococcus or Enterococcus spp. [72,73]. In most strains, the tet(O) gene is plasmid-encoded, however, some isolates do have a chromosomally encoded copy of the gene [74,75]. Tetracycline resistance in C. jejuni is also associated with the CmeABC multidrug efflux pump [54,66].
Compared to FQs, macrolides and tetracyclines, Campylobacter resistance to other antibiotics has received less attention. Aminoglycoside resistance in Campylobacter is conferred by drug modification proteins. Multiple aminoglycoside modifying enzymes, including 3′-aminoglycoside phosphotransferase types I, III, IV and VII, 3′,9-aminoglycoside adenyltransferase and 6-aminoglycoside adenyltransferase, have been described in Campylobacter [46]. In general, β-lactam antibiotics have limited efficacy against Campylobacter spp. and resistance to this class of antibiotics appears to be mediated by both intrinsic resistance and β-lactamase production [46,76].
Campylobacter exhibits intrinsic resistance to a variety of antibiotics, including bacitracin, novobiocin, rifampin, streptogramin B, trimethoprim and vancomycin [77,78]. Although the mechanisms of this intrinsic resistance are unclear, it is likely to be mediated, in part, by low permeability of the Campylobacter membrane and active efflux conferred by multidrug-efflux transporters [46].
Dynamics of antibiotic resistance emergence
Resistance to FQs and macrolides in Campylobacter occurs spontaneously owing to mutations in target genes. Assessed in culture media, the frequencies of emergence of FQ-resistant mutants range from approximately 10−6–10−8/cell/generation [79]. Different point mutations occur in the QRDR region of gyrA and confer varied levels of resistance to FQ antibiotics [79]. Thus, the measured frequencies of emergence of FQ resistance, vary with the concentration of antibiotics used in the media for mutant enumeration. In Campylobacter, the elevated expression of cmeABC increases the frequency of emergence of FQ-resistant mutants. This enhancing effect on mutant emergence is probably attributable to the synergistic action of CmeABC and gyrA mutations in conferring FQ resistance, allowing more mutants to grow on antibiotic-containing plates. In addition, Mfd (Mutant Frequency Decline), a transcription-repair coupling factor involved in strand-specific DNA repair, promotes the emergence of FQ-resistant mutants in Campylobacter [80]. Inactivation of the mfd gene in Campylobacter resulted in a 100-fold reduction in the number of spontaneous mutants resistant to ciprofloxacin, while overexpression of mfd increased the mutant numbers. Given the fact that Mfd does not affect the MIC of FQ antibiotics in Campylobacter, the altered mutant number is likely to be a result of the direct effect of Mfd on mutation rates.
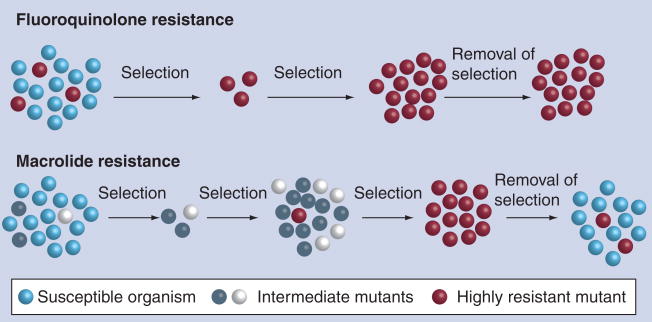
If the cell population is sufficiently large (>106), ciprofloxacin-resistant mutants will inevitably emerge when Campylobacter is exposed to FQs. This has been demonstrated both in vitro and in vivo [80]. Multiple independent studies have demonstrated the rapid development of FQ-resistant mutants in chickens originally infected with FQ-susceptible C. jejuni, but treated with enrofloxacin [50,81–84]. In the treated birds, FQ-resistant Campylobacter mutants could be detected in feces as early as 24 h after the initiation of treatment, and the FQ-resistant Campylobacter population eventually colonized the intestinal tract of the birds. Thus, treatment of Campylobacter-infected birds does not eradicate the organisms, but converts an originally FQ-susceptible population to FQ-resistant Campylobacter, by selecting for spontaneous FQ-resistant mutants from an originally FQ-susceptible population. Since contaminated poultry meat is a main source of human Campylobacter infections, the FQ-resistant Campylobacter developed in poultry can be transmitted to humans via the food chain. Owing to this concern, the FDA banned the use of FQ antimicrobials in poultry production in the USA in 2005 [201]. The development of FQ-resistant Campylobacter from antibiotic treatment was also observed in pigs infected with C. coli and human patients infected with C. jejuni [85–89]. Together, these observations indicate that Campylobacter is highly adaptable to FQ treatment. Selection of pre-existing spontaneous mutants is likely to be the key reason for the development of the FQ-resistant population (Figure 1), but de novo formation of FQ-resistant mutants during the treatment can not be totally excluded, as suggested by the results from an in vitro study, in which some FQ-resistant mutants emerged from an originally FQ-susceptible population long after the initiation of the treatment [80].
Figure 1. Model for the development and fitness of fluoroquinolone and macrolide resistance in Campylobacter.
Fluoroquinolone-resistant mutants develop rapidly during antibiotic treatment and the mutant population continues to persist even after removal of the selection pressure. Development of macrolide-resistant mutants involves a multistep process and requires prolonged exposure to the antibiotic. Once the selection pressure is removed, macrolide-resistant mutants cannot compete with macrolide-susceptible Campylobacter and will decrease in number.
In contrast to FQ resistance, the mutation frequency for macrolide resistance in Campylobacter is low (~10−10/cell/generation) and is approximately 10,000-fold lower than that of FQ resistance [58,79]. The mutants obtained by single-step selection tend to have low-to-intermediate levels (MIC = 8–64 μg/ml) of resistance to erythromycin [58,62,90]. These mutants, obtained by a single-step selection, either harbor mutations in the L4 and L22 proteins or have no detectible mutations, and are not stable in the absence of macrolide antibiotics [62,90]. Acquisition of the mutations in 23S rRNA appears to require stepwise selection (increase of antibiotic concentration in steps) and/or prolonged exposure to macrolide antibiotics, suggesting that other mutations or changes in Campylobacter may be required prior to the occurrence of the 23S rRNA mutations [58,62]. Although some specific changes in L4 and L22 proteins did not seem to be required for the development of 23S rRNA mutations, the contribution of mutations in other genes to the process can not be excluded. Once acquired, most 23S rRNA mutations confer a high level of resistance to erythromycin (MIC ≥ 512 μg/ml) and can be stably maintained in the absence of macrolide antibiotics [55,62]. Although macrolide resistance is generally more prevalent in C. coli isolates than in C. jejuni isolates, the comparison of a limited number of C. coli and C. jejuni strains did not show an elevated mutation frequency for erythromycin in C. coli, suggesting that C. coli is not intrinsically more mutable than C. jejuni [58].
Another unique feature of macrolide resistance in Campylobacter is the slow development of resistant mutants under antibiotic treatment. Using Campylobacter-infected chickens, Lin et al. showed that therapeutic treatment of Campylobacter-infected birds with tylosin (administered in drinking water for three consecutive days) did not select for erythromycin-resistant Campylobacter, even after three treatments. This is in clear contrast to the development of FQ resistance, which occurs rapidly in birds treated with enrofloxacin. But when tylosin was given to Campylobacter-infected birds daily as a feed additive, after several weeks of exposure, erythromycin-resistant Campylobacter emerged in the birds [58]. Similarly, Ladely et al. found that subtherapeutic use of tylosin in chickens, given continuously in feed, had a more significant impact than therapeutic use on the development of macrolide-resistant Campylobacter [91]. These in vivo findings are consistent with the low rate of spontaneous mutation to erythromycin resistance observed in vitro and suggest that a continuous exposure to macrolides for an extended period is required for the development of macrolide resistance in Campylobacter. The differences in the development of FQ resistance and macrolide resistance are illustrated in Figure 1. The low rate of emergence, the requirement for prolonged antibiotic exposure to select macrolide-resistance mutations and the significant fitness cost of erythromycin-resistant mutants (see the persistence section) may collectively contribute to the relatively low prevalence of macrolide resistance in Campylobacter compared with FQ resistance.
Transmission of antibiotic resistance determinants
In addition to the mutation-based mechanisms, Campylobacter can also acquire antibiotic-resistance determinants via HGT. Horizontal transfer of DNA between Campylobacter strains has been shown in bacterial cultures [92,93] and chicken intestine [94,95]. In bacteria, HGT is mediated by natural transformation, conjugation and transduction, all of which can occur in Campylobacter. Natural transformation and conjugation are especially well-recognized in Campylobacter and are often utilized as genetic tools to manipulate Campylobacter [96]. Conjugation is likely to play a major role in the transfer of plasmid-mediated resistance, for example, tet(O), while natural transformation may be a major mechanism for the transfer of chromosomally encoded resistance (e.g., FQ and macrolide resistance). It should be noted that all three HGT mechanisms show strain-to-strain variation in Campylobacter.
Natural transformation utilizes sophisticated mechanisms to take up free DNA from the environment. Multiple factors involved in Campylobacters natural transformation have been identified [92,97–101]. The transfer of genes encoding antibiotic-modifying enzymes in Campylobacter by natural transformation was demonstrated in several studies utilizing bacterial cultures or animal models [92,93,95]. In these studies, co-cultivation or co-colonization of Campylobacter strains carrying different antibiotic resistance determinants, such as aphA3, cat, or tet(O), generated progeny populations resistant to multiple antibiotics. A definitive role of natural transformation in mediating the transfer of antibiotic resistance determinants was shown in a recent study using bacterial co-cultures, in which a transformation-deficient mutation and DNase I treatment of bacterial cultures abolished the formation of double resistant progeny [92]. Transfer of antibiotic resistance determinants in Campylobacter co-cultures occurred quickly and was not prevented by prewashing the cultures prior to mixture, suggesting that Campylobacter may actively release DNA to the media during growth [92]. In addition to the genes encoding antibiotic resistance, point mutations responsible for FQ and macrolide resistance can also be transferred to Campylobacter by natural transformation [90,93,97].
To determine if natural transformation facilitates the emergence of FQ-resistant Campylobacter, Jeon et al. measured the frequency of emergence of spontaneous FQ-resistant mutants in culture media by DNase I treatment, which depleted the free DNA available for transformation [92]. The treatment did not affect the measured frequencies of emergence of FQ-resistant mutants from an originally FQ-susceptible population, suggesting that natural transformation does not contribute to the original emergence of FQ-resistant mutants. Additionally, in vitro and in vivo experiments showed that deficiencies in natural transformation did not affect the development of FQ-resistant Campylobacter during FQ treatment [92], suggesting that the denovo development of FQ-resistant mutants from a FQ-susceptible population is not influenced by natural transformation and is primarily owing to selection and enrichment of the spontaneous FQ-resistant mutants. However, natural transformation may contribute to the spread of FQ resistance across different Campylobacter populations, strains or species.
Multiple plasmids have been reported in Campylobacter, some of which can be transmitted by conjugation [75,98,102,103]. Many of the conjugative plasmids carry genes mediating resistance to tetracyclines [75,104] and aminoglycosides [103,105]. Although interspecies conjugative transfer of drug-resistant plasmids was reported with Campylobacter, conjugation was most successful at the intraspecies level [75,94,103,105]. In addition, intergenus conjugation from C. jejuni to E. coli was occasionally successful [103,105]. It was also reported that the transfer of a conjugative plasmid carrying tet(O) occurred between C. jejuni strains in the intestinal tract of chickens [94]. Considering the high prevalence of conjugative tet(O) plasmids in Campylobacter, it is possible that conjugation has contributed to the spread of tetracycline resistance in Campylobacter. Interestingly, a recent study reported conjugative transfer of a chromosomally encoded streptomycin resistance gene from Helicobacter pylori to C. jejuni at the intergenus level [106]. This transfer was believed to have happened by a conjugation-like mechanism, since it required physical contact of the two species and was protected from DNase I treatment.
Integrons and mobile genetic elements, such as transposons and insertional sequences, are important players for the transmission and spread of antibiotic resistance genes in bacteria [107]. However, these elements are not common in Campylobacter and do not appear to play a major role in the horizontal transfer of antibiotic resistance in Campylobacter. Class I integrons, which are the most common integrons associated with antibiotic resistance, were reported in both C. jejuni and C. coli and were found to carry aminoglycoside resistance genes (aadA2 and aacA4) [108–110].
Campylobacter-infecting bacteriophages were isolated from different Campylobacter species and various sources [111–113]. Recent work in chickens has demonstrated that bacteriophages cause genomic instability in C. jejuni and mediate interstrain transfer of large DNA fragments [114,115]. These findings suggest that Campylobacter–bacteriophage interactions may be more common than previously recognized and might play a role in HGT in Campylobacter. However, the exact role of bacteriophages in transmitting antibiotic resistance determinants between Campylobacter awaits further investigation.
Persistence & fitness of antibiotic resistant Campylobacter
Resistance-conferring mutations or determinants may affect bacterial physiology (e.g., growth rate) and consequently their adaptability in antibiotic-free environments. In the absence of antibiotic selection pressure, antibiotic-resistant Campylobacter may or may not show a fitness burden. Whether antibiotic-resistant Campylobacter persists is influenced by its ability to transmit between hosts and to compete with antibiotic-susceptible Campylobacter. This competition determines if antibiotic-resistant Campylobacter continues to prevail or decline in antibiotic-free environments.
FQ resistance mediated by gyrA mutations can be stably maintained in Campylobacter in the absence of antibiotic selection pressure [116]. FQ-resistant Campylobacter, carrying the T86I mutation in GyrA, colonized chickens persistently without losing the resistance phenotype and the resistance-associated mutation. Both in vitro culturing and chicken colonization studies suggested that FQ-resistant Campylobacter mutants do not carry a fitness burden. In fact, pairwise competition experiments indicate that FQ-resistant mutants outcompete FQ-susceptible strains in chickens [116], suggesting that, in fact, the FQ-resistant mutants possess an enhanced fitness. This fitness change is linked to the T86I mutation and does not appear to be owing to a compensatory mutation since transformation of FQ-susceptible C. jejuni strains with this mutation changed their fitness in chickens. Recently Zhang and colleagues further confirmed the link between the T86I mutation and the fitness change by creating revertants. Reversion of the T86I mutation to the wild-type allele was accompanied by the loss of the fitness advantage in chickens [Zhang Q; Pers. Comm.]. These laboratory findings are compatible with the results from several surveillance studies, in which FQ-resistant Campylobacter was found to continue to prevail in poultry from producers who had discontinued using FQ antimicrobials for up to 4 years [117–119]. Based on the results from the laboratory and surveillance studies, it is tempting to predict that once the prevalence of FQ resistance in Campylobacter is high, it will be difficult to reduce the prevalence of resistance.
An intriguing question about FQ resistance is how the resistance-associated gyrA mutations affect Campylobacter physiology and fitness. DNA gyrase controls DNA supercoiling and is important for DNA replication and transcription. It is conceivable that the resistance-conferring mutations in GyrA may alter the activities of the enzyme and affect DNA supercoiling in Campylobacter. Indeed, work using recombinant gyrases demonstrates that the mutant enzyme carrying the T86I change, which is the most common mutation observed in FQ-resistant isolates, has a greatly reduced supercoiling activity [Han J, Zhang Q; Pers. Comm.]. Using a reporter plasmid, it was further found that DNA super-coiling levels are reduced in the FQ-resistant mutant compared with the wild-type strain. These results provide compelling evidence that this resistance-conferring mutation alters the native function of DNA gyrase. Whether this alteration is sufficient to affect Campylobacter physiology and its fitness is currently under investigation [Zhang Q; Pers. Comm.].
Campylobacter mutants that show low-to-intermediate levels of erythromycin resistance and lack 23S rRNA mutations, are not stable in culture media or animal hosts and easily lose the resistance phenotype in the absence of macrolide antibiotics [62,90]. However, the macrolide-resistant mutants harboring 23S rRNA mutations are highly resistant to erythromycin and stable in terms of the resistance phenotype, and can persist in chickens in the absence of competition [55,62]. In contrast to FQ-resistant Campylobacter, erythromycin-resistant mutants show a clear fitness burden when compared with the wild-type strains. Using a chicken model, Zhang’s group conducted pairwise competitions and revealed that erythromycin-resistant mutants carrying the A2074G or A2075G mutation in 23S rRNA were rapidly outcompeted by the isogenic wild-type strains [Luangtongkum T, Zhang Q; Pers. Comm.]. This fitness cost was consistently observed in multiple chicken experiments using different pairs of strains and suggests that, in the absence of the antibiotics, the mutations conferring macrolide resistance render Campylobacter less fit in its natural host. This finding is supported by surveillance data from Denmark, where reduced use of tylosin as a growth promoter in swine has led to a significant decrease in the number of erythromycin-resistant C. coli isolated from pigs [120]. These observations suggest that removal of the selection force will quickly reduce the prevalence of macrolide-resistant Campylobacter.
Tetracycline resistance conferred by tet(O) has become highly prevalent in Campylobacter worldwide. This gene is usually carried on a plasmid, although it can be chromosomally encoded. Interestingly, recent studies conducted with poultry operations demonstrate that tetracycline-resistant Campylobacter are prevalent in both organic and conventional production systems [29,121]. Cui et al. also reported the high prevalence of tetracycline-resistant Campylobacter in organic chickens from retail stores [122]. In addition, tetracycline-resistant Campylobacter were also frequently isolated from organic dairy farms and antibiotic-free pigs [31,123]. Since the majority of these cited references originate from the USA, where organic production is regulated under the National Organic Program, these animals were not permitted to have any exposure to antibiotics following the last trimester of their gestation for mammals and following the second day of life for poultry. Additionally all forages and grains provided to these animals must be grown in an organic environment that has not been exposed to antibiotics for the 3 years prior to harvest. Different from the USA regulations, many organic certification programs in other regions of the world do allow for a limited use of antibiotics in certain circumstances. Although it is not possible to say that absolutely no antibiotic exposure occurs on the USA organic operations, the known use of such products is strictly prohibited and the local environments (including pastures) are required to meet organic standards for 3 years prior to their use for the production of organic products. Therefore, the prevalence of tetracycline-resistant Campylobacter in the USA organic production systems is unlikely to be maintained by antibiotic selection and suggests that tet(O)-containing plasmids may have coevolved with Campylobacter, such that carrying the plasmid is no longer a burden to the host.
Future perspective
Antibiotic resistance in Campylobacter continues to be a challenge for food safety and public health. Owing to the high prevalence of FQ resistance, FQ antimicrobials are losing effectiveness in the clinical treatment of human campylobacteriosis. Enhanced research efforts are needed to understand the factors affecting the transmission and persistence of FQ-resistant Campylobacter in various environments and hosts. It will also be interesting to examine how FQ resistance influences Campylobacter fitness and if withdrawal of FQ antimicrobials from animal production decreases the prevalence of FQ-resistant Campylobacter. Additionally, newer FQs that are effective against ciprofloxacin-resistant Campylobacter and novel treatment schemes that avoid the selection of FQ-resistant mutants, should be evaluated. Macrolides are still the most effective antibiotics against Campylobacter infections, but the rising trend of erythromycin resistance in C. coli and C. jejuni in some regions requires prudent use of this class of antibiotics. Additional studies are needed to understand how macrolide-resistant Campylobacter emerge under selective pressure. Application of advanced approaches, such as genomics and proteomics, is expected to provide new insights into the molecular mechanisms involved in the development of macrolide resistance in Campylobacter. It has become clear that the multidrug efflux pump, CmeABC, plays an important role in mediating antibiotic resistance in Campylobacter, but the contributions of other efflux transporters to antibiotic resistance remains to be elucidated. In addition, the natural functions of these efflux transporters in Campylobacter physiology await further investigation. Novel approaches that target drug efflux transporters or block the emergence and transmission of resistance determinants can be explored to control antibiotic-resistant Campylobacter.
Executive summary
Epidemiology
▪ Campylobacter is increasingly resistant to clinically important antibiotics, which has become a major concern for public health.
▪ Campylobacter isolates resistant to fluoroquinolone (FQ) and tetracycline are highly prevalent in many countries. Although macrolide resistance is relatively low and stabilized in Campylobacter jejuni, there is a trend for increased prevalence of macrolide-resistant Campylobacter in certain regions of the world and the trend is especially clear in Campylobacter coli isolates recovered from swine and turkey.
Resistance mechanisms
▪ FQ and macrolide resistance in Campylobacter is mediated by point mutations in gyrA and 23S rRNA, respectively. tet(O) is the only tet gene currently identified in Campylobacter and confers resistance to the tetracycline class of antibiotics.
▪ As an efflux pump, CmeABC reduces the intracellular concentration of antibiotics, functions synergistically with other resistance mechanisms and contributes to the resistance to multiple antimicrobials.
Emergence of antibiotic resistance
▪ Resistance-associated gyrA mutations occur spontaneously at a relatively high frequency in Campylobacter and FQ treatment rapidly selects for FQ-resistant mutants. The C257T change in gyrA is the most frequently observed mutation in FQ-resistant isolates and confers high-level resistance to FQs.
▪ In Campylobacter, the rate of spontaneous mutation to macrolide resistance is substantially lower than that observed for FQ resistance and the development of stable macrolide-resistant mutants requires stepwise selection and prolonged exposure to the antibiotics.
Transmission of antibiotic resistance determinants
▪ Campylobacter acquires resistance determinants by mutation and horizontal gene transfer. Natural transformation, conjugation and transduction can all occur in Campylobacter and are likely to contribute to the spread of antibiotic resistance determinants.
Persistence & fitness of antibiotic resistance
▪ The gyrA mutation conferring FQ resistance does not incur a fitness cost in Campylobacter. Once FQ-resistant Campylobacter is prevalent, it may be difficult to reverse the resistance trend because FQ resistance can persist even in the absence of antibiotic selection.
▪ Erythromycin-resistant Campylobacter carries a significant fitness burden and removal of the selective pressure will quickly reduce the prevalence of macrolide resistance.
Acknowledgments
Financial & competing interests disclosure
The work conducted in Zhang’s laboratory is supported by the NIH (grant no. RO1DK063008) and National Research Initiative Competitive grants (grant no. 2005–51110–03273 and 2006–34211–17310) from the US Department of Agriculture Cooperative State Research, Education and Extension Services. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Website
201. US FDA. Final decision of the commissioner, Docket No. 2000N-1571. Withdrawal of the approval of the new animal drug application for enrofloxacin in poultry. Department of Health and Human Services. www.fda.gov/oc/antimicrobial/baytril.html
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Ruiz-Palacios GM. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken . Clin Infect Dis. 2007;44(5):701–703. doi: 10.1086/509936. [DOI] [PubMed] [Google Scholar]
- 2.Humphrey T, O’Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol. 2007;117(3):237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Altekruse SF, Tollefson LK. Human campylobacteriosis: a challenge for the veterinary profession . J Am Vet Med Assoc. 2003;223(4):445–452. doi: 10.2460/javma.2003.223.445. [DOI] [PubMed] [Google Scholar]
- 4.Allos BM. Campylobacter jejuni infections: update on emerging issues and trends . Clin Infect Dis. 2001;32(8):1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 5.Blaser MJ, Engberg J. Clinical aspects of Campylobacter jejuni and Campylobacter coli infections. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3. ASM Press; Washington DC, USA: 2008. pp. 99–121. [Google Scholar]
- 6.Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis. 2001;7(1):24–34. doi: 10.3201/eid0701.010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.▪.Moore JE, Barton MD, Blair IS, et al. The epidemiology of antibiotic resistance in Campylobacter. Microbes Infect. 2006;8(7):1955–1966. doi: 10.1016/j.micinf.2005.12.030. Excellent review on the global prevalence of antibiotic-resistant Campylobacter. [DOI] [PubMed] [Google Scholar]
- 8.Alfredson DA, Korolik V. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol Lett. 2007;277(2):123–132. doi: 10.1111/j.1574-6968.2007.00935.x. [DOI] [PubMed] [Google Scholar]
- 9.Gaudreau C, Gilbert H. Antimicrobial resistance of Campylobacter jejuni subsp jejuni strains isolated from humans in 1998 to 2001 in Montreal, Canada . Antimicrob Agents Chemother. 2003;47(6):2027–2029. doi: 10.1128/AAC.47.6.2027-2029.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta A, Nelson JM, Barrett TJ, et al. Antimicrobial resistance among Campylobacter strains, United States, 1997–2001 . Emerg Infect Dis. 2004;10(6):1102–1109. doi: 10.3201/eid1006.030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nachamkin I, Ung H, Li M. Increasing fluoroquinolone resistance in Campylobacter jejuni, Pennsylvania, USA, 1982–2001. Emerg Infect Dis. 2002;8(12):1501–1503. doi: 10.3201/eid0812.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papavasileiou E, Voyatzi A, Papavasileiou K, Makri A, Andrianopoulou I, Chatzipanagiotou S. Antimicrobial susceptibilities of Campylobacter jejuni isolates from hospitalized children in Athens, Greece, collected during 2004–2005. Eur J Epidemiol. 2007;22(1):77–78. doi: 10.1007/s10654-006-9080-3. [DOI] [PubMed] [Google Scholar]
- 13.Gallay A, Prouzet-Mauleon V, Kempf I, et al. Campylobacter antimicrobial drug resistance among humans, broiler chickens, and pigs, France . Emerg Infect Dis. 2007;13(2):259–266. doi: 10.3201/eid1302.060587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakanen AJ, Lehtopolku M, Siitonen A, Huovinen P, Kotilainen P. Multidrug resistance in Campylobacter jejuni strains collected from Finnish patients during 1995–2000. J Antimicrob Chemother. 2003;52(6):1035–1039. doi: 10.1093/jac/dkg489. [DOI] [PubMed] [Google Scholar]
- 15.Krausse R, Ullmann U. In vitro activities of new fluoroquinolones against Campylobacter jejuni and Campylobacter coli isolates obtained from humans in 1980 to 1982 and 1997 to 2001. Antimicrob Agents Chemother. 2003;47(9):2946–2950. doi: 10.1128/AAC.47.9.2946-2950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luber P, Wagner J, Hahn H, Bartelt E. Antimicrobial resistance in Campylobacter jejuni and Campylobacter coli strains isolated in 1991 and 2001–2002 from poultry and humans in Berlin, Germany. Antimicrob Agents Chemother. 2003;47(12):3825–3830. doi: 10.1128/AAC.47.12.3825-3830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucey B, Cryan B, O’Halloran F, Wall PG, Buckley T, Fanning S. Trends in antimicrobial susceptibility among isolates of Campylobacter species in Ireland and the emergence of resistance to ciprofloxacin. Vet Rec. 2002;151(11):317–320. doi: 10.1136/vr.151.11.317. [DOI] [PubMed] [Google Scholar]
- 18.Pezzotti G, Serafin A, Luzzi I, Mioni R, Milan M, Perin R. Occurrence and resistance to antibiotics of Campylobacter jejuni and Campylobacter coli in animals and meat in northeastern Italy. Int J Food Microbiol. 2003;82(3):281–287. doi: 10.1016/s0168-1605(02)00314-8. [DOI] [PubMed] [Google Scholar]
- 19.Isenbarger DW, Hoge CW, Srijan A, et al. Comparative antibiotic resistance of diarrheal pathogens from Vietnam and Thailand, 1996–1999 . Emerg Infect Dis. 2002;8(2):175–180. doi: 10.3201/eid0802.010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putnam SD, Frenck RW, Riddle MS, et al. Antimicrobial susceptibility trends in Campylobacter jejuni and Campylobacter coli isolated from a rural Egyptian pediatric population with diarrhea . Diagn Microbiol Infect Dis. 2003;47(4):601–608. doi: 10.1016/s0732-8893(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 21.Tjaniadi P, Lesmana M, Subekti D, et al. Antimicrobial resistance of bacterial pathogens associated with diarrheal patients in Indonesia . Am J Trop Med Hyg. 2003;68(6):666–670. [PubMed] [Google Scholar]
- 22.Chu YW, Chu MY, Luey KY, Ngan YW, Tsang KL, Kam KM. Genetic relatedness and quinolone resistance of Campylobacter jejuni strains isolated in 2002 in Hong Kong . J Clin Microbiol. 2004;42(7):3321–3323. doi: 10.1128/JCM.42.7.3321-3323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma H, Unicomb L, Forbes W, et al. Antibiotic resistance in Campylobacter jejuni isolated from humans in the Hunter Region, New South Wales . Commun Dis Intell. 2003;27:S80–S88. [PubMed] [Google Scholar]
- 24.Goodchild C, Dove B, Riley D, Morris AJ. Antimicrobial susceptibility of Campylobacter species . N Z Med J. 2001;114(1145):560–561. [PubMed] [Google Scholar]
- 25.▪.Gibreel A, Taylor DE. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother. 2006;58(2):243–255. doi: 10.1093/jac/dkl210. Excellent review on macrolide resistance in Campylobacter. [DOI] [PubMed] [Google Scholar]
- 26.Belanger AE, Shryock TR. Macrolide-resistant Campylobacter: the meat of the matter . J Antimicrob Chemother. 2007;60(4):715–723. doi: 10.1093/jac/dkm300. [DOI] [PubMed] [Google Scholar]
- 27.Guevremont E, Nadeau E, Sirois M, Quessy S. Antimicrobial susceptibilities of thermophilic Campylobacter from humans, swine, and chicken broilers. Can J Vet Res. 2006;70(2):81–86. [PMC free article] [PubMed] [Google Scholar]
- 28.Levesque S, Frost E, Michaud S. Comparison of antimicrobial resistance of Campylobacter jejuni isolated from humans, chickens, raw milk, and environmental water in Quebec. J Food Prot. 2007;70(3):729–735. doi: 10.4315/0362-028x-70.3.729. [DOI] [PubMed] [Google Scholar]
- 29.Luangtongkum T, Morishita TY, Ison AJ, Huang S, McDermott PF, Zhang Q. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl Environ Microbiol. 2006;72(5):3600–3607. doi: 10.1128/AEM.72.5.3600-3607.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Englen MD, Hill AE, Dargatz DA, Ladely SR, Fedorka-Cray PJ. Prevalence and antimicrobial resistance of Campylobacter in US dairy cattle . J Appl Microbiol. 2007;102(6):1570–1577. doi: 10.1111/j.1365-2672.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- 31.Thakur S, Gebreyes WA. Prevalence and antimicrobial resistance of Campylobacter in antimicrobial-free and conventional pig production systems . J Food Prot. 2005;68(11):2402–2410. doi: 10.4315/0362-028x-68.11.2402. [DOI] [PubMed] [Google Scholar]
- 32.Bardon J, Kolar M, Cekanova L, Hejnar P, Koukalova D. Prevalence of Campylobacter jejuni and its resistance to antibiotics in poultry in the Czech Republic. Zoonoses Public Health. 2008 doi: 10.1111/j.1863-2378.2008.01176.x. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 33.McGill K, Cowley D, Moran L, et al. Antibiotic resistance of retail food and human Campylobacter isolates on the island of Ireland from 2001–2002 . Epidemiol Infect. 2006;134(6):1282–1291. doi: 10.1017/S0950268806006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Hees BC, Veldman-Ariesen MJ, de Jongh BM, Tersmette M, van Pelt W. Regional and seasonal differences in incidence and antibiotic resistance of Campylobacter from a nationwide surveillance study in The Netherlands: an overview of 2000–2004. Clin Microbiol Infect. 2007;13(3):305–310. doi: 10.1111/j.1469-0691.2006.01643.x. [DOI] [PubMed] [Google Scholar]
- 35.Andersen SR, Saadbye P, Shukri NM, Rosenquist H, Nielsen NL, Boel J. Antimicrobial resistance among Campylobacter jejuni isolated from raw poultry meat at retail level in Denmark. Int J Food Microbiol. 2006;107(3):250–255. doi: 10.1016/j.ijfoodmicro.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 36.Hakkinen M, Heiska H, Hanninen ML. Prevalence of Campylobacter spp in cattle in Finland and antimicrobial susceptibilities of bovine Campylobacter jejuni strains . Appl Environ Microbiol. 2007;73(10):3232–3238. doi: 10.1128/AEM.02579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samie A, Ramalivhana J, Igumbor EO, Obi CL. Prevalence, haemolytic and haemagglutination activities and antibiotic susceptibility profiles of Campylobacter spp isolated from human diarrhoeal stools in Vhembe District, South Africa . J Health Popul Nutr. 2007;25(4):406–413. [PMC free article] [PubMed] [Google Scholar]
- 38.Kassa T, Gebre-Selassie S, Asrat D. Antimicrobial susceptibility patterns of thermotolerant Campylobacter strains isolated from food animals in Ethiopia. Vet Microbiol. 2007;119(1):82–87. doi: 10.1016/j.vetmic.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Feizabadi MM, Dolatabadi S, Zali MR. Isolation and drug-resistant patterns of Campylobacter strains cultured from diarrheic children in Tehran . Jpn J Infect Dis. 2007;60(4):217–219. [PubMed] [Google Scholar]
- 40.Hong J, Kim JM, Jung WK, et al. Prevalence and antibiotic resistance of Campylobacter spp isolated from chicken meat, pork, and beef in Korea, from 2001 to 2006 . J Food Prot. 2007;70(4):860–866. doi: 10.4315/0362-028x-70.4.860. [DOI] [PubMed] [Google Scholar]
- 41.Senok A, Yousif A, Mazi W, et al. Pattern of antibiotic susceptibility in Campylobacter jejuni isolates of human and poultry origin . Jpn J Infect Dis. 2007;60(1):1–4. [PubMed] [Google Scholar]
- 42.Serichantalergs O, Jensen LB, Pitarangsi C, Mason CJ, Dalsgaard A. A possible mechanism of macrolide resistance among multiple resistant Campylobacter jejuni and Campylobacter coli isolated from Thai children during 1991–2000. Southeast Asian J Trop Med Public Health. 2007;38(3):501–506. [PubMed] [Google Scholar]
- 43.Shin E, Lee Y. Antimicrobial resistance of 114 porcine isolates of Campylobacter coli. Int J Food Microbiol. 2007;118(2):223–227. doi: 10.1016/j.ijfoodmicro.2007.07.040. [DOI] [PubMed] [Google Scholar]
- 44.Miflin JK, Templeton JM, Blackall PJ. Antibiotic resistance in Campylobacter jejuni and Campylobacter coli isolated from poultry in the South-East Queensland region . J Antimicrob Chemother. 2007;59(4):775–778. doi: 10.1093/jac/dkm024. [DOI] [PubMed] [Google Scholar]
- 45.Payot S, Bolla JM, Corcoran D, Fanning S, Megraud F, Zhang Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006;8(7):1967–1971. doi: 10.1016/j.micinf.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q, Plummer P. Mechanisms of antibiotic resistance in Campylobacter. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3. ASM Press; Washington DC, USA: 2008. pp. 263–276. [Google Scholar]
- 47.Payot S, Cloeckaert A, Chaslus-Dancla E. Selection and characterization of fluoroquinolone-resistant mutants of Campylobacter jejuni using enrofloxacin. Microb Drug Resist. 2002;8(4):335–343. doi: 10.1089/10766290260469606. [DOI] [PubMed] [Google Scholar]
- 48.Piddock LJ, Ricci V, Pumbwe L, Everett MJ, Griggs DJ. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes . J Antimicrob Chemother. 2003;51(1):19–26. doi: 10.1093/jac/dkg033. [DOI] [PubMed] [Google Scholar]
- 49.Bachoual R, Ouabdesselam S, Mory F, Lascols C, Soussy CJ, Tankovic J. Single or double mutational alterations of gyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb Drug Resist. 2001;7(3):257–261. doi: 10.1089/10766290152652800. [DOI] [PubMed] [Google Scholar]
- 50.▪.Luo N, Sahin O, Lin J, Michel LO, Zhang Q. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother. 2003;47(1):390–394. doi: 10.1128/AAC.47.1.390-394.2003. One of the original reports on the development of fluoroquinolone (FQ)-resistant Campylobacter from in vivo antibiotic treatment and the first study showing the synergy between gyrA mutations and CmeABC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cooper R, Segal H, Lastovica AJ, Elisha BG. Genetic basis of quinolone resistance and epidemiology of resistant and susceptible isolates of porcine Campylobacter coli strains . J Appl Microbiol. 2002;93(2):241–249. doi: 10.1046/j.1365-2672.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- 52.Parkhill J, Wren BW, Mungall K, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences . Nature. 2000;403(6770):665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 53.Ge B, McDermott PF, White DG, Meng J. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2005;49(8):3347–3354. doi: 10.1128/AAC.49.8.3347-3354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.▪.Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46(7):2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. An original report defining the key role of CmeABC in antibiotic resistance in Campylobacter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibreel A, Kos VN, Keelan M, et al. Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype . Antimicrob Agents Chemother. 2005;49(7):2753–2759. doi: 10.1128/AAC.49.7.2753-2759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrow SA, Gilpin BJ, Klena JD. Characterization of erythromycin resistance in Campylobacter coli and Campylobacter jejuni isolated from pig offal in New Zealand . J Appl Microbiol. 2004;97(1):141–148. doi: 10.1111/j.1365-2672.2004.02278.x. [DOI] [PubMed] [Google Scholar]
- 57.Mamelli L, Prouzet-Mauleon V, Pages JM, Megraud F, Bolla JM. Molecular basis of macrolide resistance in Campylobacter: role of efflux pumps and target mutations . J Antimicrob Chemother. 2005;56(3):491–497. doi: 10.1093/jac/dki253. [DOI] [PubMed] [Google Scholar]
- 58.▪.Lin J, Yan M, Sahin O, Pereira S, Chang YJ, Zhang Q. Effect of macrolide usage on emergence of erythromycin-resistant Campylobacter isolates in chickens. Antimicrob Agents Chemother. 2007;51(5):1678–1686. doi: 10.1128/AAC.01411-06. The first report describing the unique aspects associated with macrolide resistance development in Campylobacter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cagliero C, Mouline C, Cloeckaert A, Payot S. Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2006;50(11):3893–3896. doi: 10.1128/AAC.00616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roe DE, Weinberg A, Roberts MC. Mobile rRNA methylase genes in Campylobacter (Wolinella) rectus . J Antimicrob Chemother. 1995;36(4):738–740. doi: 10.1093/jac/36.4.738. [DOI] [PubMed] [Google Scholar]
- 61.Corcoran D, Quinn T, Cotter L, Fanning S. An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int J Antimicrob Agents. 2006;27(1):40–45. doi: 10.1016/j.ijantimicag.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 62.Caldwell DB, Wang Y, Lin J. Development, stability, and molecular mechanisms of macrolide resistance in Campylobacter jejuni. Antimicrob Agents Chemother. 2008;52(11):3947–3954. doi: 10.1128/AAC.00450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kurincic M, Botteldoorn N, Herman L, Smole MS. Mechanisms of erythromycin resistance of Campylobacter spp isolated from food, animals and humans . Int J Food Microbiol. 2007;120(1–2):186–190. doi: 10.1016/j.ijfoodmicro.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 64.Fouts DE, Mongodin EF, Mandrell RE, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species . PLoS Biol. 2005;3(1):e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vacher S, Menard A, Bernard E, Santos A, Megraud F. Detection of mutations associated with macrolide resistance in thermophilic Campylobacter spp. by real-time PCR. Microb Drug Resist. 2005;11(1):40–47. doi: 10.1089/mdr.2005.11.40. [DOI] [PubMed] [Google Scholar]
- 66.Gibreel A, Wetsch NM, Taylor DE. Contribution of the CmeABC efflux pump to macrolide and tetracycline resistance in Campylobacter jejuni. Antimicrob Agents Chemother. 2007;51(9):3212–3216. doi: 10.1128/AAC.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Payot S, Avrain L, Magras C, Praud K, Cloeckaert A, Chaslus-Dancla E. Relative contribution of target gene mutation and efflux to fluoroquinolone and erythromycin resistance, in French poultry and pig isolates of Campylobacter coli. Int J Antimicrob Agents. 2004;23(5):468–472. doi: 10.1016/j.ijantimicag.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 68.Cagliero C, Mouline C, Payot S, Cloeckaert A. Involvement of the CmeABC efflux pump in the macrolide resistance of Campylobacter coli. J Antimicrob Chemother. 2005;56(5):948–950. doi: 10.1093/jac/dki292. [DOI] [PubMed] [Google Scholar]
- 69.Taylor DE, Hiratsuka K, Ray H, Manavathu EK. Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466 . J Bacteriol. 1987;169(7):2984–2989. doi: 10.1128/jb.169.7.2984-2989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Connell SR, Tracz DM, Nierhaus KH, Taylor DE. Ribosomal protection proteins and their mechanism of tetracycline resistance . Antimicrob Agents Chemother. 2003;47(12):3675–3681. doi: 10.1128/AAC.47.12.3675-3681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Connell SR, Trieber CA, Dinos GP, Einfeldt E, Taylor DE, Nierhaus KH. Mechanism of Tet(O)-mediated tetracycline resistance . EMBO J. 2003;22(4):945–953. doi: 10.1093/emboj/cdg093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species . Microbiology. 2004;150(10):3507–3517. doi: 10.1099/mic.0.27112-0. [DOI] [PubMed] [Google Scholar]
- 73.Taylor DE, Garner RS, Allan BJ. Characterization of tetracycline resistance plasmids from Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 1983;24(6):930–935. doi: 10.1128/aac.24.6.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dasti JI, Gross U, Pohl S, Lugert R, Weig M, Schmidt-Ott R. Role of the plasmid-encoded tet(O) gene in tetracycline-resistant clinical isolates of Campylobacter jejuni and Campylobacter coli. J Med Microbiol. 2007;56(6):833–837. doi: 10.1099/jmm.0.47103-0. [DOI] [PubMed] [Google Scholar]
- 75.Pratt A, Korolik V. Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J Antimicrob Chemother. 2005;55(4):452–460. doi: 10.1093/jac/dki040. [DOI] [PubMed] [Google Scholar]
- 76.Li XZ, Mehrotra M, Ghimire S, Adewoye L. β-lactam resistance and β-lactamases in bacteria of animal origin. Vet Microbiol. 2007;121(3–4):197–214. doi: 10.1016/j.vetmic.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 77.Taylor DE, Courvalin P. Mechanisms of antibiotic resistance in Campylobacter species. Antimicrob Agents Chemother. 1988;32(8):1107–1112. doi: 10.1128/aac.32.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Corry JE, Post DE, Colin P, Laisney MJ. Culture media for the isolation of campylobacters . Int J Food Microbiol. 1995;26(1):43–76. doi: 10.1016/0168-1605(95)00044-k. [DOI] [PubMed] [Google Scholar]
- 79.Yan M, Sahin O, Lin J, Zhang Q. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J Antimicrob Chemother. 2006;58(6):1154–1159. doi: 10.1093/jac/dkl412. [DOI] [PubMed] [Google Scholar]
- 80.▪▪.Han J, Sahin O, Barton YW, Zhang Q. Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog. 2008;4(6):e1000083. doi: 10.1371/journal.ppat.1000083. The first report describing the key role of Mfd in the development of FQ resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.▪.McDermott PF, Bodeis SM, English LL, et al. Ciprofloxacin resistance in Campylobacter jejuni evolves rapidly in chickens treated with fluoroquinolones. J Infect Dis. 2002;185(6):837–840. doi: 10.1086/339195. One of the original reports on FQ resistance development from in vivo antibiotic treatment. [DOI] [PubMed] [Google Scholar]
- 82.van Boven M, Veldman KT, de Jong MC, Mevius DJ. Rapid selection of quinolone resistance in Campylobacter jejuni but not in Escherichia coli in individually housed broilers. J Antimicrob Chemother. 2003;52(4):719–723. doi: 10.1093/jac/dkg402. [DOI] [PubMed] [Google Scholar]
- 83.Griggs DJ, Johnson MM, Frost JA, Humphrey T, Jorgensen F, Piddock LJ. Incidence and mechanism of ciprofloxacin resistance in Campylobacter spp isolated from commercial poultry flocks in the United Kingdom before, during, and after fluoroquinolone treatment . Antimicrob Agents Chemother. 2005;49(2):699–707. doi: 10.1128/AAC.49.2.699-707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farnell MB, Donoghue AM, Cole K, Reyes-Herrera I, Blore PJ, Donoghue DJ. Campylobacter susceptibility to ciprofloxacin and corresponding fluoroquinolone concentrations within the gastrointestinal tracts of chickens . J Appl Microbiol. 2005;99(5):1043–1050. doi: 10.1111/j.1365-2672.2005.02712.x. [DOI] [PubMed] [Google Scholar]
- 85.Segreti J, Gootz TD, Goodman LJ, et al. High-level quinolone resistance in clinical isolates of Campylobacter jejuni. J Infect Dis. 1992;165(4):667–670. doi: 10.1093/infdis/165.4.667. [DOI] [PubMed] [Google Scholar]
- 86.Wretlind B, Stromberg A, Ostlund L, Sjogren E, Kaijser B. Rapid emergence of quinolone resistance in Campylobacter jejuni in patients treated with norfloxacin. Scand J Infect Dis. 1992;24(5):685–686. doi: 10.3109/00365549209054659. [DOI] [PubMed] [Google Scholar]
- 87.Ellis-Pegler RB, Hyman LK, Ingram RJ, McCarthy M. A placebo controlled evaluation of lomefloxacin in the treatment of bacterial diarrhoea in the community. J Antimicrob Chemother. 1995;36(1):259–263. doi: 10.1093/jac/36.1.259. [DOI] [PubMed] [Google Scholar]
- 88.Adler-Mosca H, Luthy-Hottenstein J, Martinetti LG, Burnens A, Altwegg M. Development of resistance to quinolones in five patients with campylobacteriosis treated with norfloxacin or ciprofloxacin. Eur J Clin Microbiol Infect Dis. 1991;10(11):953–957. doi: 10.1007/BF02005451. [DOI] [PubMed] [Google Scholar]
- 89.Delsol AA, Sunderland J, Woodward MJ, Pumbwe L, Piddock LJ, Roe JM. Emergence of fluoroquinolone resistance in the native Campylobacter coli population of pigs exposed to enrofloxacin . J Antimicrob Chemother. 2004;53(5):872–874. doi: 10.1093/jac/dkh150. [DOI] [PubMed] [Google Scholar]
- 90.Kim JS, Carver DK, Kathariou S. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl Environ Microbiol. 2006;72(2):1316–1321. doi: 10.1128/AEM.72.2.1316-1321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ladely SR, Harrison MA, Fedorka-Cray PJ, Berrang ME, Englen MD, Meinersmann RJ. Development of macrolide-resistant Campylobacter in broilers administered subtherapeutic or therapeutic concentrations of tylosin . J Food Prot. 2007;70(8):1945–1951. doi: 10.4315/0362-028x-70.8.1945. [DOI] [PubMed] [Google Scholar]
- 92.▪.Jeon B, Muraoka W, Sahin O, Zhang Q. Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob Agents Chemother. 2008;52(8):2699–2708. doi: 10.1128/AAC.01607-07. An original report defining the role of natural transformation in horizontal gene transfer of antibiotic resistance determinants in Campylobacter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilson DL, Bell JA, Young VB, Wilder SR, Mansfield LS, Linz JE. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture . Microbiology. 2003;149(12):3603–3615. doi: 10.1099/mic.0.26531-0. [DOI] [PubMed] [Google Scholar]
- 94.Avrain L, Vernozy-Rozand C, Kempf I. Evidence for natural horizontal transfer of tetO gene between Campylobacter jejuni strains in chickens. J Appl Microbiol. 2004;97(1):134–140. doi: 10.1111/j.1365-2672.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- 95.▪.Boer P, Wagenaar JA, Achterberg RP, Putten JP, Schouls LM, Duim B. Generation of Campylobacter jejuni genetic diversity in vivo. Mol Microbiol. 2002;44(2):351–359. doi: 10.1046/j.1365-2958.2002.02930.x. Report of HGT of antibiotic resistance genes in vivo. [DOI] [PubMed] [Google Scholar]
- 96.Guerry P, Yao R, Alm RA, Burr DH, Trust TJ. Systems of experimental genetics for Campylobacter species . Methods Enzymol. 1994;235:474–481. doi: 10.1016/0076-6879(94)35163-5. [DOI] [PubMed] [Google Scholar]
- 97.Wiesner RS, Hendrixson DR, DiRita VJ. Natural transformation of Campylobacter jejuni requires components of a type II secretion system . J Bacteriol. 2003;185(18):5408–5418. doi: 10.1128/JB.185.18.5408-5418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bacon DJ, Alm RA, Burr DH, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176 . Infect Immun. 2000;68(8):4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jeon B, Zhang Q. Cj0011c, a periplasmic single- and double-stranded DNA-binding protein, contributes to natural transformation in Campylobacter jejuni. J Bacteriol. 2007;189(20):7399–7407. doi: 10.1128/JB.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Larsen JC, Szymanski C, Guerry P. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81–176. J Bacteriol. 2004;186(19):6508–6514. doi: 10.1128/JB.186.19.6508-6514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Takata T, Ando T, Israel DA, Wassenaar TM, Blaser MJ. Role of dprA in transformation of Campylobacter jejuni. FEMS Microbiol Lett. 2005;252(1):161–168. doi: 10.1016/j.femsle.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 102.Tracz DM, Keelan M, Ahmed-Bentley J, Gibreel A, Kowalewska-Grochowska K, Taylor DE. pVir and bloody diarrhea in Campylobacter jejuni enteritis . Emerg Infect Dis. 2005;11(6):838–843. doi: 10.3201/eid1106.041052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gibreel A, Skold O, Taylor DE. Characterization of plasmid-mediated aphA-3 kanamycin resistance in Campylobacter jejuni. Microb Drug Resist. 2004;10(2):98–105. doi: 10.1089/1076629041310127. [DOI] [PubMed] [Google Scholar]
- 104.Velazquez JB, Jimenez A, Chomon B, Villa TG. Incidence and transmission of antibiotic resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother. 1995;35(1):173–178. doi: 10.1093/jac/35.1.173. [DOI] [PubMed] [Google Scholar]
- 105.Nirdnoy W, Mason CJ, Guerry P. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob Agents Chemother. 2005;49(6):2454–2459. doi: 10.1128/AAC.49.6.2454-2459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oyarzabal OA, Rad R, Backert S. Conjugative transfer of chromosomally encoded antibiotic resistance from Helicobacter pylori to Campylobacter jejuni. J Clin Microbiol. 2007;45(2):402–408. doi: 10.1128/JCM.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4(8):608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 108.Lee MD, Sanchez S, Zimmer M, Idris U, Berrang ME, McDermott PF. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment . Antimicrob Agents Chemother. 2002;46(11):3660–3664. doi: 10.1128/AAC.46.11.3660-3664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.O’Halloran F, Lucey B, Cryan B, Buckley T, Fanning S. Molecular characterization of class 1 integrons from Irish thermophilic Campylobacter spp. J Antimicrob Chemother. 2004;53(6):952–957. doi: 10.1093/jac/dkh193. [DOI] [PubMed] [Google Scholar]
- 110.Ekkapobyotin C, Padungtod P, Chuanchuen R. Antimicrobial resistance of Campylobacter coli isolates from swine. Int J Food Microbiol. 2008 doi: 10.1016/j.ijfoodmicro.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 111.Connerton PL, Loc Carrillo CM, Swift C, et al. Longitudinal study of Campylobacter jejuni bacteriophages and their hosts from broiler chickens . Appl Environ Microbiol. 2004;70(7):3877–3883. doi: 10.1128/AEM.70.7.3877-3883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Atterbury RJ, Connerton PL, Dodd CE, Rees CE, Connerton IF. Isolation and characterization of Campylobacter bacteriophages from retail poultry . Appl Environ Microbiol. 2003;69(8):4511–4518. doi: 10.1128/AEM.69.8.4511-4518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hansen VM, Rosenquist H, Baggesen DL, Brown S, Christensen BB. Characterization of Campylobacter phages including analysis of host range by selected Campylobacter Penner serotypes. BMC Microbiol. 2007;7:90. doi: 10.1186/1471-2180-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scott AE, Timms AR, Connerton PL, Loc CC, Adzfa RK, Connerton IF. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation . PLoS Pathog. 2007;3(8):e119. doi: 10.1371/journal.ppat.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Scott AE, Timms AR, Connerton PL, El Shibiny A, Connerton IF. Bacteriophage influence Campylobacter jejuni types populating broiler chickens . Environ Microbiol. 2007;9(9):2341–2353. doi: 10.1111/j.1462-2920.2007.01351.x. [DOI] [PubMed] [Google Scholar]
- 116.▪▪.Luo N, Pereira S, Sahin O, et al. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci USA. 2005;102(3):541–546. doi: 10.1073/pnas.0408966102. First report on the fitness of FQ-resistant Campylobacter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Price LB, Johnson E, Vailes R, Silbergeld E. Fluoroquinolone-resistant Campylobacter isolates from conventional and antibiotic-free chicken products. Environ Health Perspect. 2005;113(5):557–560. doi: 10.1289/ehp.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.▪.Price LB, Lackey LG, Vailes R, Silbergeld E. The persistence of fluoroquinolone-resistant Campylobacter in poultry production. Environ Health Perspect. 2007;115(7):1035–1039. doi: 10.1289/ehp.10050. An interesting investigation on the persistence of FQ-resistant Campylobacter after FQ withdrawal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gu W, Islam A, Siletzky R, Kathariou S. Prevalence of ciprofloxacin resistance in thermophilic Campylobacter from turkeys in North Carolina before and after the fluoroquinolone ban. Presented at: The 108th General Meeting of the American Society for Microbiology; Boston, MA, USA. 1–5 June 2008. [Google Scholar]
- 120.Aarestrup FM, McDermott PF, Wegener HC. Transmission of antibiotic resistance from food animals to humans. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3. ASM Press; Washington DC, USA: 2008. pp. 645–665. [Google Scholar]
- 121.Luangtongkum T, Morishita TY, Martin L, Choi I, Sahin O, Zhang Q. Prevalence of tetracycline-resistant Campylobacter in organic broilers during a production cycle. Avian Dis. 2008;52(3):487–490. doi: 10.1637/8181-112807-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 122.Cui S, Ge B, Zheng J, Meng J. Prevalence and antimicrobial resistance of Campylobacter spp. and Salmonella serovars in organic chickens from Maryland retail stores. Appl Environ Microbiol. 2005;71(7):4108–4111. doi: 10.1128/AEM.71.7.4108-4111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sato K, Bartlett PC, Kaneene JB, Downes FP. Comparison of prevalence and antimicrobial susceptibilities of Campylobacter spp isolates from organic and conventional dairy herds in Wisconsin . Appl Environ Microbiol. 2004;70(3):1442–1447. doi: 10.1128/AEM.70.3.1442-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]