Abstract
The tenovins and cambinol are two classes of sirtuin inhibitor that exhibit antitumor activity in preclinical models. This report describes modifications to the core structure of cambinol, in particular by incorporation of substitutents at the N1-position, which lead to increased potency and modified selectivity. These improvements have been rationalized using molecular modeling techniques. The expected functional selectivity in cells was also observed for both a SIRT1 and a SIRT2 selective analog.
Introduction
The central role that p53 plays in preventing tumor development is clear although ongoing research continues to dissect the details of how exactly this is achieved. Furthermore, a role for p53 in development, longevity, and overall fitness of an organism is starting to emerge.1 p53 acts as a transcriptional regulator, inducing the expression of a range of antiproliferative target genes. More than 50% of adult human tumors are characterized by inactivating mutations or deletions of the p53 gene. Other tumor types in which p53 is wild-type frequently have alterations in the mechanisms that control p53 activation. It is widely accepted that activation of the p53 tumor suppressor protein through the use of nongenotoxic compounds may prove therapeutically important. To date, our approach for identifying novel p53 activators has been based on a cell-based high-throughput reporter gene assay2 that we have used to identify a novel class of nongenotoxic p53 activators known as the tenovins (Scheme 1B).3
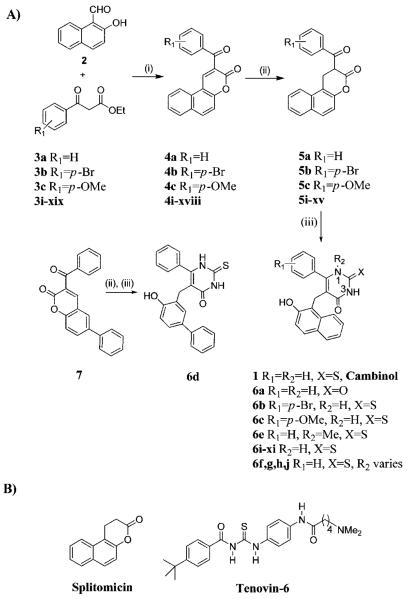
Scheme 1.
(A) Synthesis of 1 and 6a–ja and (B) Structural Comparison of Tenovin-6 and Splitomicin
a Reagents and conditions: (i) piperidine, ethanol, reflux, 2 h (4a, 86%; 4b, 87%; 4c, 95%); (ii) NaBH4, pyridine, rt, 2 h (5a, 95%; 5b, 93%; 5c, 76%); 7 was reduced in 95%; (iii) Na, ethanol, thiourea (for 1, 51%; 6b, 43%; 6c, 42%; 6d, 40%) or urea (6a, 20%) or N-methylthiourea (6e, 16%), reflux, 18 h. For substituents in compounds i–xi and 6f, g, h, and j, see Table 1; for compounds xii–xix, R1 = p-F (xii); m-I (xiii); m-NO2 (xiv); phenyl ring replaced by 2-furyl (xv); o-Cl (xvi); o-I (xvii); o-NO2 (xviii); phenyl ring replaced by 2-pyridyl (xix).
Tenovins function through the inhibition of a group of NAD+-dependent protein deacetylases known as the sirtuins (HDACa class III).3 To date, one sirtuin family member, SIRT1, is known to regulate p53 activity by deacetylating this protein at Lys382.4a–c Cells derived from SIRT1 deficient mice and cells treated with siRNAs against SIRT1 show high levels of hyperacetylated p53.4d,e Acetylation of p53 is known to augment its DNA binding ability.4f In addition, a dominant-negative SIRT1 mutant increases p53-dependent transcriptional activity.3,4c Partly due to its ability to decrease p53 function, SIRT1 is believed to represent an important target for cancer treatment with two reported sirtuin inhibitors, 1 (cambinol)5 and tenovin-6, showing antitumor activity in preclinical models.3,5 Here we report optimization studies on 1 (Scheme 1). The goal was to identify modifications to the structure of 1 that lead to an increase in potency. Additional studies aimed at exploring the effect of structural modifications on the ability of 1 to inhibit selectively SIRT1 over other members of the sirtuin family, in particular SIRT2, were planned. SIRT2 is a predominantly cytoplasmic protein that colocalizes with microtubules and has been shown to promote deacetylation of α-tubulin at Lys40 both in vitro and in vivo.6a,6b In addition, knockdown of SIRT2 via siRNA results in tubulin hyperacetylation, and SIRT2 overexpression abrogated microtubule hyperacetylation and resistance to axonal degeneration.6b Conversely, SIRT2 knockdown by using a lentiviral vector expressing small interfering RNA enhanced microtubule acetylation and resistance to axonal degeneration.6c Small molecule inhibition of SIRT2 has been linked to potential treatments for Parkinson's disease.7
The splitomicin8,3 derivative 1 (see Scheme 1B for structure of splitomicin) inhibits SIRT1 and SIRT2 in vitro with IC50 values of 56 and 59 μM, respectively, demonstrating moderate potency and no selectivity.5 Only one analog of 1 has been reported to date in which replacement of the β-naphthol ring in 1 by a phenol ring led to the loss of inhibitory activity against both SIRT1 and SIRT2.5
Synthesis of 1 and Its Analogs
An authentic sample of 1 was prepared using a previously reported route, giving 1 in 42% yield over three steps (Scheme 1).9 Initial Knoevenagel condensation of 2 (2-hydroxy-1-naphthaldehyde) with 3a (ethyl benzoylacetate) afforded 4a (3-benzoyl-5,6-benzocoumarin). Subsequent conjugate reduction of 4a using sodium borohydride in pyridine10 gave the saturated lactone 5a, which was then converted to 1 upon treatment with excess thiourea in the presence of sodium ethoxide. An analogous approach was used to prepare a set of analogs of 1 in which (i) the thiocarbonyl functionality in 1 was replaced by a carbonyl (6a); (ii) electron-withdrawing (e.g., bromine in 6b) or electron-donating (e.g., methoxy in 6c) substituents were incorporated in the para-position of the phenyl ring in 1; (iii) the β-naphthol ring in 1 was replaced by a biphenyl-4-ol group (6d), and (iv) a methyl group (R2 = Me, 6e) was incorporated at the N1 position in 1. In brief, replacement of thiourea with urea enabled the conversion of 5a to 6a, and analogs 6b and 6c were prepared from the corresponding commercially available ethyl benzoylacetates 3b and 3c, respectively. In order to prepare analog 6d, 7 (3-benzoyl-6-phenylchroman-2-one) (Scheme 1) was synthesized from 5-bromo-2-hydroxybenzaldehyde in 55% yield over 2 steps.11 Conversion of 7 to the desired analog 6d was carried out by conjugate reduction and subsequent reaction with thiourea in the presence of base.
Synthesis of N1 Substituted Analogs of 1
Analog 6e was prepared by reaction of 5a with N-methylthiourea and sodium ethoxide. Although the yield for formation of 6e was low due to the formation of additional side products,11 no evidence could be found to support formation of the N3-methyl isomer of 6e. Structural assignment of 6e was achieved using 2D [1H–13C] HMBC correlation analysis.11 The observed regiochemistry may be rationalized by initial reaction of the most nucleophilic nitrogen atom in N-methylthiourea with the most reactive carbonyl group, the aryl ketone, in 5a. The encouraging inhibition and selectivity data for SIRT2 shown by 6e (Table 1) led to the synthesis of further analogs in which different N1-aliphatic chains were incorporated (R2 in Scheme 1). Initial plans included analogs with alkyl, benzyl, and aromatic substituents at N1. However, limitations in the synthetic method restricted the set of analogs that could be prepared rapidly to four analogs (6f, g, h, and j).11
Table 1.
Inhibition (at 60 μM ± SEa (%)) of Sirtuins by the Novel Analogs 6i–xi and 6a–6j
| compd | R1 | R2 | X | SIRT1 | IC50b | SIRT2 | IC50b |
|---|---|---|---|---|---|---|---|
| 1 | H | H | S | 59.5 ± 1 | 40.7 ± 11 | 51.9 ± 1 | 47.9 ± 12 |
| 6a | H | H | O | 15.5 ± 2 | 6.5 ± 4 | ||
| 6b | p-Br | H | S | 82.3 ± 1 | 12.7 ± 2 | 9.4 ± 1 | >90 |
| 6c | p-CH3O | H | S | 6.4 ± 1 | 9.2 ± 2 | ||
| 6d | H | H | S | 37.8 ± 1 | 52.7 ± 2 | ||
| 6i | o-CH3 | H | S | 79.6 ± 1 | 43.0 ± 2 | 29.1 ± 2 | |
| 6ii | m-CH3 | H | S | 83.2 ± 4 | 44.2 ± 2 | 12.7 ± 2 | |
| 6iii | p-CH3 | H | S | 79.0 ± 2 | 44.5 ± 1 | 13.4 ± 3 | |
| 6iv | p-Cl | H | S | 13.7 ± 1 | 20.1 ± 1 | ||
| 6v | p-I | H | S | 7.7 ± 1 | 7.7 ± 1 | ||
| 6vi | p-CF3 | H | S | 15.1 ± 1 | 6.7 ± 1 | ||
| 6vii | o-Br | H | S | 19.7 ± 1 | |||
| 6viii | m-Br | H | S | 4.8 ± 1 | 6.6 ± 3 | ||
| 6ix | m-Cl | H | S | 11.3 ± 1 | 7.6 ± 6 | ||
| 6x | o-F | H | S | 89.0 ± 1 | 50.0 ± 1 | 19.5 ± 1 | |
| 6xi | m-F | H | S | 87.8 ± 1 | 38.3 ± 1 | 52.1 ± 1 | |
| 6e | H | Me | S | 29.4 ± 1 | > 90 | 80.4 ± 1 | 20.1 ± 5 |
| 6f | H | Et | S | 31.9 ± 1 | 86.8 ± 1 | 10.5 ± 3 | |
| 6g | H | allyl | S | 37.5 ± 1 | 88.3 ± 1 | 22.2 ± 1 | |
| 6h | H | n-prop | S | 25.0 ± 2 | 94.7 ± 1 | 4.8 ± 2 | |
| 6j | H | n-But | S | 16.9 ± 1 | 97.6 ± 1 | 1.0 ± 1 |
SE, standard error (n = 2).
IC50 values were determined for compounds that had over 60% inhibition at 60 μM for SIRT1 and SIRT2 (repeated at least twice).
Parallel Synthesis Approach
Encouraged by the yields and straightforward workup associated with the synthesis of analogs of 1 and the initial observation that incorporation of substituents into the phenyl ring of 1 resulted in tuning of the required biological activity (Table 1), it was decided to prepare additional analogs using a parallel synthesis approach. The commercial availability of a range of ethyl benzoylacetates enabled the rapid incorporation of different electron-withdrawing or electron-donating substituents at the required positions (Table 1). Nineteen ethyl benzoylacetates (3i–ix) were loaded with 2 into different vessels of a parallel synthesis apparatus. Ethanol and piperidine were then added, and in 18 of the 19 cases, the desired products (4i–xviii) formed upon heating in good yields (45–98%) as yellow precipitates. The 2-pyridyl-containing substrate 3xix (Scheme 1) failed to react. Subsequent reduction of 4i–xviii afforded 15 1,2-dihydroketocoumarins 5i–xv, following precipitation of the products upon addition of aqueous hydrochloric acid (2 M) to the crude reaction mixture. Analytically pure samples of 5i–xv were obtained by recrystallization from ethanol in a wide range of yields (35–95%). Interestingly, 1H NMR analysis of the ortho-substituted analog 5vii (R1 = o-Br) showed that it exists predominantly as the enol-lactone isomer.11 In the next step, a modified protocol for substrates 5i–xv was used compared to that employed in the preparation of 1. Addition of the sodium ethoxide solution as the final step and heating under reflux for 18 h was followed by removal of the solvent in parallel and dissolution of the resulting solids in water. Subsequent acidification of the aqueous solutions afforded 11 new analogs 6i–xi in moderate yields after purification by column chromatography.
In Vitro Inhibition of SIRT1 and SIRT2 by Analogs 6i–xi and 6a–6j
1 and its analogs were tested for in vitro activity against SIRT1 and SIRT2 using an established fluorescent assay system (Table 1). The IC50 values determined from our experiments with 1 were similar to those previously reported5 (41 vs 56 μM5 for SIRT1 and 48 vs 595 μM for SIRT2). One of the key structural modifications we explored was the incorporation of functional groups into the phenyl ring of 1. In brief, the introduction of a p-methoxy substituent in the phenyl ring of 1 to give 6c led to a loss of activity against both SIRT1 and SIRT2. Interestingly, only activity against SIRT2 was lost when a methyl substituent was incorporated at the para-position or any other position in the aromatic ring (6i–6iii), with these three analogs clearly demonstrating selective inhibition of SIRT1. The p-bromo-analog 6b also exhibits relatively selective inhibition of SIRT1 (IC50 = 13 μM) compared to SIRT2 (IC50 > 90 μM). The IC50 value attributed to 6b represents a 3-fold improvement in activity against SIRT1 compared to 1. Replacement of the p-bromo-substituent in 6b with chlorine, iodine, or trifluoromethyl (6iv, 6v, or 6vi, respectively) led to a significant decrease in activity against both enzymes. The position of the bromine atom in 6b was also important, as SIRT1 activity was lost when this substituent was placed at either the o-or m-position of the aromatic ring (analogs 6vii and 6viii). Incorporation of a m-chloro-substituent in analog 6ix also reduced activity against SIRT1 and SIRT2. The use of a fluorine substituent to reduce electron density without increasing steric bulk led to analogs 6x and 6xi, which have comparable activity to 1 against SIRT1. This implies that the improved activity of the p-bromo-analog 6b results from an increase in additional hydrophobic interactions rather than through a reduction in electron density associated with the aromatic ring.
Previous studies have reported that when the β-naphthol ring in 1 is replaced by a phenol ring, activity against SIRT1 and SIRT2 is lost.5 The observed reduction in activity of the biphenyl-4-ol-containing analog 6d against SIRT1 compared to 1 further supports the view that the β-naphthol ring in 1 is important for activity against this enzyme, although the observed activity against SIRT2 suggests that this isoform is more tolerant to change at this position in 1. Substitution of the thiocarbonyl functionality in 1 for a carbonyl (6a) resulted in significantly reduced activity against both enzymes.
The second key structural question that we looked to address involved the requirement for the N1-hydrogen. The presence of a N1-methyl substituent in analog 6e led to an increase in activity against SIRT2 (IC50 = 20 μM, cf. 48 μM for 1) and to a decrease in potency against SIRT1 (IC50 > 90 μM for 6e). As a result of these changes in activity compared to 1, 6e is a selective SIRT2 inhibitor in vitro. This result inspired the synthesis of four other analogs 6f, g, h, and j with different aliphatic chains at the N1-position. Interestingly, a steady improvement in potency against SIRT2 was observed as the chain length increased, culminating in the N1-butyl analog 6j being the most potent inhibitor of SIRT2 identified to date in this inhibitor class with an IC50 = 1.0 μM for SIRT2, while demonstrating poor inhibition of SIRT1 (Table 1). The majority of the synthetic intermediates prepared en route to analogs of 1 were also tested against SIRT1 at 60 μM concentration due to the structural similarity with splitomicin.11 No active compounds were identified.
Identification of a Potential Binding Mode
In order to provide a potential rationalization for the observed in vitro activity and selectivity of 1 and its analogs, automated ligand docking studies were carried out using the software GOLD.12 The reported structure of human SIRT213 was used in this study. All the SIRT2 crystal structures so far determined are characterized by a highly conserved catalytic domain of 270 amino acids that consists of a large classical Rossman-fold domain and a small zinc binding subdomain. The active site is situated at the interface between the two domains and is commonly divided into A, B, and C subpockets into which the substrate and cofactor bind. No ligand bound structures of human SIRT2 have been solved, but its homologs from yeast14 and Archaeoglobus fulgidus15 have been extensively characterized with regard to the mode of substrate and cofactor binding. To date, no crystal structure has been reported for SIRT1.
Recently reported docking studies proposed that 1 binds in the nicotinamide C-subpocket of the catalytic domain of SIRT2.16 While no direct evidence to support this binding mode was provided, our attempts to identify a binding mode for 1 using GOLD led to an analogous result. In our hands, 1 gives the lowest energy solutions when bound in the C-pocket, oriented such that the β-napthol ring is sandwiched between the two aromatic residues Phe119 and His187 with π-stacking interactions probably contributing to the calculated stability of this binding mode. It is in this pocket that the acetylated lysine of SIRT2 substrates binds. In addition, we observed that the polar component of 1, represented by the carbonyl and thiocarbonyl functionalities and the two nitrogen atoms, has the potential to form hydrogen bonds to the active site. Although our model did not include water molecules in the active site, the predicted orientation of this part of the ligand is in close agreement with that proposed previously. It is of interest that our small molecule X-ray structure of 1 in the absence of protein identifies the existence of an intramolecular hydrogen bond between the phenolic OH and the carbonyl functionality.11 When this conformation of 1 was used as the starting point for the docking studies, rather than the lowest energy conformation predicted by the PRODRG server, analogous results were obtained suggesting that loss of this hydrogen bond can be accommodated for on binding to the protein. In general, visual analysis of the highest scoring docking poses for the new analogs of 1 showed a very similar situation to that previously reported and found by us for 1 itself, with all the new inhibitors showing the same preferred binding mode with the β-naphthol ring sandwiched in the previously identified hydrophobic channel between Phe119 and His187. Modest differences in the conformation of the β-naphthol moiety of the inhibitors were observed.
Rationalization of the Observed Increase in SIRT2 Selectivity and Activity Associated with an N1-Substituent
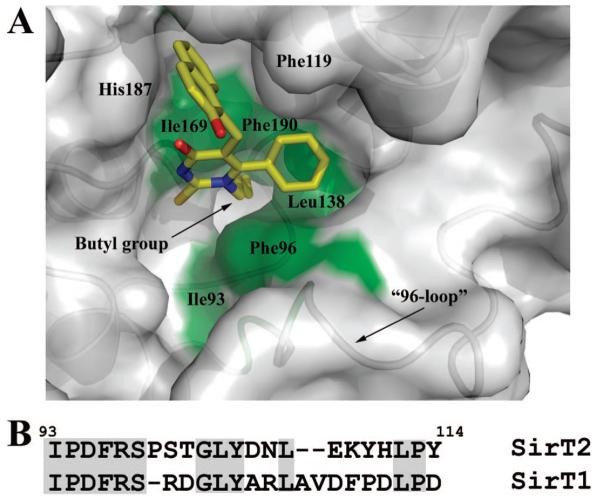
The improvement in the inhibitory activity and selectivity observed for analogs 6e and 6f–j against SIRT2 compared to 1 can be rationalized by the formation of additional hydrophobic interactions between the N1-aliphatic substituent and a previously unoccupied narrow lipophilic channel around Phe96, Leu138, and Ile169 in the SIRT2 active site (Figure 1A). The proposed binding mode of this subset of analogs was observed consistently during the docking studies. It is interesting to note that a structure of the Archaeoglobus fulgidus enzyme has been solved in which this channel is occupied by a pentaethylene glycol molecule.15 While increasing aliphatic chain lengths lead to increased SIRT2 activity at least up to a C4 chain, it is difficult to predict from computational studies what the optimal N1 side chain will be. For example, it is difficult to explain why the N1-allyl analog 6g has reduced but still significant activity. Visual analysis of this newly identified hydrophobic channel suggests that there is insufficient space to accommodate larger substituents, such as phenyl or benzyl groups in the human SIRT2 active site. However, a structure of the yeast enzyme has been elucidated which shows a nicotinamide molecule occupying this hydrophobic channel.17a Further studies are required to fully explore the range of substituents that can be accommodated.
Figure 1.
(A) Docking solution obtained for N1 substituted analog 6j. The N1-aliphatic carbon chain in 6j is proposed to insert into a narrow lipophilic channel delimited by Phe96, Leu 138, and Ile169. (B) A comparison of the amino acid sequence for the 96-loop in human SIRT1 and SIRT2.
In the absence of a crystal structure for SIRT1, the selectivity of the N1-substituted series for SIRT2 over SIRT1 was addressed via the generation of a homology model of human SIRT118 based on the crystal structure of human SIRT213 using the PHYRE server. While the majority of the active site is predicted to be highly conserved, notable differences in the loop at the base of the site potentially underlie the observed specificity. Figure 1B shows a sequence alignment of the residues which comprises this loop in the human SIRT2 and SIRT1 enzymes. This loop (the 96-loop) contains Phe96, which lines the lipophilic channel into which the N1-substituent packs. An amino acid insertion and a two amino acid deletion in SIRT2 compared to SIRT1 subsequent to Phe96 is likely to cause a structural alteration of this loop and thus cause a difference in the size and nature of the lipophilic channel. This loop also contains Arg97, which interacts with the phosphate of an NAD+ analog in the yeast enzyme.17a The yeast and human SIRT2 enzymes, despite having near identical sequences of this loop, do show conformational differences, reinforcing the concept that structural differences of this loop between human SIRT2 and SIRT1 could explain the observed selectivity.
Rationalization of the Observed Variation in Activity for Analogs Incorporating a Substituent in the Phenyl Ring of 1
A key conclusion from the SAR data (Table 1) is the increase in potency and net SIRT1 selectivity associated with the incorporation of a p-bromo-substituent (6b) into the phenyl ring of 1. Visual analysis of the docking poses for N1-substituted analogs suggested that the active site of SIRT2 is too small to accommodate a phenyl ring bearing a large p-substituent, consistent with the observed relative lack of activity against SIRT2 of the p-bromo (6b) and p-iodo (6v) analogs. Although the majority of the amino acids in this region are conserved between SIRT2 and SIRT1, again the structural variation of the 96-loop could account for the observed selectivity. Within this loop, Tyr104 points directly into the pocket that is predicted to accommodate the phenyl ring of 1. Due to the predicted structural alteration of the 96-loop between SIRT2 and SIRT1, this pocket has the potential to be larger in SIRT1 compared to SIRT2, with the pocket now being able to accommodate the p-bromo-substituent.
During the course of these studies, we demonstrated that it was possible to convert the essentially unselective 1 into analogs that possess either SIRT1 (6b) or SIRT2 (e.g., 6j) selectivity in vitro. Further evidence to support the selective targeting of SIRT1 and SIRT2 by these analogs comes from cell-based studies. In a previous report,5 Bedalov and co-workers detected increased levels of p53 and acetylated p53 in NCI H460 cancer cells treated with 1 in the presence of the genotoxic agent etoposide (10 μM). Here we tested our more selective SIRT1 inhibitor 6b for this effect in MCF-7 breast adenocarcinoma cells (p53 wild type) and compared it to 1. As previously observed for 1, in the presence of etoposide (10 μM), the levels of p53, and especially the levels of Lys382 acetylated p53, increased with 20 and 50 μM concentrations of 1 (Figure 2). The levels of p53 and acetylated p53 decreased again upon exposure to 1 at higher concentrations of 200 μM, probably as a result of toxicity. Treatment of MCF-7 cells with different concentrations of 6b alone did not increase the levels of p53 in the absence of etoposide, as was the case for all the analogs of 1 we have tested in this assay to date.11 However, in the presence of etoposide (10 μM), compound 6b increased total p53 levels and the levels of p53 acetylated at Lys382 more clearly than 1 (Figure 2).
Figure 2.
Levels of p53 and acetylated p53 in MCF-7 breast adenocarcinoma cells treated with different concentrations of cambinol (1) and 6b in the presence of etoposide. The black line represents the position of the 51KDa molecular weight marker. Samples were first analyzed with the K382-acetylated p53 antibody and subsequently reloaded and analyzed with the antibody against total p53.
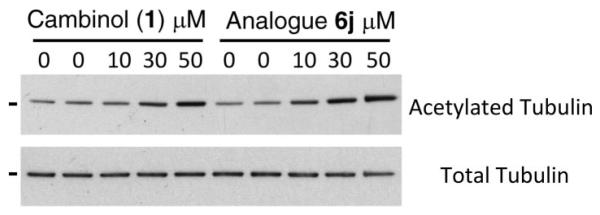
As expected for a selective SIRT1 inhibitor, 6b did not lead to an increase in the levels of acetylated α-tubulin (Figure S1311). As a control, and in agreement with previous reports, treatment of H1299 cells with the unselective inhibitor 1 did lead to an increase in levels of acetylated tubulin (Figure 3). When this experiment was repeated, this time using our improved and SIRT2 selective inhibitor 6j, an increase in the levels of acetylated α-tubulin was achieved compared to the effect of 1 across a concentration range of 10–50 μM (Figure 3 and Figure S14). This experiment could not be carried out at higher concentrations due to toxicity associated with the use of 6j at these concentrations. While an improvement in the ability of 6j to increase levels of acetylated α-tubulin in cells compared to 1 was observed, the effect was less than may have been expected given the 50-fold difference in IC50 of 6j and 1 against SIRT2 observed in vitro. Issues relating to cell permeability, compartmentalization of the compound, or its metabolism may explain this observation and are currently being assessed.
Figure 3.
Western blot analysis of the levels of α-tubulin acetylated at K40 and total α-tubulin in H1299 cells treated with increasing concentrations of cambinol (1) and analog 6j. All samples were also treated with trichostatin A, an inhibitor of class I and II HDACs, to reduce the background effect of these deacetylases. The black line represents the position of the 51KDa molecular weight marker. Samples were first analyzed with the K40-acetylated α-tubulin antibody and subsequently reloaded and analyzed with the antibody against total α-tubulin. For a more detailed statistical analysis of this experiment, see Supporting Information Figure S14.
Conclusion
A recent report has described 1 as a potential lead for cancer chemotherapy due to its ability to inhibit the function of the class III histone deacetylases known as the sirtuins. Here we describe the results of a SAR study we have carried out on 1. These studies have indicated that inhibition of the sirtuins is sensitive to changes in the phenyl ring of 1 leading to the identification of an analog 6b with improved activity and increased selectivity for SIRT1. In addition, we have demonstrated that N1-functionalization leads to an increase in potency against SIRT2 in vitro that is not seen for SIRT1, resulting in the identification of a range of SIRT2 selective analogs. In both cases, we have rationalized the observed selectivity through the use of computational methods providing evidence in support of a previously proposed binding mode for 1. The identification of an interesting lipophilic channel, the targeting of which may aid selective SIRT2 inhibitor development, is also reported. Studies on the activity of 6b and 6j in cells are also consistent with the observed in vitro selectivity, implying that the novel analogs identified in this work may also be of interest in the treatment of cancer and neurodegenerative diseases.
General Experimental Procedures
Chemistry
All chemicals and solvents were purchased from Aldrich (U.K.) or Alfa-Aesar and used without further purification. All the reactions were carried out under a positive pressure of nitrogen or argon in flame or oven-dried glassware. Ethanol was dried over Mg/I2; pyridine was dried over KOH pellets. Thin layer chromatography (TLC) analysis was performed on silica precoated SIL G-25 UV254 sheets (layer: 0.25 mm silica gel with fluorescent indicator UV254, Alugram, U.K.). Compounds were visualized by UV light (UV lamp, model UVGL-58, Mineralight LAMP, Multi-band UV-254/365 nm) and stained with potassium permanganate. Flash column chromatography was carried out on silica gel (40–63 μm, Fluorochem, U.K.). Melting points were measured with an electrothermal 9100 capillary melting point apparatus and are uncorrected. Fourier Transform infrared spectra (FT IR) were acquired on a Perkin-Elmer Paragon 1000 FT spectrometer. Absorption maxima are reported in wavenumbers (cm−1). Unless otherwise stated, 1H NMR spectra were measured at room temperature (298 K) on Bruker DPX 400 (1H = 400 MHz) and Bruker Avance 300 (1H = 300.1 MHz) instruments. Deuterated solvents were used and 1H NMR chemical shifts were internally referenced to CHCl3 (7.26 ppm) in chloroform-d1 solution, to CD2HSO2CHD2 (2.50 ppm) in dimethylsulfoxide-d6 solution, and to CD3COCHD2 (2.09 ppm) in acetone-d6. Chemical shifts are expressed as δ in units of ppm. 13C NMR spectra were recorded in the same conditions and in the same solvents using the PENDANT sequence mode on a Bruker DPX 400 (13C = 100 MHz). Data processing was carried out using TOPSPIN 2 NMR version (Bruker UK, Ltd.). In 1H NMR assignment the multiplicity used is indicated by the following abbreviations: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad, br s = broad singlet. Signals of protons and carbons were assigned, as far as possible, by using the following two-dimensional NMR spectroscopy techniques: [1H, 1H] COSY, [1H, 13C] COSY (HSQC: heteronuclear single quantum coherence) and long-range [1H, 13C] COSY (HMBC: heteronuclear multiple bond connectivity). Mass spectra (electrospray mode, ES; chemical ionization mode, CI) were recorded on a high performance orthogonal acceleration reflecting TOF mass spectrometer operating in positive and negative mode, coupled to a Waters 2975 HPLC.
Synthesis of 1 and Analogs 6b and 6c
1 was prepared via 4a and 5a as described in Scheme 1 according to the following generalized protocols. Analytical data for additional compounds made using these protocols are also provided.
General Procedure for Preparation of 4a–4c
To a solution of 2-hydroxy-1-naphthaldehyde (2) (0.86 g, 5 mmol) in ethanol (10 mL) were added ethyl benzoyl acetate analogs (3a–c, 1.06 g, 5 mmol). Piperidine (15 drops) was added, and the reaction was heated under reflux for 2 h. After cooling to room temperature, the product was collected by filtration and washed with ethanol.
2-Benzoyl-3H-benzo[f]chromen-3-one (4a).19,20
Yielded 1.30 g (4.33 mmol, 86%) as a yellow powder. Mp 208–210 °C (lit.19,20 209 °C). 1H NMR (CDCL3, 300 MHz): δ = 8.93 (s, 1H, H-1), 8.27 (d, 1H, J = 8.2 Hz, H-10), 8.12 (d, 1H, J = 9.0 Hz, H-6), 7.98–7.90 (m, 3H, H-7 + 2 × ArH), 7.77–7.70 (m, 1H, H-9), 7.67–7.59 (m, 2H, H-8, ArH), 7.56–7.47 (m, 3H, H-5 + 2 × ArH). LRMS [ES]+: m/z 323.06 [M + Na]+ (100%).
2-(5′-Bromobenzoyl)-3H-benzo[f]chromen-3-one (4b).20
Yielded 600 mg (1.58 mmol, 93%) as a yellow powder. Mp 245–248 °C (lit.20 246–247 °C). 1H NMR (CDCl3, 400 MHz): δ = 8.98 (s, 1H, H-1), 8.29 (d, 1H, J = 8.3 Hz, H-10), 8.13 (d, 1H, J = 9.0 Hz, H-6), 7.96 (d, 1H, J = 8.2 Hz, H-7), 7.81–7.73 (m, 3H, H-9, AA′BB′ system, J = 8.7 Hz, H-3′, H-7′), 7.64 (m, 3H, H-8, AA′BB′ system, J = 8.7 Hz, H-4′, H-6′), 7.51 (d, 1H, J = 9.0 Hz, H-5). LRMS [ES]+: m/z 401.07 [M + Na]+ (100%).
2-(5′-Methoxybenzoyl)-3H-benzo[f]chromen-3-one (4c).19,20
Yielded 1.45 g (4.39 mmol, 95%) as a white-red powder. Mp 207–210 °C (lit.19,20 209–210 °C). 1H NMR (400 MHz, CDCl3): δ = 8.86 (s, 1H, H-1), 8.26 (d, 1H, J = 8.4 Hz, H-10), 8.10 (d, 1H, J = 9.0 Hz, H-6), 7.98–7.89 (m, 3H, H-7, AA′BB′ system, J = 8.8 Hz, H-3′, H-7′), 7.76–7.70 (m, 1H, H-9), 7.65–7.58 (m, 1H, H-8), 7.53 (d, 1H, J = 9.0 Hz, H-5), 6.97 (d, 2H, AA′BB′ system, J = 8.8 Hz, H-4′, H-6′), 3.83 (s, 3H, CH3O). LRMS [ES]+: m/z 353.07 [M + Na]+ (100%).
General Procedure for Preparation of 5a–5c
To a stirring solution of 2-benzoylketocoumarin (4a–c, 500 mg, 1.56 mmol) in dry pyridine (10 mL) was added NaBH4 (59 mg, 1.56 mmol). The reaction was stirred at room temperature for 2 h. The reaction mixture was poured into cold 2 M hydrochloric acid (90 mL). The resulting white precipitate was collected by filtration, washed with HCl (aq, 2 M), and recrystallized from ethanol.
2-Benzoyl-1,2-dihydrobenzo[f]chromen-3-one (5a).21
Yielded 440 mg (1.45 mmol, 95%) as a yellow powder. Mp 158–160 °C (lit.21 158–160 °C). 1H NMR (CDCl3, 300 MHz): δ = 8.03–7.98 (m, 2H, ArH), 7.90–7.85 (m, 2H, H-10, H-7), 7.81 (d, 1H, J = 9.0 Hz, H-6), 7.67–7.61 (m, 1H, H-9), 7.59–7.52 (m, 1H, H-8), 7.54–7.45 (m, 3H, ArH), 7.29 (d, 1H, J = 9.0 Hz, H-5), 4.82 (dd, 1H, 3J = 10.2 Hz, 3′J = 6.8 Hz, H-2), 3.85 (dd, 1H, 2J = 16.7 Hz, 3J = 10.2 Hz, H-1), 3.62 (dd, 1H, 2J = 16.7 Hz, 3J = 6.8 Hz, H-1). LRMS [ES]+: m/z 325.10 [M + Na]+ (100%).
2-(5′-Bromobenzoyl)-1,2-dihydrobenzo[f]chromen-3-one (5b)
Yielded 892 mg (2.35 mmol, 94%) as a white powder. Mp 221–223 °C. 1H NMR (CDCl3, 400 MHz): δ = 7.91–7.84 (m, 4H, H-7, H-10, AA′BB′ system, J = 8.6 Hz, H-3′, H-7′), 7.82 (d, 1H, J = 8.8 Hz, H-6), 7.65 (d, 2H, AA′BB′ system, J = 8.6 Hz, H-2′, H-4′), 7.62–7.54 (m, 1H, H-9), 7.52–7.45 (m, 1H, H-8), 7.28 (d, 1H, J = 8.8 Hz, H-5), 4.74 (dd, 1H, 3J = 10.5 Hz, 3′J = 6.7 Hz, H-2), 3.83 (dd, 1H, 2J = 16.6 Hz, 3J = 10.5 Hz, H-1), 3.62 (dd, 1H, 3J = 16.6 Hz, 2J = 6.7 Hz, H-1). LRMS [CI]+: m/z 337.06 [M + H]+ (100%).
2-(5′-Methoxybenzoyl)-1,2-dihydrobenzo[f]chromen-3-one (5c)
Yielded 400 mg (1.20 mmol, 76%) as a white-red powder. Mp 175–176 °C. 1H NMR (400 MHz, CDCl3): δ = 7.99 (d, 2H, AA′BB′ system, J = 9.0 Hz, H-3′, H-7′), 7.90–7.84 (m, 2H, H-10, H-7), 7.80 (d, 1H, J = 9.0 Hz, H-6), 7.60–7.53 (m, 1H, H-9), 7.51–7.44 (m, 1H, H-8), 7.27 (d, 1H, J = 9.0 Hz, overlapped with solvent signal, H-5), 6.97 (d, 2H, AA′BB′ system, J = 9.0 Hz, H-4′, H-6′), 4.77 (dd, 1H, 3J = 10.5 Hz, 3′J = 6.8 Hz, H-2), 3.88 (s, 3H, CH3O), 3.83 (dd, 1H, 2J = 16.6 Hz, 3J = 10.5 Hz, 1-H), 3.60 (dd, 1H, 3J = 16.6 Hz, 2J = 6.8 Hz, H-1). LRMS [ES]+: m/z 355.12 [M + Na]+ (100%).
General Procedure for the Synthesis of 1 and Its Analogs 6b and 6c
To a solution of NaOEt (10 mL, 2 M), prepared by dissolving Na metal in dry ethanol, were added thiourea (15.6 equiv) and different 2-benzoyl-1,2-dihydrocoumarins (5a–c, 200 mg, 1 equiv), and the reaction was heated under reflux for 24 h. After removing the solvent in vacuo, the residue was dissolved in the minimum amount of distilled water and the products were precipitated upon addition of 2 M aqueous HCl. The solid was collected by filtration and purified by recrystallization from ethanol.
6-Phenyl-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (1, Cambinol).9
Yielded 120 mg (51%) as a white powder after recrystallization from ethanol. Mp 253–255 °C (lit.S4 253 °C). IR (NaCl, thin layer) νmax/cm−1: 3111 (OH), 2804 (CH2), 1628 (C=O), 1556 (NH), 1494 (C–N), 1212 (C=S, CSNH), 877 and 766 (C–HAr). 1H NMR (400 MHz, DMSO-d6): δ = 12.55 (br s, 1H, NH), 12.30 (br s, 1H, NH), 9.44 (s, 1H, OH), 7.64 (d, 1H, J = 7.7 Hz, H-5″), 7.48 (d, 1H, J = 8.8 Hz, H-4″), 7.44–7.11 (m, 8H, H-6″, H-7″, H-8″ + 5 × ArH), 6.90 (d, 1H, J = 8.8 Hz, H-3″), 3.90 (s, 2H, CH2). 13C NMR (100 MHz, DMSO-d6): δ = 173.8 (C=S), 162.2 (C=O), 152.7 (C2″), 150.1 (C6), 133.1 (C8″a), 131.6 (C1′), 129.4 (C4′), 128.5 (C5″, C3′), 128.0 (C5″), 128.0 (C4″a), 127.8 (C2′, C6′), 127.5 (C4″), 125.5 (C7″), 122.8 (C6″), 121.9 (C8″), 118.3 (C3″), 116.4 (C1″a), 115.2 (C5), 21.5 (CH2). LRMS [ES+]: m/z 382.97 [M + Na]+ (100%). LRMS [ES]−: m/z 358.78 [M − H]− (100%). HRMS [ES]−: m/z calcd for C21H15N2O2S 359.0854 [M − H]−, found 359.0858 (−0.9 ppm). A sample of 1 was further recrystallized from ethanol to give crystals of sufficient size for small molecule X-ray crystallographic analysis. Crystallographic data for cambinol was deposited at the Cambridge Crystallographic Data Centre and allocated the deposit number CCDC 704011.
6-(4′-Bromophenyl)-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6b)
Yielded 100 mg (0.22 mmol, 43%) as a white powder after recrystallization from ethanol. Mp 282–287 °C. IR (NaCl, thin layer) νmax/cm−1: 3413 (OH), 1699 and 1634 (C=O), 1547 (NH), 1445 (C–N), 1189 (C=S, CSNH), 1158, 1124, and 1112 (C–O and C=S), 1070 (C–O), 971, 827, and 742 (C–HAr), 668 (C–Br). 1H NMR (400 MHz, DMSO-d6): δ = 12.56 (br s, 1H, NH), 12.22 (br s, 1H, NH), 9.42 (s, 1H, OH), 7.71 (d, 1H, J = 7.5 Hz, H-5″), 7.47 (d, 1H, J = 8.8 Hz, H-4″), 7.44 (d, 1H, J = 8.7 Hz, H-8″), 7.32 (d, 2H, AA′BB′ system, J = 8.4 Hz, H-2′, H-6′), 7.30–7.22 (m, 1H, H-7″), 7.20–7.14 (m, 1H, H-6″), 6.98 (d, 2H, AA′BB′ system, J = 8.4 Hz, H-3′, H-5′), 6.87 (d, 1H, J = 8.7 Hz, H-3″), 3.91 (s, 2H, CH2). 13C NMR (100 MHz, DMSO-d6): δ = 173.8 (C=S), 161.9 (C=O), 152.6 (C2″), 148.8 (C6), 132.9 (C1′), 130.6 (C8″a), 130.4 (C3′), 130.1 (C2′), 128.0 (C5″), 127.9 (C4″a), 127.4 (C4″), 125.5 (C7″), 122.7 (C6″), 121.8 (C8″), 117.9 (C3″), 116.3 (C1″), 115.7 (C5), 20.9 (CH2). LRMS [ES]+: m/z 462.96 [M + Na]+ (100%). LRMS [ES]−: m/z 436.93 [M − H]− (100%). HRMS [ES]−: m/z calcd for C21H12N2O2S81Br 436.9960 [M − H]−, found 436.9970 (+ 0.2 ppm); m/z calcd for C21H12N2O2S79Br 438.9945 [M − H]−, found 438.9946 (+ 0.1 ppm).
6-(4′-Methoxyphenyl)-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6c)
Yielded 95 mg (0.24 mmol, 42%) as a white powder after recrystallization from ethanol. Mp 262–265 °C. IR (NaCl, thin layer) νmax/cm−1: 3425 (OH), 2930 (CH2), 1652 and 1628 (C=O), 1576 (NH), 1457 (C–N), 1253 (OH), 1213, 1186, and 1023 (C–O and C=S), 736 (C–HAr). 1H NMR (400 MHz, DMSO-d6): δ = 12.52 (br s, 1H, NH), 12.25 (br s, 1H, NH), 9.50 (s, 1H, OH), 7.70–7.62 (m, 1H, H-8″), 7.51 (d, 1H, J = 8.8 Hz, H-4″), 7.35 (d, 1H, J = 8.3 Hz, H-5″), 7.26–7.14 (m, 4H, H-6″, H-7″,AA′BB′ system, J = 8.7 Hz, H-2′, H-6′), 6.94 (d, 1H, J = 8.8 Hz, H-3″), 6.86 (d, 2H, AA′BB′ system, J = 8.7 Hz, H-3′, H-5′), 3.92 (s, 2H, CH2), 3.75 (s, 3H, CH3O). 13C NMR (100 MHz, DMSO-d6): δ = 173.8 (C=S), 162.4 (C=O), 160.1 (C4′), 152.7 (C2″), 150.0 (C6), 133.1 (C1′), 130.2 (C2′, C7′), 128.1 (C4″), 127.5 (C5″), 125.5 (C7″), 123.8 (C8″a), 122.8 (C6″), 121.9 (C8″), 118.5 (C3″), 116.6 (C1″a), 115.0 (C5), 113.3 (C3′, C5′), 55.2 (CH3O), 21.7 (CH2). LRMS [ES]+: m/z 412.99 [M + Na]+ (100%). LR MS [ES]−: m/z 388.73 [M − H]− (100%). HRMS [ES]−: m/z calcd for C22H17N2O3S 389.0960 [M − H]−, found 389.0965 (+1.4 ppm).
Synthesis of 6a
To a solution of NaOEt (10 mL, 2 M), prepared by dissolving Na metal in dry ethanol, were added urea (15.6 equiv) and 2-benzoyl-1,2-dihydrocoumarins 5a (200 mg, 1 equiv), and the reaction was heated under reflux for 24 h. After removing the solvent in vacuo, the mixture was dissolved in distilled water, and the products precipitated upon addition of 2 M aqueous HCl. The solid was collected by filtration and purified by recrystallization from ethanol or chloroform.
6-Phenyl-5-[2″-hydroxynaphthyl-(1″)-methyl]-1H-pyrimidine-2,4-dione (6a)
Yielded 40 mg (0.11 mmol, 20%) as a white powder after recrystallization from CHCl3. Mp 292–295 °C. IR (NaCl, thin layer) νmax/cm−1: 3417 and 3105 (OH), 2923 (CH2), 1653 (C=O), 1576 (NH), 1456 (C–N), 1119 (C–O, C=S). 1H NMR (400 MHz, DMSO-d6): δ = 11.33 (br s, 1H, NH), 10.90 (br s, 1H, NH), 9.64 (s, 1H, OH), 7.65 (dd, 1H, 3J = 7.8 Hz, 4J = 1.8 Hz, H-5″), 7.50 (d, 1H, J = 8.8 Hz, H-4″), 7.46–7.35 (m, 3H, H-8, 2 × ArH), 7.35–7.28 (m, 2H, 2 × ArH), 7.19–7.08 (m, 3H, H-7″, H-6″ + 1 × ArH), 6.92 (d, 1H, J = 8.8 Hz, H-3″), 3.90 (s, 2H, CH2). 13C NMR (100 MHz, DMSO-d6): δ = 166.0 (C=O), 152.8 (C2″), 150.3 (C=O), 133.0 (C1′), 132.6 (C8″a), 129.4 (5″), 128.4 (C5′, C3′), 128.2 (C4″a), 128.1, 127.5 (C4″), 125.4 (C7″), 122.8 (C8″), 121.9 (C6″), 118.8 (C3″), 117.3 (C1″a), 109.3 (C5), 21.2 (CH2). LRMS [ES]+: m/z 367.04 [M + Na]+ (100%). LR MS [ES]−: m/z 342.81 [M − H]− (100%). HRMS [ES]−: m/z calcd for C21H16N2O3Na 367.1059 [M − H]−, found 367.1056 (−0.7 ppm).
2-Benzoyl-6a-phenylchromen-2-one (7)
To a solution of 2-hydroxy-5-phenylbenzaldehyde22,23 (220 mg, 1.1 mmol) in ethanol (6 mL) was added ethyl benzoyl acetate 3a (211 mg, 1.1 mmol). Piperidine (5 drops) was added, and the reaction was heated under reflux for 2 h. After cooling to room temperature, the product was collected by filtration and washed with ethanol. The reaction yielded 235 mg (0.72 mmol, 66%) as a white-yellow powder. Mp 166–168 °C. 1H NMR (400 MHz, CDCl3): δ = 8.14 (s, 1H, H-1), 7.93–7.84 (m, 3H, ArH), 7.77 (s, 1H, J = 2.1 Hz, H-7), 7.66–7.56 (m, 3H, ArH), 7.54–7.45 (m, 5H, ArH), 7.45–7.37 (m, 1H, ArH). LRMS [ES]+: m/z 349.07 [M + Na]+ (100%).
6-Phenyl-5-[2″-hydroxybiphenyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6d)
To a stirring solution of 7 (213 mg, 0.6 mmol) in dry pyridine (6 mL) was added NaBH4 (25 mg, 0.65 mmol). The reaction was stirred at room temperature for 2 h. The reaction mixture was poured into cold 2 M hydrochloric acid (20 mL). The resulting white precipitate was collected by filtration, washed with HCl (aq, 2 M), and recrystallized from ethanol. The reaction yielded 200 mg (0.61 mmol, 94%) as a white powder. 1H NMR (400 MHz, CDCl3): δ = 8.00–7.94 (m, 2H, ArH), 7.67–7.60 (m, 1H, ArH), 7.56–7.47 (m, 5H, ArH), 7.47–7.38 (m, 3H, ArH), 7.38–7.30 (m, 1H, ArH), 7.18 (d, 1H, J = 8.4 Hz, ArH), 4.75 (dd, 1H, 3J = 10.5 Hz, 3′J = 6.4 Hz, H-2), 3.60 (dd, 1H, 2J = 16.6 Hz, 3J = 9.6 Hz, H-1), 3.25 (dd, 1H, 2J = 16.6 Hz, 3J = 6.4 Hz, H-1). LRMS [ES]+: m/z 350.12 [M + Na]+ (100%). To a solution of NaOEt (10 mL, 2 M), prepared by dissolving Na metal in dry ethanol, were added thiourea (15.6 equiv) and the white solid (170 mg, 1 equiv), and the reaction was heated under reflux for 24 h. After removing the solvent in vacuo, the residue was dissolved in the minimum amount of distilled water and the product was precipitated upon addition of 2 M aqueous HCl. The solid was collected by filtration. The reaction yielded 80 mg (40%) as a white powder after recrystallization from ethanol. Mp 253–255 °C. IR (NaCl, thin layer) νmax/cm−1: 3417 (OH), 1628 (CO), 1119 (C=S). 1H NMR (400 MHz, DMSO-d6): δ = 12.59 (br s, 1H, NH), 12.46 (br s, 1H, NH), 9.49 (s, 1H, OH), 7.49–7.34 (m, 9H, ArH), 7.31–7.22 (m, 2H, ArH), 6.99 (d, 1H, J = 2.1 Hz, ArH), 6.80 (d, 1H, J = 8.3 Hz, H-3″), 3.41 (s, 2H, CH2). 13C NMR (100 MHz, DMSO-d6): 174.5 (C=S), 161.9 (C=O), 154.5 (C2″), 151.4 (C6), 140.4, 131.6, 130.8, 129.9, 128.8, 128.3, 128.2, 126.2, 126.0, 126.0, 125.2, 115.1, 113.0, 25.3 (CH2). LRMS [ES]−: m/z 385.06 [M − H]− (100%). HRMS [ES]−: m/z calcd for C23H17N2O2S 385.1011 [M − H]−, found 385.1013 (+ 0.6 ppm).
General Protocol for Synthesis of Analogs 6e–j
To a stirring solution of N-substituted thiourea (15.6 equiv) in NaOEt (10, mL, 2 M) was added 5a (1 equiv) in one portion, and the reaction was stirred at reflux for 18 h. After cooling to room temperature, the solvent was removed in vacuo and the remaining residue was dissolved in the minimum amount of distilled water and acidified with 2 M aq HCl. The aqueous layer was extracted with DCM (3 × 50 mL), the organic layers were collected and dried over MgSO4, and the solvent was removed in vaquo. The products were purified by flash column chromatography (EtOAc/hexane) and recrystallized from ethanol.
6-Phenyl-5-[2″-hydroxynaphthyl-(1″)-methyl]-1-methyl-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6e)
Yielded 40 mg (0.10 mmol, 16%) as a white powder after recrystallization from CHCl3. Mp 254–257 °C. IR (KBr) νmax/cm−1: 3348 (OH), 2804 (CH2), 1628 (C=O), 1556 (NH), 1431 (CSNH), 1212, (C=S), 877, 766 and 702 (C–HAr). 1H NMR (400 MHz, DMSO-d6): δ = 12.80 (br s, 1H, NH), 9.22 (br, 1H, OH), 7.65 (d, 1H, J = 7.8 Hz, H-5″), 7.51–7.47 (m, 2H, H-4″, H-8″), 7.30–7.11 (m, 5H, H-6″, H-7″ + 3 × ArH, H-3′, H-4′, H-5′), 6.97 (d, 2H, J = 7.0 Hz, ArH, H-2′, H-6′), 6.85 (d, 1H, J = 8.8 Hz, H-3″), 3.82 (s, 2H, CH2), 3.16 (s, 3H, CH3). 13C NMR (100 MHz, DMSO-d6): δ = 175.2 (C=S), 160.6 (C=O), 152.7 (C2″), 152.0 (C6), 133.1 (C8″a), 132.2 (C1′), 128.9 (C3′,C5′), 128.3 (C2′, C6′), 128.0 (C4′), 127.9 (C5″), 127.7 (C4″a), 127.2 (C4″), 125.5 (C7″), 122.7 (C8″), 121.7 (C6″), 118.5 (C3″), 117.9 (C1″a), 116.2 (C5), 40.2 (CH3), 22.0 (CH2). LRMS [ES]+: m/z 397.08 [M+Na]+ (100%); LRMS [ES]−: m/z 373.05 [M − H]− (100%); HRMS [ES]−: m/z calcd for C22H17N2O2S 373.1011 [M − H]−, found 373.1010 (−0.2 ppm).
6-Phenyl-5-[2″-hydroxynaphthyl-(1″)-methyl]-1-ethyl-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6f)
Yielded 30 mg (0.07 mmol, 20%) as a white powder after column chromatography (EtOAc/hexane, 1:4) and recrystallization from ethanol. Mp 256–258 °C. IR (KBr) ν max/cm−1: 3676, 2988, and 2972 (OH), 2902 (CH2, CH3), 1633 (C=O), 1437 (C–N), 1394 and 1241 (OH), 1217 and 1103 (C=S), 1076, 1057, and 1028 (C–O), 812, 767, and 741 (C–HAr). 1H NMR (DMSO-d6, 400 MHz): δ = 12.75 (br, 1H, NH), 9.20 (br, 1H, OH), 7.65 (d, 1H, J = 7.1 Hz, H-5″), 7.51–7.45 (m, 2H, H-4″, H-8″), 7.27–7.17 (m, 5H, H-6″, H-7″, H-3′, H-4′, H-5′), 6.98 (d, 2H, J = 7.1 Hz, H-2′, H-6′), 6.84 (d, 1H, J = 8.6 Hz, H-3″), 3.86 (br, 2H, CH2, H-1‴), 3.77 (s, 2H, CH2, H-1″), 0.92 (t, 3H, J = 7.0 Hz, CH3). 13C NMR (DMSO-d6, 100 MHz): 175.5 (C=S), 161.4 (C=O), 153.6 (C2″), 152.6 (C6), 132.7 (C8″a), 131.6 (C1′), 129.1 (C4′), 128.9 (C3′, C5′), 128.9 (C4″a), 128.3 (C2′, C6′), 127.9 (C5″), 127.2 (C4″), 126.5 (C7″), 123.6 (C8″), 122.8 (C6″), 118.9 (C3″), 117.2 (C1″a), 116.1 (C5), 46.9 (C1‴), 23.0 (C1″), 13.7 (C2‴). LRMS [ES]+: m/z 411.33 [M + Na]+ (100%). LRMS [ES]−: m/z 387.16 [M − H]− (100%). HRMS [ES]+: m/z calcd for C23H20N2O2SNa 411.1143 [M + Na]+, found 411.1149 (+1.3 ppm).
6-Phenyl-5-[2″-hydroxynaphthyl-(1″)-methyl]-1-allyl-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6g)
Yielded 20 mg (0.05 mmol, 15%) as a white powder after column chromatography (EtOAc/hexane, 1:4) and recrystallization from ethanol. Mp 175–177 °C. IR (KBr) ν max/cm−1: 3676 and 2988 (OH), 2902 (CH2), 1626 (C=O), 1479 (CSNH), 1406, 1394, and 1242 (OH), 1075, 1057, and 1028 (C–O), 892, 823, and 747 (C–HAr), 701 (C–H). 1H NMR (DMSO-d6, 300 MHz): δ = 12.82 (br s, 1H, NH), 9.22 (br, 1H, OH), 7.65 (d, 1H, J = 7.1 Hz, H-5″), 7.52–7.43 (m, 2H, H-4″, H-8″), 7.28–7.18 (m, 5H, H-6″, H-7″, H-3′, H-4′, H-5′), 6.93 (d, 2H, J = 7.0, H-2′, H-6′), 6.85 (d, 1H, J = 8.8 Hz, H-3″), 5.61–5.50 (m, 1H, H-2‴), 4.98 (dd, 1H, 3J = 10.5 Hz, 4J = 1.2 Hz, H-3‴), 4.64 (dd, 1H, 3J = 17.3 Hz, 4J = 1.2 Hz, H-3‴), 4.50 (m, 2H, H-1‴), 3.78 (s, 2H, H-1″). 13C NMR (DMSO-d6, 100 MHz): δ = 174.8 (C=S), 160.7 (C=O), 152.7 (C2″), 151.7 (C6), 133.0 (C8″a), 131.9 (C2‴), 131.4 (C1′), 128.9 (C3′, C5′), 128.4 (C2′, C6′), 128.1 (C5″), 128.0 (C4′), 127.8 (C4″a), 127.2 (C4″), 125.5 (C7″), 122.5 (C8″), 121.7 (C6″), 117.8 (C3″), 117.1 (C3‴), 116.5 (C1″a), 116.1 (C5), 52.5 (C1‴), 22.19 (C1″). LRMS [ES]+: m/z 423.12 [M + Na]+ (100%). HRMS [ES]+: m/z calcd for C24H20N2O2NaS 423.1143 [M + Na]+, found 423.1142 (−0.3 ppm).
6-Phenyl-5-[2″-hydroxynaphthyl-(1″)-methyl]-1-propyl-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6h)
Yielded 16 mg (0.04 mmol, 10%) as a white powder after column chromatography (EtOAc/hexane, 1:4) and recrystallization from ethanol. Mp 227–229 °C. IR (KBr) ν max/cm−1: 3676 and 2988 (OH), 2969, and 2902 (CH3, CH2), 1630 (C=O), 1482 (CSNH), 1432 (C–N), 1206, 1131, and 1108 (C=S), 1075, 1066, and 1057 (C–O), 880, 820, and 767 (C–HAr), 745, 701, and 666 (C–H). 1H NMR (DMSO-d6, 400 MHz): δ = 12.77 (br s, 1H, NH), 9.21 (s, 1H, OH), 7.65 (d, 1H, J = 8.8 Hz, H-5″), 7.55–7.45 (m, 2H, H-4″, H-8″), 7.29–7.12 (m, 5H, H-6″, H-7″, H-3′, H-4′, H-5′), 6.97 (d, 2H, J = 7.0 Hz, H-2′, H-6′), 6.84 (d, 1H, J = 8.8 Hz, H-3″), 3.78 (s, 2H, H-1″), 3.69 (br, 2H, H-1‴), 1.41 (m, 2H, H-2‴), 0.43 (t, 3H, J = 7.4 Hz, H-3‴). 13C NMR (DMSO-d6, 100 MHz): δ = 174.8 (C=S), 160.4 (C=O), 152.7 (C2″), 151.7 (C6), 133.1 (C8″a), 131.7 (C1′), 129.3 (C4′), 128.9 (C5′, C3′), 128.6 (C2′, C6′), 128.1 (C5″), 128.0 (C4″), 127.8 (C4″a), 125.6 (C7″), 122.6 (C8″), 121.8 (C6″), 119.0 (C1″a), 117.9 (C3″), 116.2 (C5), 52.1 (C1‴), 22.1 (C1″), 20.3 (C2‴), 10.5 (C3‴). HRMS [ES]+: m/z calcd for C24H22N2O2NaS 425.1300 [M + Na]+, found 425.1307 (+0.2 ppm).
6-Phenyl-5-[2″-hydroxynaphthalyl-(1″)-methyl]-1-butyl-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6j)
Yielded 29 mg (0.07 mmol, 7%) as a white powder after column chromatography (EtOAc/hexane 1:9) and recrystallization from ethanol. Mp 201–203 °C. IR (KBr) ν max/cm−1: 3667 and 3213 (OH), 2972 and 2902 (CH3, CH2), 1645 (C=O), 1487 (CSNH), 1428 (C–N), 1394 and 1233 (OH), 1204, 1179, 1129, and 1104 (C=S), 1067 and 1057 (C–O), 823 and 747 (C–HAr), 705 and 756 (C–H). 1H NMR (DMSO-d6, 400 MHz): δ = 12.75 (s, 1H, NH), 9.20 (s, 1H, OH), 7.65 (d, 1H, J = 8.2 Hz, H-5″), 7.51–7.45 (m, 2H, H-8″, H-4″), 7.24–7.14 (m, 5H, H-7″, H-6″, H-3′, H-4′, H-5′), 6.98 (d, 2H, J = 7.0 Hz, H-2′, H-6′), 6.84 (d, 1H, J = 8.7 Hz, H-3″), 3.79 (s overlapped with br, 4H, H-1″, H-1‴), 1.40 (br, 2H, H-2‴), 0.90–0.80 (m, 2H, H-3‴), 0.50 (t, 3H, J = 7.3 Hz, H-4‴). 13C NMR (DMSO-d6, 100 MHz): δ = 174.6 (C=S), 160.4 (C=O), 152.7 (C2″), 151.7 (C6), 133.1 (C8″a), 131.6 (C1′), 128.9 (C3′, C5′), 128.1 (C2′, C6′), 128.0 (C4′), 128.0 (C5″), 127.9 (C4″a), 127.6 (C4″), 125.6 (C7″), 122.6 (C8″), 121.8 (C6″), 119.1 (C3″), 117.9 (C1″a), 116.2 (C5), 50.3 (C1‴), 28.6 (C2‴), 22.0 (C1″), 18.9 (C3‴), 12.9 (C4‴). LRMS [ES]+: m/z 439.05 [M + Na]+ (100%). LRMS [ES]−: m/z 415.16 [M − H]− (100%). HRMS [ES]+: m/z calcd for C25H24N2O2SNa 439.1456 [M + Na]+, found 439.1473 (+3.8 ppm).
General Procedure for the Parallel Synthesis of Analogs 6i–6xi.
Thiourea (15.6 equiv) and intermediates 5i–xv (100 mg, 1 equiv) were mixed into different vessels of the parallel synthesis apparatus. 2 M NaOEt stock solution (5 mL), previously prepared by dissolving Na metal in dry ethanol, was added to each reaction vessel and the reaction was heated under reflux for 18 h. After removing the solvent in vacuo, the crude reaction mixtures were dissolved in the minimum amount of distilled water and the products precipitated upon addition of 2 M aqueous HCl. The solids were collected by filtration and purified by column chromatography (EtOAc/hexane) and recrystallized from CHCl3. Reactions using 5xii–5xv were unsuccessful.
6-(2′-Methylphenyl)-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6i)
Yielded 115 mg (0.30 mmol, 57%) as a white powder after column chromatography (EtOAc/hexane, 1:4) and recrystallization from CHCl3. Mp 170–172 °C. IR (KBr) νmax/cm−1: 2988 (OH), 2902 (CH3, CH2), 1627 (C=O), 1554 (NH), 1453 (CSNH), 1394 and 1255 (OH), 1211 (C=S), 1075 and 1057 (C–O), 819, 808, and 744 (C–HAr), 729 (C–H). 1H NMR (DMSO-d6, 300 MHz): δ = 12.60 (br s, 1H, NH), 12.17 (br s, 1H, NH), 9.21 (br s, 1H, OH), 7.63 (d, 1H, J = 8.1 Hz, H-5″), 7.43 (d, 1H, J = 8.8 Hz, H-4″), 7.39 (d, 1H, J = 7.4 Hz, H-8″), 7.26–7.08 (m, 3H, H-6″, H-7″ + ArH), 7.03–6.99 (m, 2H, ArH), 6.90 (d, 1H, J = 7.1 Hz, ArH), 6.78 (d, 1H, J = 8.8 Hz, H-3″), 3.88 (q, 2H, CH2), 1.62 (s, 3H, CH3). 13C NMR (100 MHz, DMSO-d6): δ = 173.3 (C=S), 162.4 (C=O), 152.6 (C2′), 150.0 (C6), 135.6 (C2′), 132.9 (C1′), 131.0 (C8″a), 129.2 (CAr), 129.0 (CAr), 128.4 (C5″), 128.0 (C4″), 127.8 (C4″a), 127.2 (CAr), 125.4 (C7″), 125.1 (CAr), 122.6 (C8″), 121.8 (C6″), 117.8 (C3″), 116.0 (C5), 115.9 (C1″a), 20.1 (CH2), 18.36 (CH3). LRMS [ES]+: m/z 397.12 [M + Na]+ (100%). HRMS [ES]+: m/z calcd for C20H18N2O2NaS 397.0987 [M + Na]+, found 397.0987 (+0.1 ppm).
6-(3′-Methylphenyl)-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6ii)
Yielded 54 mg (0.14 mmol, 51%) as a white powder after column chromatography (EtOAc/hexane, 1:4) and recrystallization from CHCl3. Mp 235–237 °C. IR (KBr) νmax/cm−1: 3767 and 2988 (OH), 2902 (CH3, CH2), 1646 (C=O), 1563 (NH), 1465 (CSNH), 1403, 1394, and 1233 (OH), 1211, 1198, and 1128 (C=S), 1066, 1044, and 1017 (C–O), 993 (C–H), 823 and 747 (C–HAr). 1H NMR (DMSO-d6, 300 MHz): δ = 12.55 (br s, 1H, NH), 12.22 (br s, 1H, NH), 9.41 (br s, 1H, OH), 7.65 (d, 1H, J = 7.9 Hz, H-5″), 7.47 (d, 1H, J = 8.8 Hz, H-4″), 7.36 (d, 1H, J = 8.5 Hz, H-8″), 7.26–7.09 (m, 4H, H-7″, H-6″ + 2 × ArH), 7.00–6.96 (m, 1H, ArH), 6.93–6.85 (m, 2H, H-3″ + 1 × ArH), 3.91 (s, 2H, CH2), 2.18 (s, 3H, CH3). 13C NMR (100 MHz, DMSO-d6): δ = 173.8 (CdS), 162.4 (C=O), 152.7 (C2″), 150.9 (C6), 136.9 (C3′), 133.0 (C8″a), 131.5 (C1′), 129.9 (C2′), 128.9 (CAr), 128.3 (C4″a), 128.0 (C4″), 127.6 (C5″), 127.4 (CAr), 126.2 (CAr), 125.4 (C7″), 122.7 (C8″), 121.8 (C6″), 118.2 (C3″), 116.5 (C1″a), 115.0 (C5), 21.3 (CH2), 20.2 (CH3). LRMS [ES]+: m/z 397.10 [M + Na]+ (100%). HRMS [ES]+: m/z calcd for C20H18N2O2NaS 397.0987 [M + Na]+, found 397.0988 (+0.1 ppm).
6-(4′-Methylphenyl)-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6iii)
Yielded 56 mg (0.15 mmol, 50%) as a white powder after column chromatography (EtOAc/hexane, 1:4) and recrystallization from CHCl3. Mp 220–221 °C. IR (KBr) νmax/cm−1: 3663 and 2988 (OH), 2902 (CH3, CH2), 1630 (C=dO), 1540 (NH), 1515 (CSNH), 1445 (C–N), 1405, 1349, and 1250 (OH), 1226 and 1208 (C=S), 1066 and 1028 (C–O), 818, 744, and 729 (C–HAr). 1H NMR (DMSO-d6, 300 MHz): δ = 12.52 (br s, 1H, NH), 12.26 (br s, 1H, NH), 9.46 (br s, 1H, OH), 7.65 (d, 1H, J = 7.4 Hz, H-5″), 7.50 (d, 1H, J = 8.7 Hz, H-4″), 7.35 (d, 1H, 3J = 7.7 Hz, H-8″), 7.22–7.08 (m, 6H, H-7″, H-6″ + 4 × ArH), 6.92 (d, 1H, J = 8.7 Hz, H-3″), 3.89 (s, 2H, CH2), 2.29 (s, 3H, CH3). 13C NMR (100 MHz, DMSO-d6): δ = 173.8 (C=S), 162.2 (C=O), 152.7 (C2″), 150.1 (C6), 139.1 (C4′), 133.0 (C8″a), 128.7 (C4″a), 128.5 (CAr), 128.4 (CAr), 128.0 (C5″), 127.4 (C4″), 125.7 (C7″), 122.8 (C8″), 121.8 (C6″), 118.4 (C3″), 116.5 (C1″a), 115.1 (C5), 21.1 (CH2), 20.9 (CH3). LRMS [ES]+: m/z 397.09 [M + Na]+ (100%). HRMS [ES]+: m/z calcd for C20H18N2O2NaS 397.0987 [M + Na]+, found 397.0989 (+0.1 ppm).
6-(4′-Chlorophenyl)-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6iv)
Yielded 20 mg (0.05 mmol, 18%) as a white powder after column chromatography (EtOAc/hexane, 1:2) and recrystallization from CHCl3. Mp 287–289 °C. IR (KBr) νmax/cm−1: 3277 and 3030 (OH), 2941 (CH2), 1702 and 1633 (C=O), 1545 (NH), 1448 (C–N), 1219 and 1204 (C=S, CSNH), 1114, 1127, and 1090 (C=S), 1090 and 1014 (C–O), 837 and 820 (C–HAr). 1H NMR (400 MHz, DMSO-d6): δ = 12.57 (br s, 1H, NH), 12.23 (br s, 1H, NH), 9.38 (s, 1H, OH), 7.64 (d, 1H, J = 7.5 Hz, H-5″), 7.49–7.41 (m, 2H, H-4″, H-8″), 7.30–7.22 (m, 1H, H-7″), 7.21–7.14 (m, 3H, H-6″ overlapped with AA′BB′ system, J = 8.3 Hz, H-3″, H-5″), 7.04 (d, 2H, J ) 8.3 Hz, AA′BB′ system, H-2′, H-6′), 6.85 (d, 1H, J = 8.7 Hz, H-3″), 3.91 (s, 2H, CH2). 13C NMR (100 MHz, DMSO-d6): δ = 174.2 (C=S), 160.6 (C=O), 152.7 (C2″), 151.6 (C6), 133.8 (C4′), 133.0 (C8″a), 132.9 (C1′), 130.4 (C3′,C5′), 128.4 (C5″), 127.9 (C2′,C6′), 127.9 (C4″a), 127.8 (C4″), 125.9 (C7″), 123.1 (C8″), 122.3 (C6″), 118.4 (C3″), 116.8 (C1″a), 115.9 (C5), 20.9 (CH2). LRMS [ES]+: m/z 416.97 [M – H]+ (100%). HRMS [ES]+: m/z calcd for C21H15N2O2NaSCl 417.0440 [M + Na]+, found 417.0443 (+3.1 ppm).
6-(4′-Iodophenyl)-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydropyrimidin-1H-4-one (6v)
Yielded 13 mg (0.02 mmol, 22%) as a white powder after column chromatography (EtOAc/ hexane, 1:2) and recrystallization from CHCl3. Mp > 250 °C (decomposes). IR (KBr) νmax/cm−1: 3090 (OH), 1628 (C=O), 1560 (NH), 1427 (C–N), 1208 (C=S, CSNH), 1154 and 1126 (C–O, C=S), 1008 (C–O), 850, 814, and 754 (C–HAr). 1H NMR (400 MHz, DMSO-d6): δ = 12.55 (br s, 1H, NH), 12.21 (br s, 1H, NH), 9.38 (s, 1H, OH), 7.64 (d, 1H, J = 8.4 Hz, H-5″), 7.52–7.39 (m, 4H, H-4″, H-8″, AA′BB′ system, J = 8.4 Hz, H-2′, H-6′), 7.28–7.15 (m, 2H, H-7″, H-6″), 6.86–6.83 (m, 3H, H-3″,AA′BB′ system, J = 8.4 Hz, H-3′, H-5′), 3.88 (s, 2H, CH2). LRMS [ES]+: m/z 508.02 [M + Na]+ (100%).
6-(4′-Trifluoromethylphenyl)-5-[2″-hydroxynaphthyl-(1″)methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6vi)
Yielded 33 mg (0.077 mmol, 25%) as a white powder after column chromatography (EtOAc/hexane, 1:2) and recrystallization from CHCl3. Mp > 250 °C (decomposes). IR (KBr) νmax/cm−1: 2970 (OH), 2912 (CH2), 1691, 1549, and 1428 (C–N), 1635 (C=O), 1515 (CSNH), 1409 (OH), 1321, 1286 and 1167 (C–F), 1131 and 1112 (C=S), 1065 and 1017 (C–O), 860 and 702 (C–H), 821, 778, and 743 (C–HAr). 1H NMR (400 MHz, DMSO-d6): δ = 12.67 (br s, 1H, NH), 12.28 (br s, 1H, NH), 9.40 (br s, 1H, OH), 7.65 (d, 1H, J = 8.3, H-5″), 7.52–7.35 (m, 4H, H-4″, and H-8″ overlapped with AA′BB′ system, J = 8.0 Hz, H-2′, H-6′), 7.17 (m, 1H, H-7″), 7.17–7.12 (m, 3H, H-6″ overlapped with AA′BB′ system, J = 8.0 Hz, H-3′, H-5′), 6.80 (d, 1H, J = 8.8 Hz, H-3″), 3.95 (s, 2H, CH2 13C NMR (100 MHz, DMSO-d6): δ = 173.9 (C=S), 161.9 (C=O), 152.2 (C2″), 148.8 (C6), 135.3 (C1′), 132.9 (C8″a), 129.3 (C4′), 128.8 (C3′, C5′), 128.0 (C4″), 127.9 (C5″), 127.8 (C4″a), 125.6 (C7″), 124.1 (C2′, C6′), 122.6 (C6″), 121.9 (C8″), 119.8 (CF3), 117.6 (C3″), 116.2 (C1″a), 116.0 (C5), 20.54 (CH2). LRMS [ES]+: m/z 451.05 [M + Na]+ (100%).
6-(2′-Bromophenyl)-5-[2″-hydroxynaphthyl-(1″-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6vii)
Yielded 30 mg (0.07 mmol, 20%) after purification by column chromatography and recrystallization from CHCl3. Mp 224–226 °C. IR (KBr) νmax/cm−1: 3055 (OH), 2927 (CH2), 1627 (C=O), 1548 (N–H), 1436 (C–N), 1196 (C=S, CSNH), 1132 and 1117 (C–O, C=S), 1027 (C–O), 813 and 740 (C–HAr), 678 (C–Br). 1H NMR (acetone-d6, 300 MHz): δ = 11.61 (br, 1H, NH), 11.23 (br, 1H, NH), 8.93 (br, 1H, OH), 7.74–7.60 (m, 1H, H-5′), 7.61–7.52 (m, 2H, H-4″, H-8″), 7.38–7.11 (m, 6H, H-6″, H-7″ + 4 × ArH), 6.88 (d, 1H, J = 8.8 Hz, H-3″), 4.13 (d, 1H, AB system, J = 15.6 Hz, CH2), 3.98 (d, 1H, AB system, J = 15.6 Hz, CH2). 13C NMR (DMSO-d6, 100 MHz): 175.3 (C=S), 164.5 (C=O), 154.6 (C2″), 150.6 (C6), 134.4 (C1′), 133.4 (C8″a), 133.6 (CAr), 131.9 (CAr), 132.5 (CAr), 129.9 (C4″a), 129.1 (C4″), 129.0 (CAr), 128.3 (C5″), 126.7 (C7″), 123.6 (C8″), 123.3 (C2′), 123.1 (C6″), 119.7 (C3″), 117.0 (C1″a), 117.0 (C5), 21.4 (CH2). LRMS [ES]+: m/z 460.92, 462.92 [M + Na]+. LRMS [ES]−: m/z 437.01, 439.01 [M – H]−. HRMS [ES]+: m/z calcd for C21H15N2O2SNa79Br 460.9935 [M + Na]+, found 460.9941 (+ 1.3 ppm); m/z calcd for C21 H15N2O2SNa81Br 462.9915 [M + Na] , found 462.9903 (−2.6 ppm).
6-(3′-Bromophenyl)-5-[2″-hydroxynaphthyl-(1″-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6viii)
Yielded 15 mg (0.07 mmol, 26%) as a white powder after column chromatography (EtOAc/hexane, 1:2) and recrystallization from CHCl3. Mp 237–239 °C. IR (KBr) νmax/cm−1: 3114 and 3081 (OH), 2935 (CH2), 1713 and 1629 (C=O), 1561 (NH), 1462 and 1439 (C–N), 1199 (C=S, CSNH), 1119 (C–O, C=S), 820, 806, and 746 (C–HAr), 697 (C–Br). 1H NMR (400 MHz, DMSO-d6): δ = 12.58 (br s, 1H, NH), 12.21 (br s, 1H, NH), 9.36 (s, 1H, OH), 7.63 (d, 1H, J = 7.7 Hz, H-5″), 7.51–7.47 (m, 2H, H-4″, H-8″), 7.40–7.35 (m, 1H, ArH), 7.31–7.25 (m, 1H, H-7″), 7.20–7.15 (m, 1H, H-6″), 7.12–6.99 (m, 3H, ArH), 6.85 (d, 1H, J = 8.8 Hz, H-3″), 3.91 (s, 2H, CH2). 13C NMR (100 MHz, acetone-d6): δ = 175.3 (C=S), 164.7 (C=O), 154.3 (C2″), 150.5 (C6), 134.8 (C1′), 134.3 (C8″a), 134.0 (CAr), 132.7 (CAr), 131.2 (CAr), 130.0 (C4″a), 129.3 (CAr), 129.2 (C4″), 128.8 (C5″), 126.6 (C7″), 123.8 (C8″), 123.3 (C6″), 122.7 (C3′), 120.3 (C3″), 117.9 (C1″a), 116.0 (C5), 22.2 (CH2). LRMS [ES]−: m/z 436.99 [M – H]−, 439.00 [M – H]−. HRMS [ES]−: m/z calcd for C21H14N2O2S79Br 436.9959 [M – H]−, found 436.9966 (+1.5 ppm); m/z calcd for C21H14N2O2S81Br 438.9939 [M – H]−, found 438.9948 (+2.1 ppm).
6-(4′-Chlorophenyl)-5-[2″-hydroxynaphthyl-(1″)-ylmethyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6ix)
Yielded 18 mg (0.004 mmol, 32%) as a white powder after column chromatography (EtOAc/hexane, 1:2) and recrystallization from CHCl3. Mp 229–232 °C. IR (KBr) ν max/cm−1: 3086 (OH), 2941 (CH2), 2363, 1635 (C=O), 1556 (NH), 1439 (C–N), 1257 and 1213 (C=S, CSNH), 1129 (C–O), 1041 and 1027 (C–O), 819, 750 (C–HAr). 1H NMR (acetone-d6, 400 MHz): δ = 11.59 (br s, 1H, NH), 11.22 (br s, 1H, NH), 9.25 (s, 1H, OH), 7.68 (d, 1H, J = 7.6 Hz, H-5″), 7.57 (d, 1H, J = 8.8 Hz, H-4″), 7.53–7.40 (m, 4H, H-8″ + 3 × ArH), 7.23–7.08 (m, 3H, H-7″, H-6″ + 1 × ArH), 7.00 (d, 1H, J = 8.8 Hz, H-3″), 3.92 (s, 2H, CH2). 13C NMR (acetone-d6, 100 MHz): δ = 175.0 (C=S), 164.7 (C=O), 154.3 (C2″), 150.6 (C6), 134.7 (C3′), 134.5 (C1′), 133.5 (C8″a), 131.1 (CAr), 131.0 (CAr), 130.0 (C4″a), 129.9 (CAr), 129.3 (C4″), 129.2 (C5″), 128.3 (CAr), 126.6 (C7″), 123.8 (C8″), 123.3 (C6″), 120.1 (C3″), 117.8 (C1″), 116.0 (C5), 22.1 (CH2). LRMS [ES]−: m/z 393.12 [M – H]− (100%). LRMS [ES]+: 417.10 [M + Na]+ (100%). HRMS [ES]−: m/z calcd for C21H14N2O2SCl 393.0465 [M – H]−, found 393.0461 (−0.9 ppm).
6-(2′-Fluorophenyl)-5-[2″-hydroxynaphthyl-(1″)methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6x)
Yielded 17 mg (0.10mmol, 10%) as a white powder after column chromatography (EtOAc/hexane, 1:2) and recrystallization from CHCl3. Mp 197 °C (decomposes). IR (KBr) νmax/cm−1: 3316 and 2935 (OH), 2885 (CH2), 1630 (C=O), 1558 (NH), 1439 (C–N), 1269 and 1216(C–F), 1131 (C–O, C=S), 816 and 751 (C–HAr). 1H NMR (400 MHz, DMSO-d6): δ = 12.61 (br, 1H, NH), 12.35 (br, 1H, NH), 9.27 (s, 1H, OH), 7.63 (d, 1H, J = 7.3 Hz, H-5″), 7.53–7.43 (m,2H, H-4″ + 1 × ArH), 7.30–7.22 (m, 2H, H-7″ + 1 × ArH), 7.17–7.12 (m, 1H, H-6″), 7.13–7.05 (m, 1H, ArH), 7.00–6.92 (m, 2H, ArH), 6.82 (d, 1H, J = 8.8 Hz, H-3″), 3.90 (s, 2H, CH2). 13C NMR (100 MHz, DMSO-d6): δ = 173.3 (C=S), 161.9 (C=O), 159.9 (C2′), 157.4 (C6), 152.7 (C2″), 132.9 (C8″a), 131.5 (C6′), 130.3 (C4′), 128.0 (C5″), 127.8 (C4″a), 127.4 (C4″), 125.4 (C7″), 123.7 (C5′), 122.6 (C8″), 121.8 (C6″), 121.7 (C1′), 117.7 (C3″), 117.2 (C5), 115.8 (C3′), 115.1 (C1″a), 20.5 (CH2). LRMS [ES]−: m/z 377.14 [M – H]− (100%). LRMS [ES]+: m/z 401.10 [M + Na]+ (100%). HRMS [CI]+: m/z calcd for C21H16N2O2SF 379.0917 [M + H]+, found 379.0911 (−1.5 ppm).
6-(3′-Fluorophenyl)-5-[2″-hydroxynaphthyl-(1″)-methyl]-2-thioxo-2,3-dihydro-1H-pyrimidin-4-one (6xi)
Yielded 18 mg (0.10 mmol, 10%) as a white powder after column chromatography (EtOAc/hexane, 1:2) and recrystallization from CHCl3. Mp 150 °C (decomposes). IR (KBr) νmax/cm−1: 3378, 3126, 3059 (OH), 2941 (CH2), 1630 (C=O), 1554 (NH), 1439 (C–N), 1268 (C–F), 1199 (C=S, CSNH), 1158 and 1119 (C–O, C=S), 821, 792, and 746 (C–HAr). 1H NMR (400 MHz, DMSO-d6): δ = 12.58 (br s, 1H, NH), 12.24 (br s, 1H, NH), 9.41 (s, 1H, OH), 7.64 (d, 1H, J = 7.5 Hz, H-5″), 7.48–7.45 (m, 2H, H-4″, H-8″), 7.30–6.99 (m, 4H, H-7″, H-6″ + 2 × ArH), 6.93–6.68 (m, 3H, H-3″ + 2 × ArH), 3.92 (s, 2H, CH2). 13C NMR (100 MHz, DMSO-d6): δ × = 173.8 (C=S), 162.6 (C3′), 162.1 (C=O), 159.5 (C6), 152.7 (C2″), 134.1 (C1′), 133.0 (C8″a), 129.7 (CAr), 128.1 (C5″), 128.0 (C4″a), 127.5 (C4″), 125.6 (C7″), 124.4 (C6′), 122.8 (C8″), 121.9 (C6″), 117.9 (C3″), 116.3 (C1″a), 116.0 (CAr), 115.7 (C5), 115.4 (CAr), 20.8 (CH2). HRMS [ES]+: m/z calcd for C21H15N2O2NaSF 401.0736 [M + Na]+, found 401.0735 (−0.2 ppm).
Molecular Docking Studies
The SIRT2 structure (PDB ID: 1J8F) was used for the docking studies, which were performed using the program GOLD.12 Hydrogens were added to the protein structure using the program Reduce,24 and small molecule structures were generated using the PRODRG server.25 A homology model of SIRT1 was generated using the PHYRE server. All visualization and analysis was performed using Pymol.26
Biology Protocols. SIRT1 and SIRT2 Inhibition Assay
Compounds were tested for inhibition of SIRT1 and SIRT2 using the human recombinant SIRT1 and SIRT2 enzymes provided with the Fluor de Lys fluorescence-based assay kit (AK555, AK556, BIOMOL, Plymouth Meeting, U.S.A.). All the other required reagents were provided with the kit. All kit components were stored at −78 °C for highest stability. Positive controls were provided by the known SIRT1/T2 inhibitors tenovin-63 and cambinol.5 Fresh dilutions of cambinol analogues were prepared in DMSO (Aldrich), added to the assay buffer, and pipeted (10 μL) into a 96 well white microplate. Enzyme (15 μL, 0.07 U/μL for SIRT1, 0.03 U/μL for SIRT2) and Fluor de Lys SIRT1 or SIRT2 (15 μM) plus NAD+ (1 mM) in assay buffer were added. After incubating for 1 h at 37 °C, a developer solution (50 μL developer solution added to each well, developer solution contains 38 μL buffer, 10 μL developer and 2 μL nicotinamide) was added (50 μL) to each well and the microplate was incubated for a further 45 min at room temperature. Plates were read in a Spectra max Gemini XS fluorimeter with an excitation wavelength of 355 nm and an emission wavelength of 460 nm.
Cell Culture and Western Blotting
Human cancer cell lines MCF-7 were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, U.K.), and H1299 cells were cultured in RPMI. Both were supplemented with 10% fetal calf serum (FCS, Hyclone, U.K.) and gentamycin (complete medium). Cells were seeded at a concentration of 2 × 105 (MCF-7) and 6 × 104 (H1299) in 6 well collagen precoated plates (TPP, Helena Biosciences, U.K.) and incubated in a humidified atmosphere containing 5% CO2–95% air at 37 °C for 42 h (MCF-7) and 24 h (H1299). Different concentrations of target compounds in DMSO were added to MCF-7, and they were incubated for a further 6 h. Different concentrations of target compounds in DMSO were also added to H1299 cells, with 40 nM trichostatin (TSA) also added and incubated for a further 24 h. After lysing the cells with 1 × LDS sample buffer (100 μL per well for MCF-7 and 200 μL for× H1299) (Invitrogen, U.K.), the protein concentration was assessed with a BCA protein assay kit (Pierce, U.K.) and the concentration of protein equalized with 1 × LDS sample buffer (the total amount of protein loaded in gels in Figures S14 and S15 was 2.0 μg; Figure S16, 2.3 μg). Proteins were separated with 4–12% bis-tris gels (Invitrogen, USA) and electrophoretically transferred to PVDF transfer membranes (Millipore, U.K.). Membranes were blocked with Marvel nonfat milk (45 min, 5% solution in PBS/0.1% tween) and immunoblotted using a rabbit polyclonal antibody against p53 acetylated at K382 (Biolegend, U.K.) and DO.1 (anti total p53, in-house produced) for the MCF-7 membranes. PC-10 mouse monoclonal antibody (in house produced) was used to detect PCNA as a loading control. Anti-K40 acetylated tubulin (SIGMA) and anti α-tubulin (SIGMA) were used for the H1299 membranes. All the primary antibodies were diluted in Marvel nonfat milk (5% solution in PBS/0.1% tween). The secondary antibody used against antiacetylated p53 was a HRP-tagged polyclonal swine antirabbit IgG (DAKO, U.K.). The secondary antibody used against the remaining primary antibodies was a HRP-tagged polyclonal rabbit antimouse IgG (DAKO, U.K.). After incubation with primary (1 h) and secondary antibodies (45 min), bound antibody was visualized with enhanced chemiluminescence (ECL) Western blotting developer (Amersham, U.K.) in a darkroom.
Supplementary Material
Acknowledgment
We are grateful to support from Cancer Research UK (funding for F.M., S.L., M.H., and J.C.) and the Royal Society (University Research Fellowship for N.J.W.). We would like to thank Tomas Lebl for NMR interpretation and Chris Lawson for help in determining the purity of the analogs using HPLC techniques.
Footnotes
Abbreviations: SIRT1, sirtuin 1; SIRT2, sirtuin 2; HDAC, histone deacetylase; HMBC, heteronuclear multiple-bond correlation.
Supporting Information Available: Detailed experimental procedures for the parallel synthesis and characterization of all intermediates along with HPLC analysis of all analogs of 1; selected NMR spectra; small molecule X-ray crystal structure analysis of 1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Vousden KH, Lane DP. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 2.Berkson RG, Hollick JJ, Westwood NJ, Woods JA, Lane DP, Lain S. Pilot screening programme for small molecule activators of p53. Int. J. Cancer. 2005;115:701–710. doi: 10.1002/ijc.20968. [DOI] [PubMed] [Google Scholar]
- 3.Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy AR, Appleyard V, Murray KE, Baker L, Thompson A, Mathers J, Holland SJ, Stark MJ, Pass G, Woods J, Lane DP, Westwood NJ. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:377–388. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]; (b) Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]; (d) Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]; (f) Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, Bedalov A. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 6.(a) Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]; (b) North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]; (c) Suzuki K, Koike T. Mammalian Sir2-related protein (SIRT) 2-mediated modulation of resistance to axonal degeneration in slow Wallerian degeneration mice: A crucial role of tubulin deacetylation. Neuroscience. 2007;147(3):599–612. doi: 10.1016/j.neuroscience.2007.04.059. [DOI] [PubMed] [Google Scholar]
- 7.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 8.Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. USA. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whamhoff H, Korte F. Heterocycles by capture reactions of opened acyllactones. IV. A simple synthesis of 5H-1benzopyrano and 12H-naphtho[1′,2′:5′,6′]pyrano[2,3-d]pyrimidines. Chem. Ber. 1967;100:1324–1330. [Google Scholar]
- 10.Kadin SB. Reduction of conjugated double bonds with Sodium Borohydride. J. Org. Chem. 1966;31:620–622. [Google Scholar]
- 11.See Supporting Information.
- 12.GOLD v. 3.0. Cambridge Crystallographic Data Centre; Cambridge, U.K.: http://www.ccdc.cam.ac.uk. [Google Scholar]
- 13.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 2001;8:621–625. doi: 10.1038/89668. PDB ID: 1J8F. [DOI] [PubMed] [Google Scholar]
- 14.Zhao K, Chai X, Clements A, Marmorstein R. Structure and autoregulation of the Yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 2003;10:864–871. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- 15.Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of sir2 enzymes. Mol. Cell. 2004;13:639–648. doi: 10.1016/s1097-2765(04)00082-6. PDB ID: 1S7G. [DOI] [PubMed] [Google Scholar]
- 16.Neugebauer RC, Uchiechowska U, Meier R, Hruby H, Valkov V, Verdin E, Sippl W, Jung M. Structure-activity studies on Splitomicin derivatives as sirtuin inhibitors and computational prediction of binding. J. Med. Chem. 2008;51:1203–1213. doi: 10.1021/jm700972e. [DOI] [PubMed] [Google Scholar]
- 17.(a) PDB ID: 2HJH; (b) PDB ID: 2OD7.
- 18.For previous reports of studies involving homology models of SIRT1, see: Huhtiniemi T, Wittekindt C, Laitinen T, Leppänen J, Salminen A, Poso A, Lahtela-Kakkonen M. Comparative and pharmacophore model for deacetylases SIRT1. J Comput. Aided Mol. Des. 2006;20:589–599. doi: 10.1007/s10822-006-9084-9. Huhtiniemi T, Suuronen T, Rinne VM, Wittekindt C, Lahtela-Kakkonen M, Jarho E, Wallen EAA, Salminen A, Poso A, Leppänen J. Oxadiazole-carbonylaminothioureas as SIRT1 and SIRT2 Inhibitors. J. Med. Chem. 2008;51:4377–4380. doi: 10.1021/jm800639h. Kiviranta PH, Salo HS, Leppänen J, Rinne VM, Kyrylenko S, Kuusisto E, Suuronen T, Salminen A, Poso A, Kakkonen ML, Wallen EAA. Characterisation of the binding properties of SIRT2 inhibitors with a N-(3-phenylpropenoyl)-glycine tryptamide backbone. Bioorg. Med. Chem. 2008;16:8054–8062. doi: 10.1016/j.bmc.2008.07.059.
- 19.Herkstroeter W, Specht D, Farid S. The triplet state of methyl 1,2-diphenylcyclopropene-3-carboxylate. J. Photochem. 1983;21:325–342. [Google Scholar]
- 20.Andreichikov YS, Tokmakova TN. Five membered 2,3-dioxo heterocycles. IV. J. Org. Chem. USSR (Eng. Transl) 1987;23:880–885. [Google Scholar]
- 21.Wamhoff H, Schorn G, Korte F. Zur Synthese und umlagerung biund tricyclischer α-acyl-δ-lactone. Chem. Ber. 1967;100:1296–1304. [Google Scholar]
- 22.Morris GA, Nguyen ST. A general route to pyridine-modified salicylaldehydes via Suzuki coupling. Tethraedron Lett. 2001;42:2093–2096. [Google Scholar]
- 23.Fahmy AM. Rev. Roum. Chim. 1985;V30(8):749–752. [Google Scholar]
- 24.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Schuettelkopf W, Van Aalten DMF. Acta Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.