Abstract
Neurodegenerative diseases affect millions of patients annually and are a significant burden on the health care systems around the world. While there are symptomatic remedies for patients suffering from various neurodegenerative diseases, there are no cures as of today. Cell death by apoptosis is a common hallmark of neurodegeneration. Therefore, deciphering the molecular pathways regulating this process is of significant value to scientists' endeavor to understand neurodegenerative disorders. Efforts along these lines have uncovered a number of molecular pathways that regulate neuronal apoptosis. Recently, a family of proteins known as histone deacetylases (HDACs) has been linked to regulation of cell survival as well as death. The focus of this review is to summarize our current understanding of the role of HDACs and in particular a subgroup of proteins in this family classified as class IIa HDACs in the regulation of neuronal cell death. It is apparent based on the information presented in this review that although very similar in their primary sequence, members of this family of proteins often have distinct roles in orchestrating apoptotic cell death in the brain.
Keywords: Histone deacetylase (HDAC), neuronal apoptosis, neurodegeneration, cell cycle, cerebellar granule neuron (CGN
2. INTRODUCTION
Histone Deacetylases (HDACs) are a family of enzymes that are involved in the regulation of gene transcription in higher organisms. The main function of these enzymes is to remove the acetyl moieties from lysine residues located within the N-terminal tail of histones, causing compactation of chromatin around histone proteins and repression of transcription (1-3). Histone acetyl transferases (HATs) reverse the action of HDACs by adding acetyl groups to these lysine residues. Negatively charged acetyl moieties added to histones repel the negatively charged phosphate backbone of DNA. This repulsion makes DNA accessible to various transcription initiation factors and facilitates transcription. The orchestration between HDAC and HAT activities results in post translational modifications in chromatin structure that lead to both global and regional regulation of gene transcription in Eukaryotic cells (1).
Eighteen distinct HDACs have been described as of today and are categorized into three classes based on their homology to yeast HDACs (1, 2). Class I HDACs (HDAC1, 2, 3 and 8) mainly contain a deacetylase domain and are primarily localized in the nucleus of cells. Members of this class are ubiquitously expressed throughout various developmental stages and different tissue and cell types. Contrary to class I HDACs, class II HDACs (HDAC4, 5, 6, 7, 9 and 10) have tissue and stage specific expression and are able to shuttle in and out of the nucleus. Class II HDACs are sub-classified into class IIa (HDAC4, 5, 7 and 9) and class IIb (HDAC 6 and 10) based on their primary structure (2). Class IIa HDACs contain a large N-terminal regulatory domain involved in protein-protein interactions in addition to a C-terminal catalytic domain. The N-terminus regulatory domain of these HDACs has important roles in the recruitment of various cofactors. Class IIa HDACs are abundantly expressed in the brain and have recently been shown to have important roles in regulation of neuronal apoptosis (2, 4-6), however, the other functions of these HDACs in the brain are currently unclear. Class IIb HDACs are different from class IIa HDACs in that they contain two separate catalytic domains. Moreover, as opposed to class IIa HDACs that are highly expressed in brain, HDAC6 is mainly expressed in testis and HDAC10 expression is primarily detected in liver, spleen and kidney (2).
Class III HDACs, Sirtuins (SIRTs 1-7), are mammalian counterparts of yeast SIR2 HDACs. Sirtuins are not homologous to class I or class II HDACs (2). Although serving as HDACs in yeast these proteins do not deacetylate histones in mammalian cells and are hence not considered to be classical HDACs.
3. CONTRADICTORY RESULTS USING HDAC INHIBITORS
The pharmacological inhibition of HDACs has been widely used to ascertain the role of HDACs in control of neurodegeneration. However, these studies have yielded contradictory results. It has been reported that administration of HDAC inhibitors in C.elegans, drosophila and murine models of polyglutamine expansion diseases rescues the pathological hallmark of the disease (7-10). Rodent models of other neurodegenerative conditions such as Amyotrophic Lateral Sclerosis (ALS), ischemia and Parkinsonism induced by 1-methyl 4-phenyl 1,2,3,6-tetrahydropyridine (MPTP) have also been responsive to treatments with HDAC inhibitors (11-14). Similarly, HDAC inhibitors were found to be neuroprotective in a number of in vitro paradigms of neurodegeneration (15-18). Contradictory to these studies, HDAC inhibitors have been reported to actively induce apoptosis in cerebellar granule neurons by several laboratories (5, 19, 20). It is noteworthy to mention that, the most commonly used pharmacological inhibitors of HDACs inhibit all HDACs effectively (21). These inhibitors lack specificity for individual HDACs. Therefore, the effect of HDAC inhibitors may depend on the roles, contributions, and ratio of individual HDAC members present within a given cell. Additionally, because HDAC inhibitors inhibit all HDACs effectively, current HDAC inhibitors are not useful for deciphering the role of individual HDACs. Consequently, this review focuses on research that characterizes the role of individual HDACs in the regulation of neurodegeneration with specific emphasis on class IIa HDACs.
4. IMPORTANCE OF HDAC IN REGULATION OF NEURONAL FUNCTIONS
Given that HDACs are involved in regulation of non-histone proteins and also act at the chromosome level to regulate gene transcription, it is not surprising that these multi-complex enzymes are involved in various cellular processes such as differentiation (22), DNA replication (23) and cell cycle progression (24). A large number of HDACs have been demonstrated to have important functions in neurons. Much information gained from the use of pharmacological HDAC inhibitors is available and has been reviewed previously (21) . The purpose of this section is to present information available on the role of individual HDAC proteins in neurons and in the nervous system excluding their roles in regulation of neurodegeneration which will be discussed in other sections in detail.
4.1. Involvement of class I HDACs in regulation of neuronal differentiation
Amongst class I HDACs that are ubiquitously expressed in various tissue and cell types, HDAC1 and HDAC2 have been shown to be involved in determination of neuronal fate. HDAC1 mediates neuronal differentiation through its interaction with the cell cycle modulating protein, retinoblastoma (Rb) (25). There is also accumulating evidence that HDAC2 is involved in nerve growth factor (NGF)-induced differentiation of PC12 (pheochromocytoma) cel lines into neuronal cells (26). In this report authors describe that DNA methyl transferase 3b (Dnmt3b) is an inducer of neuronal differentiation of the PC12 cell line (26). Bai et al., show that the N-terminal domain of Dnmt3b is involved in mediation of neuronal differentiation through recruitment of HDAC2. Besides demonstrating that HDAC2 is recruited by Dnmt3b through co-immunoprecipitation and co-sedimentation experiments, the authors also show that HDAC inhibitors are able to hinder NGF induced neuronal differentiation of PC12 cells and that Dnmt3b exhibits elevated HDAC activity after NGF treatment (26). Additionally, repressor element 1(RE-1)-silencing transcription factor (REST) recruitment of HDAC2 has been shown to be required for NGF induced PC12 cell differentiation (27, 28). It has been suggested that REST represses neuronal-specific gene expression in non-neuronal cell types by binding RE-1 which is a critical element for the silencing of neuronal genes (29-33). CoREST, one of co-repressors of REST is shown to exist in tight association with HDAC1 and HDAC2 (27). HDAC1 and HDAC2 have also been implicated in oligodendrocyte differentiation (34). The myelin transcription factor 1 (Myt1) which is a modulator of proliferation and differentiation of oligodendrocytes, the myelin-forming cell of the CNS (35), was shown to be in the same complex as the co-repressor Sin3B, HDAC1 and HDAC2 (34)
4.2. Involvement of class II HDACs in regulation of neuronal functions
Class II HDACs have been classified to class IIa (HDACs 4, 5, 7 and 9) and class IIb HDACs (HDACs 6 and 10) based on their structures. Among class IIa HDACs, HDACs 4, 5, and a splice variant of HDAC9, histone deacetylase related protein (HDRP), have been shown to be involved in regulation of neurodegeneration which will be discussed in detail later (4-6, 36-40). HDAC5, HDAC9 and the two class IIb HDACs have also been demonstrated to regulate neuronal functions other than neurodegeneration which will be discussed here.
4.2.1. Class IIa HDACs
HDAC5 has been shown to be involved in long term memory integration in Aplysia (41), and in effectiveness of antidepressants in a rodent model of depression (42). In Aplysia, Phe-Met-Arg-Phe-amide (FMRFamide) inhibitory transmitter is a key player in domination of long term depression. FMRFamide displays its inhibitory effects by facilitating c-AMP response element binding protein (CREB) 1 displacement by CREB2 and recruitment of the co-repressor HDAC5 (41). The chronic administration of a tricyclic antidepressant, imipramine, increases histone acetylation at (brain derived neurotrophic factor) BDNF transcripts III and IV promoters (42). This hyperacetylation was demonstrated to be associated with selective downregualtion of HDAC5. Not surprisingly, the authors showed that forced expression of HDAC5 inhibits imipramine's ability to alleviate depression-like behavior in rodent models (42).
HDAC9 has been demonstrated to couple neuronal activity to muscle chromatin acetylation (43). The authors showed that overexpression of HDAC9 in denervated muscle inhibits the upregulation of activity-dependent genes and chromatin acetylation in association with myocyte enhancer factor 2 (MEF2) and class I HDACs. Moreover, HDAC9-null mice were demonstrated to be supersensitive to denervation-induced changes in gene expression and have delayed perinatal downregulation of myogenin, an activator of AChR genes (43).
4.2.2. Class IIb HDACs
HDAC6 and HDAC10 are two members of class II HDACs that have been further classified as class IIb HDACs. These two HDACs differ from the rest of class II HDACs because of their distinctive structure. HDACs 6 and 10 contain two deacetylase domains. Although both catalytic domains of HDAC6 are active, HDAC10 contains one active and one nonfunctional enzymatic domain (44). While HDAC10 has not yet been extensively studied in the context of neurons, it is known that this HDAC interacts with HDACs 1-5 and HDAC7 (1). The ability of HDAC10 to bind various HDACs whose involvement in neuronal processes have been solidified, suggests that this HDAC may also play a role in regulation of different neuronal functions via recruiting other HDACs (21). HDAC6 activity has been linked to neurodegenerative disease. In relevance to Huntington's disease (HD), Iwata and colleagues showed that the autophagic degradation of aggregated Huntingtin, the affected protein in HD, requires HDAC6 (45). This HDAC also has the ability to associate with the spinocerebellar ataxia type 3 protein, Ataxin 3. Ataxin 3 is a neurodegenerative protein and has been shown to posses the ability to regulate aggresome formation (46). HDAC6 has also been described as a microtubule-associated deacetylase and as a component of the aggresome (47). It was demonstrated that HDAC6 plays a crucial role in regulation of misfolded protein-induced stress since it has the ability to recruit misfolded protein cargo to dynein motors for transport (47). This is of particular interest in regard to neurodegeneration since build-up of misfolded and aggregated proteins is thought to be a key contributor to cell death in several neurodegenerative diseases (48, 49).
5. INVOLVEMENT OF CLASS IIA HDACS IN REGULATION OF NEURODEGENERATION
5.1. Role of HDAC and MEF2 interaction in regulation of neuronal apoptosis
It is well established that class IIa HDACs are involved in transcriptional regulation. However, these HDACs do not have the ability to bind DNA directly and therefore are recruited to the target promoter via interaction with other proteins (2, 50, 51). A group of well-studied interactors of HDACs are the MEF2 family of proteins. All class IIa HDACs contain a highly conserved MEF2 binding domain located in their N-termini (2). The functional role of this interaction has been widely studied in the context of muscle differentiation. Class IIa HDACs bind MEF2 in the nucleus and inhibit the expression of MEF2 regulated genes whose products are responsible for induction of the fetal myocyte gene program. Various external stimuli cause phosphorylation of HDACs via activation of the calcium calmodulin kinase (CaMK) signaling pathways. Phosphorylation of HDACs at two highly conserved N-terminal serine residues by CaMK and/or protein kinase D creates a docking site for the protein, 14-3-3. As a result of HDAC and 14-3-3 binding, HDACs are exported out of the nucleus. Cytoplasmic localization of HDACs frees MEF2 transcription factors in the nucleus to begin transcriptional activation of genes that affect muscle differentiation (1, 2, 52, 53). Besides being a master regulator of myogenesis, it is well established that the MEF2 family of proteins play important roles in the regulation of neuronal survival and differentiation (54-59). Among MEF2 family members, MEF2A and MEF2D are highly expressed and active in cerebellar granule neurons (CGNs) (60). Although all class IIa HDACs are known to contain a MEF2 binding site, HDAC5 is the only class IIa HDAC whose interaction with MEF2 has been demonstrated to have functional significance in regulation of neuronal cell death (36). Linseman et al., showed that depolarization-mediated MEF2 activity and CGN survival are compromised by forced expression of HDAC5, which would normally repress MEF2 function (36). The functional significance of the interaction between MEF2 family members and other class IIa HDACs in the context of neuronal survival is not known and needs additional study.
5.2. Role of HDAC4 and HDAC5 in regulation of neurodegeneration
Altered HDAC4 regulation has been linked to a number of neurodegenerative disorders. However, it is unclear from these studies whether it plays a harmful or beneficial role with respect to disease pathogenesis. Evidence for the association of HDAC4 in Parkinson's disease (PD) stems from the observation that HDAC4 is a downstream target of Parkin, an E3 ubiquitin ligase involved in early onset PD (38, 61). Parkin maturation, seen in early onset PD, causes build up of the E3 SUMO ligase, Ran binding protein 2 (RanBP2). RanBP2 sumoylates HDAC4 causing an increase in HDAC4 deacetylase activity and gene repression (38). HDAC4 has also been shown to be a component of protein aggregate complexes in neurodegenerative disorders. The discovery of HDAC4 as a major component of intranuclear inclusions produced in neuronal intranuclear inclusion disease (NIIND) (39) and in Lewy Bodies (40), protein aggregates found in the brains of PD patients, further supports the involvement of HDAC4 in regulation of neurodegenerative diseases. In addition, crystallization of the N-terminal domain of HDAC4 has revealed a conserved glutamine rich domain (62). Interestingly, proteins with regions containing a high content of glutamine have been observed to increase susceptibility to Amyloid formation associated with Alzheimer's disease (63, 64).
HDAC4 and 5 subcellular localization, which is hypothesized to control class IIa HDAC activity, is regulated by neuronal activity in hippocampal neurons and in CGNs (4, 6, 36, 65). Localization of both HDAC4 and 5 appears to be calcium regulated in both of the aforementioned cell culture models. In CGNs, both HDAC4 and 5 are localized in the cytoplasm during treatments with depolarizing media and removal of this media causes their nuclear transport as does treatment with CaMK inhibitors (4, 6, 36). However, in hippocampal neurons, these HDACs act differently in response to spontaneous electrical activity. Such a signal was shown to be sufficient for nuclear export of HDAC4 but not that of HDAC5 (65). HDAC5 nuclear export was demonstrated to be induced following stimulation of calcium flux through NMDA receptors or L-type calcium channels. Moreover, shuttling of HDAC4 and 5 in and out of the nucleus is shown to be blocked by pharmacological inhibition of CaMK (65). Therefore, localization of both HDAC4 and 5 are regulated by neuronal activity, which is a crucial process for survival of neurons. Moreover, HDAC5 may play a role in HD as its nuclear import increases in diseased neurons from the brain of HD patients.
5.3. Contradictory results for the role of HDAC4 in activity deprivation-induced CGN apoptosis
The role of HDAC4 in the regulation of CGN survival is currently contested. This is due to the contradicting results observed by our lab (6) and the lab of Tso-Pang Yao (4). Bolger and Yao reported that the intracellular trafficking of HDAC4 promotes neuronal cell death (4). In contrast, our lab has shown that HDAC4 translocation from the cytosol (where HDAC4 resides in depolarizing media, containing high potassium (HK)) to the nucleus in non-depolarizing media, containing low potassium (LK), leads to higher survival of CGNs (6). One of the major points agreed upon by both of these reports is the fact that HDAC4 is localized in the cytoplasm in normal CGNs and translocates to the nucleus when cells are treated with non-depolarizing media. Another common point made by both groups is the fact that HDAC4 expression is not dynamically regulated by neuronal activity dependent apoptosis in CGNs. However, the papers disagree in their answer to the question of whether HDAC4 overexpression leads to neuronal survival or cell death in non-depolarizing media. Bolger and Yao show that overexpression of wild-type HDAC4 in original media containing serum and in LK cause <5% increase in cell death when compared with GFP transfected cells. However, the work done by our lab shows that overexpression of HDAC4 in LK condition completely saves the cells against LK induced apoptosis. In control GFP infected cells, CGNs undergo about 40-60% cell death when treated in LK condition, however, those overexpressing HDAC4 show complete survival (6). Significant experimental differences exist between the two studies. In the Bolger and Yao study, relatively immature (2days after plating) CGN cultures were used in contrast to our CGN cultures which are used 6 – 7 days after plating. Additionally, Bolger and colleagues transfected cells while plating and later assessed viability on day 2, 6 hours after treatment with HK and LK medium. In our lab, cells were infected on day 5 after plating, and viability of cells were examined 24 after LK treatment on day 7. We have also verified our results using various truncation mutants of HDAC4. We believe that the contradiction between these two papers can be explained by the age of cultures used. Moreover, it has been reported that HDAC4-deficient mice are severely runted, display skeletal abnormalities, and die within 2 weeks of birth (66). In our lab, examination of the cerebellum of HDAC4−/− mice and wild-type littermates at P1, P3 and P7 revealed a delay in the formation of folia with less developed fissures in the mutant. Moreover, the pattern of foliation and shape of the lobes, particularly the posterior lobes, is altered. Based on the importance of HDAC4 for normal brain development, we believe that the moderate increase in death of HDAC4 overexpressing cells in immature cultures tested by Bolger et al., might be due to interrupting of the development of these cultures at an early stage (day 2). It is known that CGNs mature in culture after plating and become even more responsive to cell death induced by LK condition as they age. Moreover, the neuroprotection by HDAC4 was further confirmed in our lab, as we found that in addition to protecting CGNs against activity withdrawal induced apoptosis, HDAC4 also protects cultured cortical neurons from 6-hydroxydopamine-induced neurotoxicity and HT22 neuroblastoma cells from death induced by oxidative stress induced by homocysteic acid.
Another contradiction with regard to these two papers is the effect of HDAC inhibitors on CGN cultures. Bolger and Yao show that treatment of mature CGNs with the HDAC inhibitors Trichostatin A (TSA) or Trapoxin A reduces the extent of cell death in LK. The authors assert that the reduction in cell death caused by HDAC inhibitors is evidence for the pro-apoptotic role of HDAC4 in CGNs. In contrast to the results found by Bolger and Yao, our lab and several other laboratories have shown that the pharmacological inhibition of HDACs has deleterious effects on CGN survival (5, 19, 67). In order to further verify these findings, we repeated Bolger and Yao treatment of young CGNs with HDAC inhibitors, and not only were we unable to observe any rescue by these treatments; we in fact noticed the same toxicity seen in mature cultures (unpublished results, Majdzadeh and D'Mello)
6. DISTINCT AND OFTEN OPPOSING ROLE FOR HDAC FAMILY MEMBERS
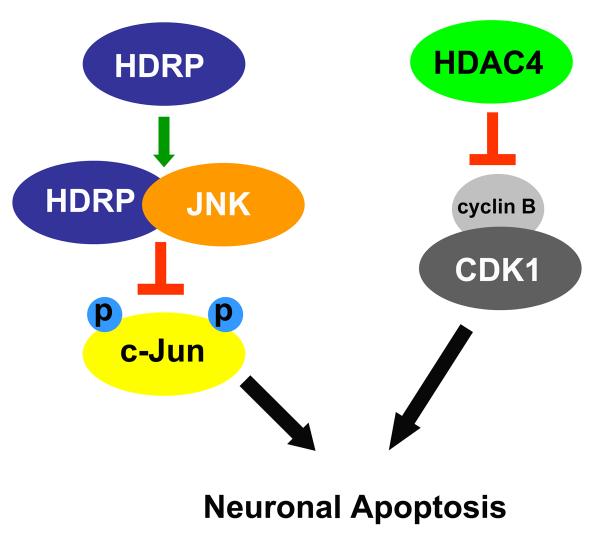
When examining the role of HDACs in various cellular processes one should be aware that these proteins cannot be considered mutually exclusive as they can interact with each other and also have common interaction partners. HDAC4, 5, 9 and the splice variant of HDAC9 (HDRP) for example, have been shown to contain a conserved MEF binding site functioning in cardiac myocytes (2). Through direct interaction, these HDACs inhibit MEF2 transcriptional activity. The interaction between MEF2 and these HDACs in nervous system has not been shown as extensively. Among the aforementioned HDACs, there's evidence that HDAC5 binds to and inhibits MEF2 in CGNs (36). The same group has also shown that MEF2 plays a survival-promoting role in CGNs. Overexpression of HDAC5 induces apoptosis in CGNs as the result of inhibiting the pro-survival role of MEF2 (36). No interaction between HDAC4, HDAC9/HDRP and MEF2 has been defined in CGNs. However, our lab has shown that overexpression of HDAC4 and HDRP protect CGNs against activity withdrawal induced apoptosis (5, 6). The mechanism by which HDAC4 and HDRP mediate their neuroprotection seems to be distinct (Figure 1). HDRP binds to and inhibits Jun N-terminal kinase (JNK), an apoptosis inducing MAP kinase. HDRP also inhibits the expression of the pro-apoptotic protein, c-Jun, by affecting c-Jun promoter acetylation (5). HDRP is an N-terminal truncate of HDAC9 and therefore lacks a deacetylase domain. However, by recruiting HDAC1 to c-Jun promoter, HDRP inhibits c-Jun expression. Although high homology exists between HDAC4 and HDRP (2), our lab has shown that HDAC4 mediates neuroprotection via a distinct mechanism than that utilized by HDRP. HDAC4 suppresses cell cycle progression by inhibiting the activity of cyclin dependent kinase 1 (CDK1) (6). Activation of CDK1 has been shown to play an essential role in the death of CGNs caused by activity withdrawal (68, 69). Taken together these results suggest that although high sequence similarities exist among HDAC4, HDAC5 and HDRP (Figure 2), they exert different roles in regulation of neuronal apoptosis. HDAC5 has pro-apoptotic properties, while HDAC4 and HDRP are neuroprotective proteins.
Fig. 1. HDAC4 and HDRP -mediated neuroprotection.
HDRP and HDAC4 mediate neuronal survival via distinct mechanisms. While HDRP binds to and inhibits the pro-apoptotic protein, JNK, therefore inhibiting c-Jun induction and phosphorylation, HDAC4 inhibits cell cycle progression by inhibiting CDK1 activity.
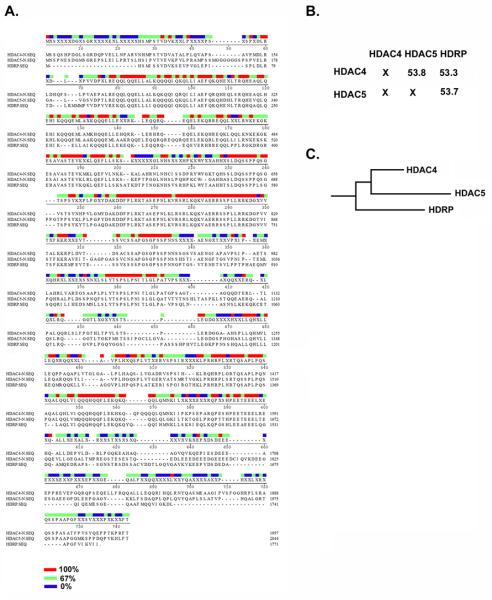
Fig. 2. HDAC4, HDAC5 and HDRP sequence similarities.
(A-B) The N-terminal protein sequences of human HDAC4 (653 aa), HDAC5 (682 aa) and HDRP are compared in this figure. (A) Protein sequences were aligned using the Jotun Hain method. Red, green and blue bars show 100%, 67% and 0% similarities amongst the three sequences respectively. (B) Percent similarities amongst the protein sequences are reported. This is a gross measure of similarity between sequences, derived by taking the matches over the matches, mismatches and gaps. (C) Phylogenic tree, showing the evolutionary relationship amongst HDAC4, HDAC5 and HDRP. Actual distances are not exact and may be greater than shown. MegAlign software was used for the generation of this data.
Besides having differing roles in the maintenance of CGN survival, HDACs have been shown to have opposing contributions in a C.elegan model of Huntington polyglutamine toxicity. In C.elegans, expression of a human huntingtin fragment with an extended polyglutamine tract (Htn-Q150) causes progressive neurodegeneration (10). Bates et al, have described that HDA-1 (C.elegan homolog of mammalian HDAC1 and HDAC2) suppresses neurotoxicity in Htn-Q150 animals whereas HDA-3 (C.elegan homolog of mammalian HDAC3) promotes it. To show that the role of HDA-1 and 3 in governing neurodegeneration was specific to the polyglutamine expansion animal model, the authors performed similar experiments in animals with a cold-sensitive mutation in a sodium channel, deg-1(rt70), which causes temperature-dependent necrotic degeneration. Manipulation of neither HDA-1 nor HDA-3 was able to alter necrotic neurodegeneration suggesting that the differential contributions of these HDACs were specific to polyglutamine expansion model and not necrotic cell death (10). More importantly, is the finding that although HDA-1 and 3 have opposing effects in regard to the extent of toxicity in C.elegans, they cannot function exclusively. Bates et al. reported that in HDA-1 compromised animals, HDA-3 knock down does not inhibit the extent of degeneration suggesting that these HDACs function as multi-complex enzymes and one is directly or indirectly required for the function of another (10).
The opposing roles of HDACs are not confined to the context of neurodegeneration. Several in vitro studies have also shown that HDACs behave differently and their localization pattern can vary in response to assorted stimuli. An expression screen intended to reveal modulators of class II HDAC phosphorylation has shown that class II HDACs respond differently to activators of HDAC phosphorylation (70). These studies showed that HDAC4 and 7 respond to a wide range of HDAC kinase activators and reside in the cytoplasm as the result of the consequent phosphorylations, while HDAC5 response is considerably more limited and stringent.
Another piece of evidence citing an intrinsic difference among HDACs comes from a study attempting to characterize upstream regulators of HDACs. The specificity of calcium calmodulin kinases, believed to phosphorylated all class IIa HDACs, are now beginning to be unraveled. Along these lines, one study has revealed that the delta B isoform of CaMK II (CaMKIIdeltaB), known to promote cardiac hypertrophy, transmits signals specifically to HDAC4 but not other class II HDACs in cardiac cells (71).
7. IMPORTANCE OF THE CATALYTIC DEACETYLASE DOMAIN
It is well known that HDACs regulate global gene expression by dictating the availability of DNA to transcription factors. In addition, a great deal of work has focused on the ability of HDACs to deacetylase non-histone proteins. However, little has been published regarding the function of the non-deacetylase encoding N-terminal region of class IIa HDACs. HDRP, an isoform of HDAC9 which lacks the catalytic domain, was the first member of HDAC family to be described as a neuroprotective protein (5). In CGNs, HDRP forced expression protects neurons against activity withdrawal-induced apoptosis (5). Moreover, CGNs cultured from HDAC9/HDRP deficient mice are more susceptible to cell death in culture. Acute knockdown of HDRP using antisense oligos also has deleterious effects on the survival of CGNs. Although the intrinsic deacetylase domain of HDAC9 is not necessary for HDRP-mediated neuroprotection, we found that pharmacological inhibition of HDACs inhibits this neuroprotection (5). This phenomenon is explained by the fact that HDAC activity is needed because HDRP recruits HDAC1 to the c-Jun promoter where HDAC1 facilitates deacetylation and repression. A similar finding was reported with regard to HDAC10. HDAC10 was shown to repress transcription once localized in the nucleus independent of its deacetylase domain (44). It has been shown that HDAC10 possesses the ability to bind various HDACs such as HDACs 1-5 and 7 suggesting that HDAC10, similar to HDRP, can recruit other HDACs to facilitate transcriptional repression (1).
Since HDAC4 and HDRP have high homology and both HDACs protect CGNs against neuronal cell death (albeit by distinct mechanisms) we wanted to know whether HDAC4 required a deacetylase domain for neuroprotection (5, 6). While an HDAC4 isoform containing only an N-terminus regulatory domain has not been found to occur naturally, we constructed an HDAC4 N-terminus truncation mutant (HDAC4-N) and tested its ability to protect CGNs (6). Very interestingly, we found that this construct is as effective as the full length HDAC4 in mediating neuroprotection in CGNs. However, in constrast to HDRP, we believe that HDAC4-N does not mediate its neuroprotection through the recruitment of other HDACs, since the inhibition of global HDAC activity using TSA does not reduce the extent of protection exerted by HDAC4. We therefore believe that the importance of the deacetylase domain in CGN protection my not be general. We hypothesize that, HDACs may in some cases recruit the deacetylase domain of other HDAC family members or may mediate their function through protein-protein interactions independent of deacetylation all together.
8. ROLE OF HDAC IN CELL CYCLE REGULATION AND NEURONAL APOPTOSIS
We propose that HDAC4 mediated neuronal protection is mediated through inhibition of CDK1 activity. Thus, we detail the role of HDACs in cell cycle regulation. Several HDACs have been implicated in differentiation and tumor cell growth (22, 24). This implies that HDACs may be playing a role in the regulation of cell cycle. The activation of numerous cell cycle regulating proteins has been observed in both in vitro and in vivo paradigms of neurodegeneration (72-74), as a result literature that relates HDAC function and cell cycle progression will be reviewed here.
Mature neurons are post-mitotic and rest at the G0 phase of the cell cycle. It is believed that once activated in neurons, cell cycle regulating proteins are rewired to induce cell death as opposed to cell proliferation. The first evidence suggesting a role for HDACs in governing the cell cycle came from studies revealing that HDAC inhibitors can serve as potent inducers of growth arrest, differentiation, and/or apoptotic cell death of transformed cells (1, 22, 24). However, the use of broad spectrum HDAC inhibitors does not advance our understanding of the role of individual HDACs in cell cycle regulation. We therefore, focus on studies that have used non-pharmacological methods to study HDACs. HDACs 1, 3 and 4 have been shown by independent groups to play roles in cell cycle regulation in non-neuronal systems (24, 25, 75-77).
Progression through the cell cycle is dependent on expression of cell cycle regulating proteins that are involved in cell growth and DNA replication. Members of the E2F family of transcription factors have been shown to possess important and conserved roles in cell cycle progression in several organisms examined (78-80). Eight members of the E2F family of proteins have been characterized to date (81). E2F activity is largely regulated by the tumor suppressor protein, Rb (82-84). E2F-1, 2, and 3 contain highly conserved domains for binding both cyclin A/CDK2 and Rb (81). E2F-1 was the first E2F family member to be discovered due to its ability to directly bind Rb (81). Binding of E2F-1 to Rb results in inhibition of E2F-1-meidated transcription. This inhibitory effect is relieved in mid to late G1, when Rb is phosphorylated by CDKs. Phosphorylation of Rb frees E2F-1 and causes activation of E2F-1 regulated gene transcription which results in cell cycle progression (78, 79). Moreover E2F-1 activation has been established as a contributing factor in a number of neurodegenerative in vitro and in vivo models. Accumulated E2F-1 protein has also been observed in the post mortem brains of patients affected with neurodegenerative disease (85).
It has been shown that some E2F family members including E2F-1 are acetylated by p300 and CBP (24). P300 and CBP are HATs that counteract HDAC function in the cell (1). Acetylation of E2F-1 was shown to enhance transactivation of an E2F-responsive promoter. However, the reversal of this acetylation by HDAC1 suggests that HDAC activity can serve as a mechanism in controlling the cell cycle (24). In addition, another group reported that HDAC1 is present in Rb-recruited transcription repression complexes. Nicolas et al., showed that RbAp48, an Rb-associated protein that can directly interact with histone H4 (86-88), and E2F-1 associate directly in the presence of Rb and HDAC1 to mediate transcriptional repression (25).
Another HDAC whose role in regulation of cell cycle has been uncovered is HDAC3. Panteleeva and colleagues reported HDAC3 as an Rb/E2F binding protein (75). They demonstrated that HDAC3 undergoes a caspases dependent degradation during apoptosis. The degradation of HDAC3 might be the underlying reason for the instability of E2F-responsive elements (E2F-Res) in apoptotic versus neuroprotective conditions (75).
Amongst class II HDACs, HDAC4 has been the most closely linked to cell cycle progression. Research investigating the role of HDAC4 in DNA damaged cells has yielded interesting findings regarding cell cycle progression, HDAC4 and the p53 response pathway. As discussed previously, HDAC4 shuttles in the nucleus in response to apoptotic stimuli. Similar localization takes place when cells undergo DNA damage (76). In DNA damaged cells, transcription is controlled by genes down stream of the tumor suppressor protein, p53. DNA damage causes a robust increase in p53 levels. The elevated levels of p53 act as a negative regulator of key proteins involved in cell cycle progression. Basile et al., showed that HDAC4 is recruited to G2/M promoters upon DNA damage in a p53 dependent manner. Another group also suggested a role for HDAC4 in cell cycle progression with the finding that the silencing of HDAC4 via RNA interference decreased levels of 53BP1, a protein that is recruited to nuclear foci upon DNA damage, and inhibited the DNA damage-induced G2 delay in HeLa cells (77).
Moreover, in our laboratory, we have shown that HDAC4 inhibits cell cycle progression and protects neurons through inhibition of CDK1 activity. We demonstrated that forced expression of HDAC4 suppresses cell cycle progression of both HEK293T and HT22 cells (6). Consistent with these findings, we reported decreased CDK1 activity in brain of HDAC4 deficient mice (6). This finding is in agreement with the previous finding that HDAC4 suppresses proteins regulating the G2/M transition in cell cycle progression (76).
9. CONCLUDING REMARKS
Here, we have presented a review citing the roles of HDACs in the regulation of a number of neuronal processes with a focus on class IIa HDACs and their role in the maintenance of neuronal survival. We have discussed that although very similar in primary structure, these HDACs can have opposing roles in governing neuronal functions. Broad spectrum pharmacological inhibitors of HDACs inhibit all HDACs effectively. Consequently, several studies spanning numerous fields that include cardiac myocyte differentiation and neurodegeneration have revealed contradicting results from the use of HDAC inhibitor (21, 89). Considering that members of the HDAC family of proteins may play opposing roles in the regulation of cellular processes, the use of these global inhibitors to decipher the role of individual HDACs is problematic. It is therefore important to ascertain the role of each HDAC individually in order to obtain an un-muddled picture of HDAC function. Class IIa HDACs are known to act via protein-protein interactions through their N-terminal domain. Our lab has reported that the class IIa HDACs, HDAC4 and HDRP, are able to behave as guardians aginst cerebellar granule neuron activity withdrawal-induced apoptosis even in the absence of their deacetylase domain. Therefore, the importance of the deacetylase domain with regard to HDAC function is an unanswered question. It is conceivable that the protein-protein interactions indeed play the most important roles in the ability of class IIa to regulate gene transcription. Therefore, characterization of new interacting proteins and deciphering the functional significance of these interactions is a crucial step toward understanding HDAC function in the regulation of neuronal processes.
10. ACKNOWLDEGMENTS
The authors would like to acknowledge National Institutes of Health grants NS40408 and NS047201 to SRD.
Abbreviations
- AChR
acetyl choline receptor
- ALS
amyotrophic lateral sclerosis
- BDNF
brain derived neurotrophic factor
- CaMK
calcium calmodulin kinase
- CDK
cyclin dependent kinase
- CGN
cerebellar granule neuron
- CREB
c-AMP response element binding protein
- Dnmt3b
DNA methyl transferase 3b
- E2F-Res
E2F responsive element
- FMRFamide
phe-met-arg-phe amide
- HAT
histone acetyl transferase
- HD
Huntington's disease
- HDAC
histone deacetylase
- HDRP
histone deacetylase related protein
- HK
high potassium
- HN
hippocampal neurons
- JNK
Jun N-terminal kinase
- LK
low potassium
- MEF
myocyte enhancer factor
- MPTP
methyl 4-phenyl 1,2,3,6-tetrahydropyridine
- Myt1
myelin transcription factor
- NGF
nerve growth factor
- NIIND
neuronal intranuclear inclusion disease
- PD
Parkinson's disease
- RanBP2
Ran binding protein 2
- Rb
retinoblastoma
- RE-1
repressor element 1
- REST
RE-1 silencing transcription factor
- SIRT
sirtuins
- TSA
trichostatin A
11. REFERENCES
- 1.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–49. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–93. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 3.Yang XJ, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol. 2005;25:2873–84. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J Neurosci. 2005;25:9544–53. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison BE, Majdzadeh N, Zhang X, Lyles A, Bassel-Duby R, Olson EN, D'Mello SR. Neuroprotection by histone deacetylase-related protein. Mol Cell Biol. 2006;26:3550–64. doi: 10.1128/MCB.26.9.3550-3564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majdzadeh N, Wang L, Morrison BE, Bassel-Duby R, Olson EN, D'Mello S. HDAC4 inhibits cell cycle progression and protects neurons from cell death. Submitted to Developmental Neurobiology journal. 2007 doi: 10.1002/dneu.20637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–43. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 8.Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, Steffan JS, Marsh JL, Thompson LM, Lewis CM, Marks PA, Bates GP. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2041–6. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardian G, Browne SE, Choi DK, Klivenyi P, Gregorio J, Kubilus JK, Ryu H, Langley B, Ratan RR, Ferrante RJ, Beal MF. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington's disease. J Biol Chem. 2005;280:556–63. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 10.Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–8. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardian G, Yang L, Cleren C, Calingasan NY, Klivenyi P, Beal MF. Neuroprotective effects of phenylbutyrate against MPTP neurotoxicity. Neuromolecular Med. 2004;5:235–41. doi: 10.1385/NMM:5:3:235. [DOI] [PubMed] [Google Scholar]
- 12.Ren M, Leng Y, Jeong M, Leeds PR, Chuang DM. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J Neurochem. 2004;89:1358–67. doi: 10.1111/j.1471-4159.2004.02406.x. [DOI] [PubMed] [Google Scholar]
- 13.Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, Moroni F, Chiarugi A. Pharmacological Inhibition of Histone Deacetylases by Suberoylanilide Hydroxamic Acid Specifically Alters Gene Expression and Reduces Ischemic Injury in the Mouse Brain. Mol Pharmacol. 2006;70:1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 14.Petri S, Kiaei M, Kipiani K, Chen J, Calingasan NY, Crow JP, Beal MF. Additive neuroprotective effects of a histone deacetylase inhibitor and a catalytic antioxidant in a transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2006;22:40–9. doi: 10.1016/j.nbd.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Jeong MR, Hashimoto R, Senatorov VV, Fujimaki K, Ren M, Lee MS, Chuang DM. Valproic acid, a mood stabilizer and anticonvulsant, protects rat cerebral cortical neurons from spontaneous cell death: a role of histone deacetylase inhibition. FEBS Lett. 2003;542:74–8. doi: 10.1016/s0014-5793(03)00350-8. [DOI] [PubMed] [Google Scholar]
- 16.Ryu H, Lee J, Olofsson BA, Mwidau A, Dedeoglu A, Escudero M, Flemington E, Azizkhan-Clifford J, Ferrante RJ, Ratan RR. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci U S A. 2003;100:4281–6. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanai H, Sawa A, Chen RW, Leeds P, Chuang DM. Valproic acid inhibits histone deacetylase activity and suppresses excitotoxicity-induced GAPDH nuclear accumulation and apoptotic death in neurons. Pharmacogenomics J. 2004;4:336–44. doi: 10.1038/sj.tpj.6500269. [DOI] [PubMed] [Google Scholar]
- 18.Chen PS, Peng GS, Li G, Yang S, Wu X, Wang CC, Wilson B, Lu RB, Gean PW, Chuang DM, Hong JS. Valproate protects dopaminergic neurons in midbrain neuron/glia cultures by stimulating the release of neurotrophic factors from astrocytes. Mol Psychiatry. 2006;11:1116–25. doi: 10.1038/sj.mp.4001893. [DOI] [PubMed] [Google Scholar]
- 19.Salminen A, Tapiola T, Korhonen P, Suuronen T. Neuronal apoptosis induced by histone deacetylase inhibitors. Brain Res Mol Brain Res. 1998;61:203–6. doi: 10.1016/s0169-328x(98)00210-1. [DOI] [PubMed] [Google Scholar]
- 20.Boutillier AL, Trinh E, Loeffler JP. Selective E2F-dependent gene transcription is controlled by histone deacetylase activity during neuronal apoptosis. J Neurochem. 2003;84:814–28. doi: 10.1046/j.1471-4159.2003.01581.x. [DOI] [PubMed] [Google Scholar]
- 21.Morrison BE, Majdzadeh N, D'Mello SR. Histone deacetylases: Focus on the nervous system. Cell Mol Life Sci. 2007 doi: 10.1007/s00018-007-7035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–6. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 23.Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274:23027–34. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 24.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000;275:10887–92. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 25.Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem. 2000;275:9797–804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
- 26.Bai S, Ghoshal K, Datta J, Majumder S, Yoon SO, Jacob ST. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol Cell Biol. 2005;25:751–66. doi: 10.1128/MCB.25.2.751-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–65. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 28.Iwase S, Januma A, Miyamoto K, Shono N, Honda A, Yanagisawa J, Baba T. Characterization of BHC80 in BRAF-HDAC complex, involved in neuron-specific gene repression. Biochem Biophys Res Commun. 2004;322:601–8. doi: 10.1016/j.bbrc.2004.07.163. [DOI] [PubMed] [Google Scholar]
- 29.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–57. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 30.Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci U S A. 1996;93:9881–6. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–3. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 32.Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–42. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 33.Paquette AJ, Perez SE, Anderson DJ. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc Natl Acad Sci U S A. 2000;97:12318–23. doi: 10.1073/pnas.97.22.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem. 2005;93:1444–53. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen JA, Berndt JA, Hudson LD, Armstrong RC. Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells. Mol Cell Neurosci. 2004;25:111–23. doi: 10.1016/j.mcn.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Linseman DA, Bartley CM, Le SS, Laessig TA, Bouchard RJ, Meintzer MK, Li M, Heidenreich KA. Inactivation of the myocyte enhancer factor-2 repressor histone deacetylase-5 by endogenous Ca(2+) //calmodulin-dependent kinase II promotes depolarization-mediated cerebellar granule neuron survival. J Biol Chem. 2003;278:41472–81. doi: 10.1074/jbc.M307245200. [DOI] [PubMed] [Google Scholar]
- 37.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 38.Um JW, Min do S, Rhim H, Kim J, Paik SR, Chung KC. Parkin Ubiquitinates and Promotes the Degradation of RanBP2. J Biol Chem. 2006;281:3595–603. doi: 10.1074/jbc.M504994200. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi-Fujigasaki J, Arai K, Funata N, Fujigasaki H. SUMOylation substrates in neuronal intranuclear inclusion disease. Neuropathol Appl Neurobiol. 2006;32:92–100. doi: 10.1111/j.1365-2990.2005.00705.x. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi-Fujigasaki J, Fujigasaki H. Histone deacetylase (HDAC) 4 involvement in both Lewy and Marinesco bodies. Neuropathol Appl Neurobiol. 2006;32:562–6. doi: 10.1111/j.1365-2990.2006.00733.x. [DOI] [PubMed] [Google Scholar]
- 41.Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–93. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 42.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 43.Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci. 2005;8:313–21. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- 44.Guardiola AR, Yao TP. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J Biol Chem. 2002;277:3350–6. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- 45.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–92. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 46.Burnett BG, Pittman RN. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc Natl Acad Sci U S A. 2005;102:4330–5. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–38. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 48.Thomas PJ, Qu BH, Pedersen PL. Defective protein folding as a basis of human disease. Trends Biochem Sci. 1995;20:456–9. doi: 10.1016/s0968-0004(00)89100-8. [DOI] [PubMed] [Google Scholar]
- 49.Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R, Mayer RJ, Layfield R. Inhibition of the ubiquitin-proteasome system in Alzheimer's disease. Proc Natl Acad Sci U S A. 2000;97:9902–6. doi: 10.1073/pnas.170173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D. Functional significance of histone deacetylase diversity. Curr Opin Genet Dev. 2001;11:162–6. doi: 10.1016/s0959-437x(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 51.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–7. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 52.Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97:4070–5. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97:14400–5. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDermott JC, Cardoso MC, Yu YT, Andres V, Leifer D, Krainc D, Lipton SA, Nadal-Ginard B. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol Cell Biol. 1993;13:2564–77. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ikeshima H, Imai S, Shimoda K, Hata J, Takano T. Expression of a MADS box gene, MEF2D, in neurons of the mouse central nervous system: implication of its binary function in myogenic and neurogenic cell lineages. Neurosci Lett. 1995;200:117–20. doi: 10.1016/0304-3940(95)12092-i. [DOI] [PubMed] [Google Scholar]
- 56.Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15:5727–38. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin X, Shah S, Bulleit RF. The expression of MEF2 genes is implicated in CNS neuronal differentiation. Brain Res Mol Brain Res. 1996;42:307–16. doi: 10.1016/s0169-328x(96)00135-0. [DOI] [PubMed] [Google Scholar]
- 58.Krainc D, Bai G, Okamoto S, Carles M, Kusiak JW, Brent RN, Lipton SA. Synergistic activation of the N-methyl-D-aspartate receptor subunit 1 promoter by myocyte enhancer factor 2C and Sp1. J Biol Chem. 1998;273:26218–24. doi: 10.1074/jbc.273.40.26218. [DOI] [PubMed] [Google Scholar]
- 59.Okamoto S, Krainc D, Sherman K, Lipton SA. Antiapoptotic role of the p38 mitogen-activated protein kinase-myocyte enhancer factor 2 transcription factor pathway during neuronal differentiation. Proc Natl Acad Sci U S A. 2000;97:7561–6. doi: 10.1073/pnas.130502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li M, Linseman DA, Allen MP, Meintzer MK, Wang X, Laessig T, Wierman ME, Heidenreich KA. Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J Neurosci. 2001;21:6544–52. doi: 10.1523/JNEUROSCI.21-17-06544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, Dejean A. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. Embo J. 2002;21:2682–91. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo L, Han A, Bates DL, Cao J, Chen L. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc Natl Acad Sci U S A. 2007;104:4297–302. doi: 10.1073/pnas.0608041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koo EH, Lansbury PT, Jr., Kelly JW. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc Natl Acad Sci U S A. 1999;96:9989–90. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perutz MF. Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 65.Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–9. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 66.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–66. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 67.Boutillier AL, Trinh E, Loeffler JP. Constitutive repression of E2F1 transcriptional activity through HDAC proteins is essential for neuronal survival. Ann N Y Acad Sci. 2002;973:438–42. doi: 10.1111/j.1749-6632.2002.tb04679.x. [DOI] [PubMed] [Google Scholar]
- 68.Konishi Y, Lehtinen M, Donovan N, Bonni A. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol Cell. 2002;9:1005–16. doi: 10.1016/s1097-2765(02)00524-5. [DOI] [PubMed] [Google Scholar]
- 69.Konishi Y, Bonni A. The E2F-Cdc2 cell-cycle pathway specifically mediates activity deprivation-induced apoptosis of postmitotic neurons. J Neurosci. 2003;23:1649–58. doi: 10.1523/JNEUROSCI.23-05-01649.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang S, Bezprozvannaya S, Li S, Olson EN. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc Natl Acad Sci U S A. 2005;102:8120–5. doi: 10.1073/pnas.0503275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Little GH, Bai Y, Williams T, Poizat C. Nuclear calcium/calmodulin-dependent protein kinase IIdelta preferentially transmits signals to histone deacetylase 4 in cardiac cells. J Biol Chem. 2007;282:7219–31. doi: 10.1074/jbc.M604281200. [DOI] [PubMed] [Google Scholar]
- 72.Copani A, Uberti D, Sortino MA, Bruno V, Nicoletti F, Memo M. Activation of cell-cycle-associated proteins in neuronal death: a mandatory or dispensable path? Trends Neurosci. 2001;24:25–31. doi: 10.1016/s0166-2236(00)01663-5. [DOI] [PubMed] [Google Scholar]
- 73.Becker EB, Bonni A. Cell cycle regulation of neuronal apoptosis in development and disease. Prog Neurobiol. 2004;72:1–25. doi: 10.1016/j.pneurobio.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 74.Greene LA, Biswas SC, Liu DX. Cell cycle molecules and vertebrate neuron death: E2F at the hub. Cell Death Differ. 2004;11:49–60. doi: 10.1038/sj.cdd.4401341. [DOI] [PubMed] [Google Scholar]
- 75.Panteleeva I, Rouaux C, Larmet Y, Boutillier S, Loeffler JP, Boutillier AL. HDAC-3 Participates in the Repression of e2f-Dependent Gene Transcription in Primary Differentiated Neurons. Ann N Y Acad Sci. 2004;1030:656–60. doi: 10.1196/annals.1329.076. [DOI] [PubMed] [Google Scholar]
- 76.Basile V, Mantovani R, Imbriano C. DNA damage promotes histone deacetylase 4 nuclear localization and repression of G2/M promoters, via p53 C-terminal lysines. J Biol Chem. 2006;281:2347–57. doi: 10.1074/jbc.M507712200. [DOI] [PubMed] [Google Scholar]
- 77.Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol. 2003;160:1017–27. doi: 10.1083/jcb.200209065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–62. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 79.Nevins JR, DeGregori J, Jakoi L, Leone G. Functional analysis of E2F transcription factor. Methods Enzymol. 1997;283:205–19. doi: 10.1016/s0076-6879(97)83017-0. [DOI] [PubMed] [Google Scholar]
- 80.Lavia P, Jansen-Durr P. E2F target genes and cell-cycle checkpoint control. Bioessays. 1999;21:221–30. doi: 10.1002/(SICI)1521-1878(199903)21:3<221::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 81.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 82.Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-I, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–72. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- 83.Bandara LR, La Thangue NB. Adenovirus E1a prevents the retinoblastoma gene product from complexing with a cellular transcription factor. Nature. 1991;351:494–7. doi: 10.1038/351494a0. [DOI] [PubMed] [Google Scholar]
- 84.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–61. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 85.Lim AC, Qi RZ. Cyclin-dependent kinases in neural development and degeneration. J Alzheimers Dis. 2003;5:329–35. doi: 10.3233/jad-2003-5409. [DOI] [PubMed] [Google Scholar]
- 86.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 87.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 88.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 89.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]