Abstract
Previously we have shown that nonsyndromic cleft lip with or without cleft palate (NSCL/P)1, is strongly associated with SNPs in Interferon Regulatory Factor 6 (IRF6)2. Here, multispecies sequence comparisons identify a common SNP (rs642961, G>A) in a novel IRF6 enhancer. The A allele is significantly overtransmitted (P=1×10−11) in families with NSCL/P, in particular with cleft lip (CL) but not cleft palate. Further, there is a dosage effect of the A allele, with the relative risk for CL 1.68 for the AG genotype and 2.40 for the AA genotype. EMSA and ChIP assays demonstrate that the risk allele disrupts the binding site of transcription factor AP-2α and expression analysis in the mouse localizes the enhancer activity to craniofacial and limb structures. Our findings place IRF6 and AP-2α in the same developmental pathway and identify a high frequency variant in a regulatory element contributing substantially to a common, complex disorder.
Mutations in IRF6 cause Van der Woude syndrome (VWS), a rare Mendelian clefting disorder with lower lip pits in ~85% of the cases3. The remaining 15% of the VWS cases have CL/P with no lip pits and are clinically indistinguishable from the common, isolated or NSCL/P with a birth prevalence of ~1/700 livebirths. Significant association between single nucleotide polymorphisms (SNPs) in and around IRF6 and NSCL/P was previously shown in multiple populations2 and independently replicated4–7. One particular SNP (rs2235371, G>A) that changes valine to isoleucine at amino acid position 274 (V274I) was found to be significantly associated with NSCL/P in Asian and Amerindian populations. The associated V allele is evolutionarily conserved and its frequency is very high in European and African populations (>97%). This SNP may be a surrogate for a true etiologic variant as it is located in an approximately 140Kb-wide linkage disequilibrium (LD) block. Sequencing of the protein coding and splice site regions of IRF6 in 160 NSCL/P cases did not detect any obvious causative variants2. Based on these observations, we postulated that an etiological variant was in strong LD with the V allele and would reside in a regulatory element of IRF6 within the LD block.
To identify potential cis-regulatory elements for IRF6 we obtained and aligned genomic sequences orthologous to a 500Kb region encompassing human IRF6 from 17 vertebrate species. Sequences were first aligned to the human reference sequence and then searched for multispecies conserved sequences (MCSs). A total of 407 MCSs, with an average size of 61bp, were identified within the 500Kb examined with their distribution in protein coding and untranslated regions shown in Supplementary Table 1 online.
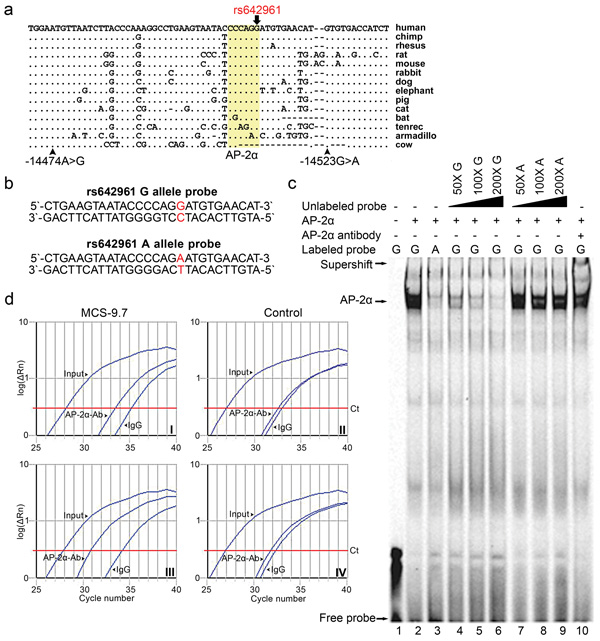
We next selected 41 non-coding MCSs contained within the 140Kb haplotype block of strong LD as most likely to contain one or more SNPs contributing to our previous association with NSCL/P. These sequences were located in the introns, 5′ and 3′ flanking sequences of IRF6 (Supplementary Table 1). The 41 MCSs ranged in size from 25bp to 168bp and were screened for potential causative variants in 184 NSCL/P cases from Iowa and the Philippines by direct sequencing (~7.5Kb of sequence in total). Overall, 18 variants were detected, of which 12 were previously identified (in dbSNP), and 6 were novel. To determine whether the frequencies of the detected variants were different in cases versus controls we sequenced an equal number of unaffected individuals from matched populations. Among 18 variants, only three SNPs (all located within a 50bp segment in MCS-9.7) (Fig. 1a) showed differences between cases and controls with P<0.05 (uncorrected for multiple comparisons as this was hypothesis generating to identify likely mutation sites). The common ancestral alleles of two of these SNPs, -14474A>G and -14523G>A, were overrepresented in cases compared with the controls from the Iowa population (χ2=6.12, df=1, P=0.01) and were in complete LD (r2=1) with each other and with the V allele of V274I. The remaining SNP rs642961, located between these two SNPs, also showed significant differences in allelic frequencies (χ2=4.8, df=1, P<0.02) and genotypic distribution (χ2=6.1, df=2, P<0.04) between cases and controls from the Iowa population. In contrast to -14474A>G and -14523G>A, the associated allele of rs642961 is the derived allele A, while its ancestral G allele is strongly conserved across 12 different vertebrates (Fig. 1a). Transcription factor binding site analysis predicted that the risk allele would alter a binding site for AP-2α, a transcription factor involved in craniofacial development8. Moreover, mutations in AP-2α cause branchio-oculo-facial syndrome9 that has overlapping features with VWS such as orofacial clefting and occasional lip pits, thus making rs642961 a good candidate for an etiological variant.
Figure 1.
Transcription factor AP-2α binds to MCS-9.7 and rs642961 disrupts its binding site. (a) Multispecies sequence alignment of the MCS-9.7 segment corresponding to nucleotides from –14470 to –14535, with respect to IRF6 translation initiation codon, that contains variants -14474A>G, rs642961 and -14523G>A and the predicted AP-2α binding site (highlighted in yellow). (b) The double-stranded IRDye-700 labeled oligonucleotide probes used in EMSA. The probes correspond to MCS-9.7 nucleotides –14495 to –14522 and differ only at the rs642961 SNP. (c) EMSA using probe alone (lane 1) and incubated with the human recombinant AP-2α protein, allele G (lane 2) and allele A (lane 3). Competition with increasing amounts of unlabeled probes with allele G (lanes 4–6) and allele A (lanes 7–9). Supershift is formed when AP-2α is pre-incubated with anti-AP-2α antibody (AP-2α-Ab) (lane 10). (d) Quantification of ChIP by real-time PCR. AP-2α-Ab and IgG immunoprecipitated chromatin fragments were amplified using primers flanking the MCS-9.7 segment with four putative AP-2α binding sites shown in Supplementary Figure 1 and a control region of a similar length devoid of predicted AP-2α binding sites. The curves represent the accumulation of PCR products given as log values (logΔR) of SYBR-Green fluorescence intensity plotted against the number of cycles. MCS-9.7 amplification plot of the immunoprecipitated chromatin from uninfected (Plot I) and ad-AP-2α-infected (Plot III) HaCaT cells. Amplification curves of the control region are shown on Plot II for the uninfected cells and on Plot IV for the infected cells.
We then assessed association between NS clefts and SNPs rs642961 (G>A) and rs2235371 (V274I) using family-based transmission disequilibrium tests (TDT) in 432 Norwegian, 479 Danish, 606 other European (Netherlands, UK, Italy) nuclear families from the EUROCRAN Project , 196 large multiplex Filipino families and 490 Filipino trios (Table 1). The CL subset consists of those families in which one or more of the affected family members have cleft lip alone, while all affected family members in the CLP subset have cleft lip and cleft palate. The CL/P subgroup is a combination of the CL and CLP subgroups. The families in the PALATE subgroup have at least one affected individual with cleft palate alone. Parent-to-offspring observed transmission values were compared with the expected transmission values using the family based association test (FBAT)10 for each SNP and haplotypes of the two SNPs. Table 1 presents the results for the CL/P group by population. We found statistically significant overtransmission of the A allele at rs642961 to affected individuals in all populations separately and combined: Norwegian (P=0.005), Danish (P=0.0001), EUROCRAN (P=0.003), All European populations (P=3×10−8), Filipino (P=6×10−6) and all populations combined (P=1×10−11).
Table 1. Association analysis (TDT), both allelic and haplotype, between NSCL/P and IRF6 SNPs rs642961 (G>A) and rs2235371 (V274I) in Norwegian, Danish, European and Filipino families as calculated in the full dataset (including intact extended kindreds).
Statistically significant P-values (i.e. P-values ≤ 0.0016) for positive associations are in bold, and for negative associations are in italics
| Population (N families) | TDT results | Haplotype TDT P-value (haplotype frequency) | |||||
|---|---|---|---|---|---|---|---|
| rs2235371 (V274I) | rs642961 | Overall | V-G | V-A | I-G | I-A | |
| P-value (inf fam, V freq a) | P-value (inf fam, A freq a) | P-value | P-value | P-value | P-value | P-value | |
| Norway (314) | 0.09 (22, 98%) | 0.005 (130, 26%) | 0.01 | 0.01 (72%) d | 0.004 (26%) | 0.35 (2%) | NA e |
| Denmark (360) | NA (8, 98%) | 1e-04 (165, 24%) | 0.003 | 5e-04 (73%) d | 3e-04 (25%) | 0.76 (2%) | NA ( <1%) e |
| EUROCRAN b (453) | 0.64 (17, 98%) | 0.003 (233, 25%) | 0.02 | 0.003 (74%) d | 0.003 (25%) | 0.72 (1%) | NA (<1%) e |
| Europec (1127) | 0.49 (47, 98%) | 3e-08 (528, 24%) | 3e-06 | 4e-07 (73%) d | 2e-07 (25%) | 0.87 (2%) | NA (<1%) e |
| Philippines (657) | 6e-12 (337, 75%) | 6e-06 (359, 32%) | 7e-11 | 0.38 (47%) | 1e-06 (31%) | 1e-09 (21%) d | 0.007(0.5%) d |
| TOTAL CL/P(1784) | 1e-11 (384, 88%) | 1e-11 (887, 27%) | 9e-16 | 0.005 (61%) d | 8e-13 (30%) | 4e-09 (11%) d | 0.004 (<1%) d |
| SUBSETS: | |||||||
| TOTAL CL(743) | 1e-07 (162, 89%) | 1e-11 (399, 30%) | 1e-11 | 0.001(58%) d | 5e-11(30%) | 1e-06 (12%) d | NA (<1%) e |
| TOTAL CLP(1041) | 1e-05 (222, 89%) | 8e-04 (488, 25%) | 3e-05 | 0.43 (63%) | 4e-04 (26%) | 5e-04 (11%) d | NA (<1%) e |
| TOTAL PALATE(N=419) | 0.78 (42, 97%) | 0.67 (179, 22%) | 0.95 | 0.76 (73%) | 0.79 (21%) | 0.89 (5%) | NA (<1%) e |
V freq: frequency of associated V allele of rs223537 (V274I) and A freq: frequency of associated A allele of rs642961. Frequencies are estimated from the founders in the family data using FBAT program.
EUROCRAN are trios from the Netherlands (CL/P=274, PALATE=63), United Kingdom (CL/P=114, PALATE=67) and Italy (CL/P=64, PALATE=23) combined.
Europe is the combination of data from EUROCRAN Project , Norway (CL/P=314, PALATE=118) and Denmark (CL/P=360, PALATE=119).
Negative association of haplotype.
NA: Not Applicable: the haplotype I-A does not exist or exists with frequency <1% and/or in less than 10 informative families.
Haplotype TDT analysis showed that rs642961 splits the V allele of V274I into two distinct haplotypes, V-G and V-A. Haplotype V-A showed strong evidence of overtransmission (P= 8×10−13) in the CL/P group, whereas haplotypes V-G, I-G, and I-A were significantly undertransmitted, i.e. negatively associated (P=0.005, P=4×10−9 and P=0.04 respectively) (Table 1). These haplotype results demonstrate a strong association with the haplotype containing the rs642961 A allele and suggest that there is not an independent association with the V allele of rs2235371 (V274I). Furthermore, the association with haplotype V-A was more strongly associated in the CL subset (P=5×10−11) than in the CLP subset (P=0.0004), and not associated in the PALATE subset (P=0.79). These patterns are consistent across populations.
We next utilized conditional haplotype analyses in the proband nuclear families derived from each extended kindred, by cleft subgroups (Table 2). First we contrasted the risk of the V-A haplotype to that of the V-G haplotype to test further whether rs2235371 has an association with clefting independent of the rs642961 association. In every population, plus combined Europe and total, V-A had a significantly increased risk over V-G in the CL/P and CL groups (i.e., OR>1.0), lending further support to the notion that the V allele of rs2245371 does not have an association with clefting independent of the rs642961 A allele association with CL/P and CL. In contrast, there were no significant differences in V-A vs V-G risk in the PALATE subgroup for any population. The CLP subgroup showed significant results in Europe but not in the Filipinos.
Table 2.
Comparison of haplotype risks for cleft phenotypic subsets, and haplotype effects odds ratios (OR), for specific rs642961 and rs2235371 (V274I) haplotypes, calculated in the proband nuclear families only
| Population | CL Haplotype V-A vs. V-G | CL Haplotype V-G vs. I-G | CLP Haplotype V-A vs. V-G | CLP Haplotype V-G vs. I-G | ||||
|---|---|---|---|---|---|---|---|---|
| LRT a P-value | V-A OR (95% CI) b [V-G reference] | LRT P-value | V-G OR (95% CI) [I-G reference] | LRT P-value | V-A OR (95% CI) [V-G reference] | LRT P-value | V-G OR (95% CI) [I-G reference] | |
| Norway (N) | 0.040 | 1.54 (1.01–2.35) | 0.237 | 2.51 (0.50–12.66) | 0.142 | 1.31 (0.91–1.88) | 0.402 | 1.54 (0.55–4.28) |
| Denmark (DK) | 0.008 | 1.58 (1.12–2.24) | 0.287 | 0.31 (0.03–3.15) | 0.002 | 1.77 (1.20–2.59) | 0.665 | 1.50 (0.24–9.45) |
| EUROCRAN | 0.001 | 1.72 (1.24–2.39) | 0.336 | 2.25 (0.20–25.22) | 0.301 | 1.16 (0.88–1.54) | 0.119 | 0.37 (0.10–1.40) |
| Europe | 3e-06 | 1.62 (1.32–2.00) | 0.413 | 1.47 (0.58–3.76) | 0.002 | 1.34 (1.11–1.62) | 0.849 | 0.93 (0.46–1.89) |
| Philippines | 0.002 | 1.50 (1.16–1.95) | 0.020 | 1.42 (1.05–1.92) | 0.618 | 1.07 (0.83–1.37) | 9e-04 | 1.55 (1.19–2.02) |
| TOTAL | 3e-08 | 1.57 (1.34–1.85) | 0.013 | 1.42 (1.07–1.88) | 0.005 | 1.24 (1.07–1.45) | 0.008 | 1.37 (1.08–1.73) |
| Population | CL/P (CL + CLP) Haplotype V-A vs. V-G | CL/P (CL + CLP) Haplotype V-G vs. I-G | PALATE Haplotype V-A vs. V-G | PALATE Haplotype V-G vs. I-G | ||||
| LRT P-value | V-A OR (95% CI) [V-G reference] | LRT P-value | V-G OR (95% CI) [I-G reference] | LRT P-value | V-A OR (95% CI) [V-G reference] | LRT P-value | V-G OR (95% CI) [I-G reference] | |
| Norway (N) | 0.013 | 1.41 (1.07–1.85) | 0.177 | 1.78 (0.75–4.20) | 0.683 | 0.91 (0.56–1.45) | 0.173 | 1.98 (0.72–5.40) |
| Denmark (DK) | 7e-05 | 1.65 (1.28–2.13) | 0.700 | 0.77 (0.20–2.91) | 0.512 | 0.86 (0.55–1.35) | 0.910 | 1.12 (0.16–7.94) |
| EUROCRAN | 0.003 | 1.38 (1.11–1.70) | 0.559 | 0.76 (0.30–1.93) | 0.469 | 1.15 (0.78–1.70) | 0.394 | 0.49 (0.09–2.67) |
| Europe | 5e-08 | 1.47 (1.28–1.69) | 0.728 | 1.10 (0.63–1.93) | 0.922 | 0.99 (0.77–1.27) | 0.517 | 1.29 (0.60–2.75) |
| Philippines | 0.009 | 1.27 (1.06–1.52) | 1e-04 | 1.47 (1.21–1.79) | 0.338 | 0.66 (0.28–1.57) | 0.493 | 0.75 (0.32–1.73) |
| TOTAL | 4e-09 | 1.39 (1.25–1.55) | 4e-04 | 1.38 (1.15–1.65) | 0.680 | 0.95 (0.75–1.21) | 0.802 | 0.93 (0.55–1.60) |
Likelihood Ratio Test (LRT) of risks of specified haplotypes: under the null, their risks are set to equal, under the alternative, the risks are assumed not equal (freely estimated), all other haplotype risks freely estimated under both null and alternative (background/nuisance parameters).
Odds Ratio of the estimated haplotype effects associated with the specified haplotype compared to the effects associated with the reference haplotype. CI, confidence interval.
Then, we contrasted haplotype V-G versus I-G to determine whether the association with rs642961 completely accounted for the IRF6 association (Table 2). For these comparisons, the only significant findings were in the Filipino population, suggesting that rs642961 is etiologic in the European populations but there may be additional alleles leading to clefting in the Filipinos. Interestingly, this finding in the Filipinos was most significant in the CLP and CL/P groups (P=0.0009 and P=0.0001 respectively), and borderline in the CL group (P=0.02). Overall, the haplotype results suggest there may be one or more additional alleles present on the V haplotype background also contributing to clefting and with perhaps a greater effect on CLP than CL. Multiple functional SNPs have previously been reported in the IRF5 gene, encoding another member of the IRF family of transcription factors, associated with systemic lupus erythematosus11. Thus, additional risk variants in IRF6 might increase the risk of clefting independently or synergistically with rs642961.
In order to assess possible dosage effects of the A allele, we used log-linear modeling to determine the Relative Risks (RR) for rs642961 genotypes within each phenotype and population (Table 3), in the proband triads. FBAT association analyses in the proband trios (Table 3) had the same patterns of significance as in the entire extended kindred dataset (Table 2), with CL subset showing highly significant association and the PALATE subgroup showing no association. The genotypic RR results suggest a dosage effect of allele A, for example, in the TOTAL combined population CL subset, the relative risk of the AG genotype is 1.68 versus 2.40 for the AA genotype, in Europe CL 1.91 and 2.29, in Filipinos 1.36 and 2.45; while the GG genotype was either not associated or strongly negatively associated with clefting (Table 3). This dosage effect trend is also seen in most of the individual populations for the CL, CLP and CL/P phenotypic subgroups, while there is no association and no allele dosage effect in the PALATE subgroup.
Table 3.
Allelic associations and genotypic relative risks (RR) for cleft phenotypic subsets and rs642961, calculated in the proband trios only
| CL TDT | CL Genotype RR (95% CI) | CLP TDT | CLP Genotype RR (95% CI) | |||||
|---|---|---|---|---|---|---|---|---|
| Population | A freq (No. inf. fam.) | P-value | AG | AA | A freq (No. inf. fam.) | P-value | AG | AA |
| Norway (N) | 0.31 (67) | 0.02 | 2.17 (1.25–3.78) | 1.99 (0.79–5.02) | 0.22 (90) | 0.15 | 1.32 (0.86–2.03) | 1.56 (0.68–3.58) |
| Denmark (DK) | 0.29 (96) | 0.02 | 1.54 (1.01–2.36) | 2.55 (1.17–5.57) | 0.20 (85) | 0.002 | 2.06 (1.32–3.23) | 2.35 (1.00–5.55) |
| EUROCRAN | 0.26 (117) | 0.001 | 2.17 (1.39–3.37) | 2.44 (1.17–5.10) | 0.22 (157) | 0.32 | 1.19 (0.85–1.68) | 1.24 (0.62–2.47) |
| Europe | 0.28 (280) | 4e-06 | 1.91 (1.46–2.49) | 2.29 (1.44–3.63) | 0.21 (332) | 0.003 | 1.41 (1.13–1.77) | 1.58 (1.01–2.47) |
| Philippines | 0.33 (174) | 0.004 | 1.36 (0.97–1.91) | 2.45 (1.45–4.13) | 0.32 (220) | 0.13 | 1.20 (0.90–1.60) | 1.37 (0.81–2.32) |
| TOTAL | 0.30 (454) | 6e-08 | 1.68 (1.37–2.07) | 2.40 (1.70–3.39) | 0.25 (552) | 0.001 | 1.33 (1.11–1.59) | 1.49 (1.06–2.10) |
| CL/P (CL + CLP) TDT | CL/P (CL + CLP) Genotype RR (95% CI) | PALATE TDT | PALATE Genotype RR (95% CI) | |||||
| Population | A freq (No. inf. fam.) | P-value | AG | AA | A freq (No. inf. fam.) | P-value | AG | AA |
| Norway (N) | 0.26 (157) | 0.009 | 1.61 (1.15–2.26) | 1.70 (0.92–2.26) | 0.19 (49) | 0.51 | 0.98 (0.58–1.65) | 1.09 (0.32–3.77) |
| Denmark (DK) | 0.24 (181) | 0.0002 | 1.76 (1.30–2.39) | 2.46 (1.38–4.37) | 0.22 (54) | 0.63 | 1.13 (0.68–1.88) | 0.19 (0.02–1.50) |
| EUROCRAN | 0.24 (274) | 0.003 | 1.51 (1.16–1.97) | 1.64 (1.00–2.69) | 0.24 (82) | 0.49 | 1.37 (0.86–2.19) | 0.84 (0.29–2.44) |
| Europe | 0.24 (612) | 1e-07 | 1.61 (1.36–1.91) | 1.87 (1.36–2.58) | 0.22 (185) | 0.84 | 1.16 (0.87–1.55) | 0.65 (0.32–1.35) |
| Philippines | 0.32 (394) | 0.002 | 1.27 (1.02–1.58) | 1.83 (1.27–2.65) | 0.29 (14) | 0.25 | 0.58 (0.19–1.79) | 0.32 (0.03–3.29) |
| TOTAL | 0.27 (1006) | 2e-09 | 1.47 (1.28–1.68) | 1.87 (1.47–2.38) | 0.22 (199) | 0.66 | 1.11 (0.84–1.47) | 0.62 (0.31–1.23) |
To determine the population impact of the risk allele between cases and controls we genotyped rs642961 in two cohorts of unbiased case collections born within defined time periods from Denmark (107 cases and 495 controls,1997–200312) and Norway (406 cases and 750 controls, 1996–200113). The A allele was more common in all cases combined than in controls from both groups. Consistent with the transmission results in the family data, the frequency of the risk allele was significantly higher in cases with cleft lip only (CLO) versus other cleft phenotypes and the CLO odds ratio was 1.99 (95% CI 1.54–2.57, P=3×10−7) (Table 4). The fraction of NSCLO cases attributable to the A allele of rs642961 is 18% in these two populations combined. Similar to the transmission results in the family data, although the risk allele showed a non-significant trend towards association with CLP (OR=1.23, 95% CI 0.97–1.57, p=0.08), it showed no association with cleft palate only (CPO) (P=0.1).
Table 4.
Association of the rs642961 A allele with various types of clefts in Norwegian and Danish cases versus controls
| Population (N) | rs642961, allele A |
||||
|---|---|---|---|---|---|
| Freq | OR | 95% CI | P-value | PAR % | |
| Norway | |||||
| CL/P and CPO (406) | 0.27 | 1.29 | 1.05–1.57 | 0.01 | 6% |
| CL/P (298) | 0.30 | 1.48 | 1.19–1.83 | 0.0005 | 10% |
| CLP (184) | 0.26 | 1.22 | 0.93–1.61 | 0.2 | 5% |
| CLO (114) | 0.36 | 1.94 | 1.44–2.62 | 0.00002 | 17% |
| CPO (108) | 0.19 | 0.82 | 0.55–1.22 | 0.3 | 0% |
| Controls (750) | 0.22 | ||||
| Denmark | |||||
| CL/P and CPO (107) | 0.27 | 1.28 | 0.91–1.79 | 0.2 | 6% |
| CL/P (70) | 0.32 | 1.67 | 1.13–2.45 | 0.009 | 13% |
| CLP (37) | 0.27 | 1.30 | 0.76–2.23 | 0.3 | 6% |
| CLO (33) | 0.38 | 2.15 | 1.28–3.61 | 0.003 | 20% |
| CPO (37) | 0.16 | 0.68 | 0.36–1.29 | 0.2 | 0% |
| Controls (495) | 0.22 | ||||
| Combined | |||||
| CL/P and CPO (513) | 0.27 | 1.28 | 1.08–1.52 | 0.004 | 6% |
| CL/P (368) | 0.30 | 1.51 | 1.26–1.82 | 0.00002 | 10% |
| CLP (221) | 0.26 | 1.23 | 0.97–1.57 | 0.08 | 5% |
| CLO (147) | 0.36 | 1.99 | 1.54–2.57 | 0.0000003 | 18% |
| CPO (145) | 0.18 | 0.79 | 0.57–1.09 | 0.13 | 0% |
| Controls (1245) | 0.22 | ||||
CL/P, Cleft Lip with or without cleft Palate; CLP, Cleft Lip with cleft Palate; CLO, Cleft Lip Only; CPO, Cleft Palate Only; OR, Odds Ratio; CI, Confidence Interval; PAR, Population Attributable Risk.
Previously, most studies have combined CLO and CLP into common etiologic and recurrence risk groups14. Recent epidemiologic evidence suggests that CLO might be separable15. In the current study there was a clear separation of risk and transmission patterns between the CLO and CLP groups based on rs642961 genotype. Further, we recently updated our large genome-wide linkage analysis16 of NSCL/P to double the sample size (now 861 multiplex families) and found genome-wide significant linkage (LOD score = 3.34) with the IRF6 region attributable to the CLO subset. We also determined the worldwide distribution of rs642961 by genotyping the samples from the CEPH diversity panel. African populations have the lowest derived allele frequency (0.11), and Native Americans have the highest (0.27) while European and Asian frequencies differ based on geographic origins (Supplementary Table 2). These frequency differences broadly mirror the observed prevalence differences in orofacial clefting across these populations and, coupled to the high frequency of this risk allele in all populations, suggest the possibility of a selective advantage for this or a linked variant in Asian and European groups17
To determine whether rs642961 affects DNA binding by transcription factor AP-2α we performed electrophoretic mobility shift assays (EMSA) using human recombinant AP-2α protein and fluorescently labeled oligonucleotide probes containing rs642961 (Fig. 1b). The oligonucleotide probe containing the G allele robustly bound the AP-2α protein, whereas an oligonucleotide probe containing the A allele did not bind at all (Fig. 1c). Increasing the amount of unlabeled G probe efficiently competed with the binding of the labeled G probe. In contrast, increasing amounts of unlabeled A probe had no effect on the binding activity of the labeled G probe, indicating a specific interaction between the G allele and AP-2α.
Further analysis of the region surrounding rs642961 revealed three additional highly conserved AP-2α binding motifs (Supplementary Fig. 1). A chromatin immunoprecipitation (ChIP) assay, coupled with quantitative real-time PCR, showed 2.6-fold enrichment for the MCS-9.7 region in AP-2α-Ab immunoprecipitated chromatin relative to IgG-precipitated chromatin from HaCaT keratinocyte cells homozygous for rs642961-G (Fig. 1d, plot I). In cells overexpressing AP-2α the MCS-9.7 chromatin enrichment was 10-fold (Fig. 1d, plot III). No difference in relative enrichment was observed for a control region devoid of AP-2α binding sites in either cells (Fig. 1d, plots II–IV). Taken together, these results demonstrate that the MCS-9.7 region is a direct target of AP-2α in vivo.
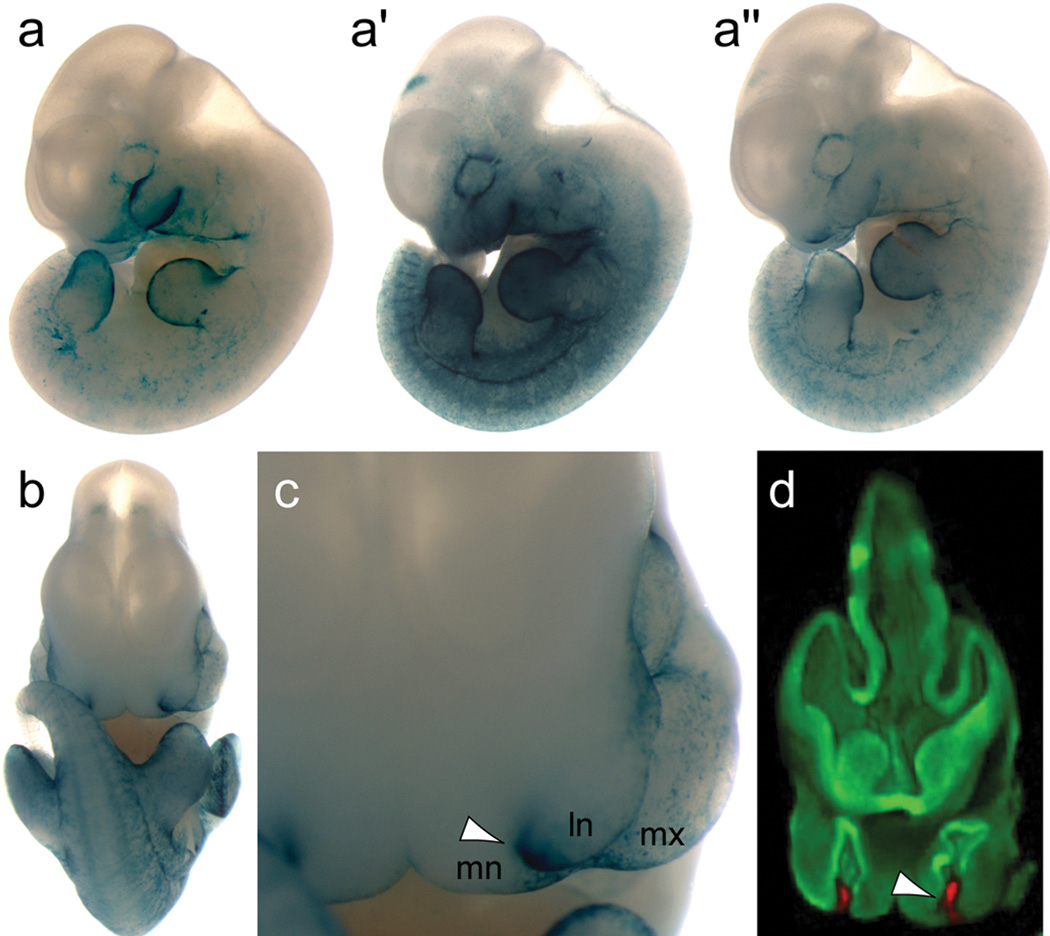
Next, we assessed the regulatory activity of MCS-9.7 in a transgenic mouse enhancer assay. A 775bp (chr1:208055673–208056447, UCSC hg18) of human genomic sequence that contains the entire MCS-9.7 region was inserted into a previously described expression vector18 in which the LacZ reporter gene is driven by a minimal mouse heat shock promoter. Eight out of nine F0 embryos reproducibly expressed LacZ in the ectoderm covering facial prominences, predominantly at the fusion sites between the prominences, and developing limbs, and in the branchial arch (Fig. 2), consistent with known sites of endogenous craniofacial Irf6 expression at this developmental stage19 and suggesting that the MCS-9.7 region functions as an enhancer element for IRF6. Three-dimensional digital imagery and virtual sections of LacZ-stained whole-mount embryos captured using optical projection tomography20 are available in Supplementary videos online.
Figure 2.
MCS-9.7 shows IRF6 enhancer activity in transgenic mouse assay. (a-a”) Lateral views of three independent transgenic embryos at embryonic day 11.5 (E11.5) expressing LacZ directed by MCS-9.7. (b) Frontal view and (c) expanded view of the orofacial region of the embryo shown in panel a”. White arrow points to LacZ expression at the fusion sites between the lateral nasal (ln), medial nasal (mn) and maxillary (mx) prominences towards the end of upper lip formation at E11.5. (d) Sagittal virtual section through the orofacial region of the embryo shown in panel a” generated with optical projection tomography. Shown in red (white arrow) is LacZ expression in ectoderm covering fusing facial prominences (ln, mn and mx). See 3D views of embryos a-a″ in Supplementary videos online
In order to asses the effects of rs642961 on IRF6 expression we performed luciferase reporter assays in human foreskin keratinocyte cell line. The risk haplotype consistently increased the luciferase expression greater than the non-associated haplotypes, but the difference did not reach statistical significance (Supplementary Fig. 2). Previous studies demonstrated that some regulatory polymorphisms do not always reflect their in vivo effects in cell culture-based assays21, particularly for developmental genes that show temporal and tissue-specific expression pattern. Although rs642961 shows minor effects on reporter gene expression in cell culture, it is possible that it has significantly larger or even opposing effects on IRF6 expression in vivo in its native chromosomal context and/or in the presence of trans-acting variants.
Although linkage and association studies are increasingly effective in identifying loci and putative genes involved in common complex traits, the progression to specific etiologic variant identification has proven difficult. Demonstrating that a specific variant is etiologic in what may be a large haplotype block with many strongly correlated SNPs, requires compelling statistical evidence coupled with convincing functional data for a specific variant. Previous sequencing of candidate genes in NSCL/P cases has disclosed rare mutations in individual families22,23 but no common causative variants have been described. In the present study we report a common single point mutation in a gene regulatory element that confers an 18% attributable risk for isolated cleft lip.
METHODS
Subjects
Written informed consent was obtained from all participants in compliance with The University of Iowa Institutional Review Board (approval nos. 199804080 and 199804081). The Danish family data consisted of: 362 NSCL/P nuclear pedigrees, with 23 affected (step) and 845 unaffected (step) siblings for a total of 1624 individuals. The Danish case control data consisted of 107 cases and 495 controls unrelated to the family data. The Norwegian family data consisted of 314 NSCL/P nuclear pedigrees, with 1 affected sibling, 1 unaffected sibling and 237 siblings with unknown affection status for a total of 1181 individuals. The Norwegian case-control data consisted of 298 cases who were the probands from the families plus 108 cases with CPO and 750 controls unrelated to the family data. The Filipino data consisted of 203 multiplex NSCL/P extended pedigrees containing 932 nuclear families for a total of 2797 individuals. For some analyses CLO and CLP subsets were analyzed from the total Filipino families where the CLO subset consists of those families in which one or more affected individual has CLO (number of CLO families=121), versus the CLP subset in which all affected family members have CL plus CP (number of CLP families=70). The EuroCran data consisted of 432 nuclear trios (263 from the Netherlands, 105 from the UK, and 64 from Italy) for a total of 1296 individuals.
Comparative genomic sequence analysis
Sequences of the genomic region encompassing the IRF6 gene for human and organisms listed in Supplementary Table 3 were compiled from publicly available whole-genome datasets. Orthologous sequences from rabbit, pig, bat, armadillo, and elephant were generated specifically for this study using targeted bacterial artificial chromosome (BAC)-based mapping and sequencing strategy24 (Supplementary Table 3). MultiPipMaker25 was used to align these sequences with the single coverage option that eliminates some matches caused by duplications and the search both strand option. MCSs were identified using the WebMCS program, which calculates a conservation score for each base in the reference sequence by analyzing windows of 25nt across MultiPipMaker-generated multispecies sequence alignment based in the algorithm developed by Margulies et al26.
Prediction of transcription factor binding motifs
Transcription Element Search System (TESS) that is linked to TRANSFAC, JASPAR, IMD, and CBIL-GibbsMat databases was used to scan MCSs for previously reported transcription factor binding sites.
Sequencing
Primers used to amplify MCSs were designed with Primer 3 (v.0.3.0) program and are available in Supplementary Table 4 online. Cycle sequencing was performed on 1µl of 1/10 diluted PCR product in 10µl reaction by using 0.25µl of ABI Big Dye Terminator sequencing reagent (v. 1.1), 0.5µl of 5μM sequencing primer, 0.5µl of DMSO, 1µl of 5X sequencing buffer and 6.75µl of ddH2O. Following a denaturation step at 96°C for 30 sec, reactions were cycle sequenced at 96°C for 10 sec, at primer Tm for 5 sec, and at 60°C for 4 min for 40 cycles. Unincorporated dyes and reaction buffers were removed by Sephadex™ G-50 columns (GE Healthcare) or magnetic beads (Agencourt) and the sequencing reaction was then injected on an ABI 3730 capillary sequencer (Applied Biosystems). Chromatograms were transferred to a Unix workstation, base called with PHRED (v.0.961028), assembled with PHRAP (v. 0.960731) scanned by POLYPHRED (v. 0.970312) and the results viewed with the CONSED program (v. 4.0)27
Genotyping and statistical analysis
Genotyping was carried out by using TaqMan® SNP Genotyping Assays on the ABI Prism 7900HT machine and analyzed with SDS 2.2 software (Applied Biosystems). The genotyping success rate was extremely high (>98%) and the Mendelian error rate was less than 2% (some of which may be due to potential deletions in this region). For the family data, SNP and haplotype TDT analyses were performed with the FBAT program (v.1.7.3), using the –e option of the program to account for multiple sibs in a family or multiple nuclear families in a pedigree. With a Bonferroni correction, the alpha level for significance of the association analyses was calculated as 0.05/32=0.0016. In order to assess possible dosage effects of the associated allele, log-linear models with genotype and imprinting effects were fit to the proband-parent triads using the SAS version of the LRT program28. Likelihood ratio tests of the model parameter effects were then used to estimate the RR (with 95% CI) for over-transmission of 1 versus 2 associated alleles (A allele) at SNP rs642961. In these same proband-parent triads plus proband siblings if available (ie proband nuclear families), we compared the risk associated with haplotype V-A to that of haplotype V-G, as well as the risks associated with haplotypes I-G vs. V-G. We also estimated the OR of the effects due to a particular haplotype with respect to a specific reference haplotype. The 95% CI associated with the OR were also estimated. The haplotype RR and OR were calculated using LRT methods, as implemented in the software Unphased29. In the case-control data, OR with Fisher’s exact P-values were calculated using SAS v.9.1.3 (Cary, NC) software. The population attributable risk percentage (PAR%) was calculated based on the following formula, PAR%= [Pe (RR-1)/RR] ×100, where Pe is the prevalence of the allele in cases, and RR is the relative risk. The OR was used as a proxy for the RR. The Danish case-control data was from the singleton cases and controls that were ascertained separately from the cleft family data. The Norwegian case-control data consisted of the probands from the cleft family data and the probands from a control family data.
EMSA
Infrared dye (IRDye-700) end-labeled and unlabeled oligonucleotide probes (Fig. 1b) were purchased from LI-COR Biosciences (Lincoln, NE) and IDT (Coralville, IA), respectively. We performed EMSAs by using the LI-COR EMSA Kit following the protocol described in Supplementary methods online.
ChIP assay
Protein cross-linked chromatin from ~2×107 Ad-AP-2α-infected and uninfected HaCaT keratinocyte cells was isolated as detailed previously30. The amount of immunoprecipitated target region was then determined by SYBR Green (Applied Biosystems) quantitative real-time PCR with primers specific for the target sequence in MCS-9.7 and control region (Supplementary Methods and Table 4). Real-time PCR was carried out in triplicate and amplification of the target amplicon was monitored as a function of increased SYBR Green fluorescence. An analysis threshold was set and the cycle threshold (Ct) computed for each sample. Fold enrichment of target sequence was calculated using the following formula (Fold enrichment = 2(Ct AP-2α-Ab IPed)−(Ct IgG IPed)).
Luciferase assay
For luciferase reporter assay we generated reporter constructs by inserting 540bp genomic segment (chr1:208055787–208056326, UCSC hg18) containing the entire MCS-9.7Kb region upstream of firefly luciferase ORF driven by the SV40 promoter. DNA samples from individuals homozygous for -14474A>G, -14523G>A, and rs642961 variants were PCR amplified and cloned into the pGL3-Basic and pGL3-Promoter vectors (Promega) in both orientations. Luciferase assays were performed in HFK cells as described in Supplementary methods. The assays were performed in triplicates in seven independent experiments on separate days. The relative luciferase activity was calculated by dividing the luminescence of firefly luciferase activity by that of the cotransfected Renilla luciferase, and pairwise comparisons of luciferase expression level from different constructs were done using 2-tailed Student’s t-test.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Akira Kinoshita, Kathy Frees, Adela Mansilla, Jamie L'Heureux, Marla Johnson, Harris Morrison, George Wehby, Nicholas Rorick, Kurt Bedell, and Linda Powers for technical assistance and Susie McConnell, Dan Benton and Melanie DeVore for their administrative assistance. We would also like to thank Al Klingelhutz for kindly providing us with HFK cell line. This work was supported by grants from the National Institutes of Health (NIH): P50 DE16215 (JCM, MLM, BCS), P30 ES05605 (JCM), R37 DE08559 (JCM, MLM), R01-DE13513 (BCS), 1 UL1 RR024979-01 (JCM, BCM), R01-CA73612 (FED), R01-HG003988 administered under Department of Energy Contract DE-AC02-05CH11231 (LAP) as well as by the Intramural Research Program of the National Human Genome Research Institute (EDG), in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (AJW), and European Commission FP5: EUROCRAN Project (Contract No. QLG1-CT-2000-01019) (MR, PAM, JL, RPS). AV was supported by the American Heart Association
REFERENCES
- 1.Jugessur A, Murray JC. Orofacial clefting: recent insights into a complex trait. Curr Opin Genet Dev. 2005;15:270–278. doi: 10.1016/j.gde.2005.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zucchero TM, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 3.Kondo S, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scapoli L, et al. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet. 2005;76:180–183. doi: 10.1086/427344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanton SH, et al. Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am J Med Genet A. 2005;137:259–262. doi: 10.1002/ajmg.a.30887. [DOI] [PubMed] [Google Scholar]
- 6.Ghassibe M, et al. Interferon regulatory factor-6: a gene predisposing to isolated cleft lip with or without cleft palate in the Belgian population. Eur J Hum Genet. 2005;13:1239–1242. doi: 10.1038/sj.ejhg.5201486. [DOI] [PubMed] [Google Scholar]
- 7.Park JW, et al. Association between IRF6 and nonsyndromic cleft lip with or without cleft palate in four populations. Genet Med. 2007;9:219–227. doi: 10.1097/GIM.0b013e3180423cca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 9.Milunsky JM, et al. TFAP2A mutations result in branchio-oculo-facial syndrome. Am J Hum Genet. 2008;82:1171–1177. doi: 10.1016/j.ajhg.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 11.Graham RR, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bille C, et al. Oral clefts and life style factors--a case-cohort study based on prospective Danish data. Eur J Epidemiol. 2007;22:173–181. doi: 10.1007/s10654-006-9099-5. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen RH, Wilcox AJ, Moen BE, McConnaughey DR, Lie RT. Parent's occupation and isolated orofacial clefts in Norway: a population-based case-control study. Ann Epidemiol. 2007;17:763–771. doi: 10.1016/j.annepidem.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell LE, et al. Guidelines for the design and analysis of studies on nonsyndromic cleft lip and cleft palate in humans: summary report from a Workshop of the International Consortium for Oral Clefts Genetics. Cleft Palate Craniofac J. 2002;39:93–100. doi: 10.1597/1545-1569_2002_039_0093_gftdaa_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 15.Harville EW, Wilcox AJ, Lie RT, Vindenes H, Abyholm F. Cleft lip and palate versus cleft lip only: are they distinct defects? Am J Epidemiol. 2005;162:448–453. doi: 10.1093/aje/kwi214. [DOI] [PubMed] [Google Scholar]
- 16.Marazita ML, et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mossey P. Epidemiology underpinning research in the aetiology of orofacial clefts. Orthod Craniofac Res. 2007;10:114–120. doi: 10.1111/j.1601-6343.2007.00398.x. [DOI] [PubMed] [Google Scholar]
- 18.Poulin F, et al. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics. 2005;85:774–781. doi: 10.1016/j.ygeno.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Knight AS, Schutte BC, Jiang R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- 20.Sharpe J, et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- 21.Cirulli ET, Goldstein DB. In vitro assays fail to predict in vivo effects of regulatory polymorphisms. Hum Mol Genet. 2007;16:1931–1939. doi: 10.1093/hmg/ddm140. [DOI] [PubMed] [Google Scholar]
- 22.Vieira AR, et al. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1:e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riley BM, et al. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci U S A. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas JW, et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz S, et al. MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margulies EH, Blanchette M, Haussler D, Green ED. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads. Am J Hum Genet. 1999;65:229–235. doi: 10.1086/302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Provenzano MJ, et al. AP-2 participates in the transcriptional control of the amyloid precursor protein (APP) gene in oral squamous cell carcinoma. Exp Mol Pathol. 2007;83:277–282. doi: 10.1016/j.yexmp.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.