Abstract
To accelerate gene isolation from plants by positional cloning, vector systems suitable for both chromosome walking and genetic complementation are highly desirable. Therefore, we developed a transformation-competent artificial chromosome (TAC) vector, pYLTAC7, that can accept and maintain large genomic DNA fragments stably in both Escherichia coli and Agrobacterium tumefaciens. Furthermore, it has the cis sequences required for Agrobacterium-mediated gene transfer into plants. We cloned large genomic DNA fragments of Arabidopsis thaliana into the vector and showed that most of the DNA fragments were maintained stably. Several TAC clones carrying 40- to 80-kb genomic DNA fragments were transferred back into Arabidopsis with high efficiency and shown to be inherited faithfully among the progeny. Furthermore, we demonstrated the practical utility of this vector system for positional cloning in Arabidopsis. A TAC contig was constructed in the region of the SGR1 locus, and individual clones with ca. 80-kb inserts were tested for their ability to complement the gravitropic defects of a homozygous mutant line. Successful complementation enabled the physical location of SGR1 to be delimited with high precision and confidence.
Molecular genetic approaches have been applied to analysis and cloning of plant genes, particularly those involved in complex biological processes such as developmental regulation and gene expression cascades (1, 2). Genes defined by mutations are isolated by positional cloning as well as by DNA tagging. For positional cloning, efforts have been devoted to producing numerous sets of DNA markers and genomic DNA libraries from various plant species by using artificial chromosomes propagated in either yeast artificial chromosome (YAC) or bacteria artificial chromosome (BAC and P1) (3–5). Therefore, an initial mapping of target gene loci using DNA markers and subsequent isolation of large, overlapping genomic DNA fragments in the target region by chromosome walking or landing have become easier in several plant species, including Arabidopsis thaliana (1) and rice (6).
Proof of successful gene identification and cloning usually requires complementation of the mutant phenotype by transformation with a wild-type allele. The major drawback of positional cloning, however, is the difficulty of narrowing down the field of candidate clones to a manageable number for complementation testing. For fine-scale mapping of a mutation locus, it is usually necessary to analyze nearly a thousand progeny (usually F2 plants) or even more if the locus falls in a “recombination cold spot,” a chromosomal region of low recombination frequency (7). Furthermore, even low levels of misscoring during mapping (because of subtlety or incomplete penetrance of the mutant phenotype) will reduce mapping precision to the point that cloning becomes impractical. In addition, even after accurate mapping, present positional cloning procedures that use YAC or BAC clones require subcloning of many small fragments into a transformation-competent vector for complementation testing. In many cases, these steps are rate-limiting hurdles to positional cloning. Therefore, to accelerate positional cloning, it is highly desirable to exploit a strategy that streamlines complementation testing.
Plant transformation-competent vectors, such as the cosmid vector pOCA18 (8) and the λ-phage vector λTI2 (9), have been developed for construction of genomic libraries with inserts of 5–25 kb that are used for genetic complementation of mutants. The low cloning capacity of these vectors, however, limits their usefulness for efficient gene isolation by positional cloning. In a previous report on an Arabidopsis genomic DNA library prepared by using a P1 phage vector (10), we suggested that if large DNA fragments could be transferred directly from P1-based clones into plants, it would greatly accelerate positional cloning of plant genes. Recently, a 150-kb human DNA fragment was transferred into the tobacco genome by using a binary-BAC vector by Agrobacterium-mediated transformation (11, 12). Here we report a vector system for constructing transformation-competent artificial chromosome (TAC) libraries. Our results show that large genomic DNA fragments of A. thaliana cloned in a TAC vector can be maintained stably in both Escherichia coli and Agrobacterium tumefaciens, transferred with a high efficiency into the Arabidopsis genome, and faithfully inherited in the transgenic progeny. We also show that TAC clones carrying ca. 80-kb genomic DNA fragments of A. thaliana complement a gravitropic mutation at the sgr1 locus.
MATERIALS AND METHODS
Construction of a TAC Vector.
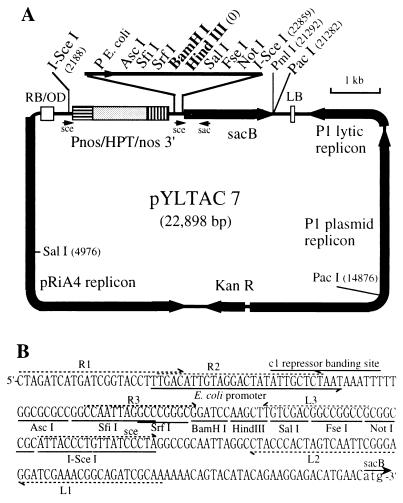
A TAC vector was constructed by using standard cloning procedures. Components of the vector were obtained from various plasmids in the minimum sizes possible. For blunt-end ligation, DNA fragments were treated with Klenow or T4 DNA polymerase. E. coli strain DH10B was used as the cloning host, and transformation of the bacterium was done by electroporation using Gene Pulser (Bio-Rad) according to the supplier’s protocol. To create an initial backbone plasmid (pYL1), a 6.3-kb KpnI-DraIII fragment carrying the P1 plasmid replicon of the P1 vector pAd10sacBII (4) was ligated to a 1.8-kb DraIII-SspI fragment from pACYC177 (New England Biolabs) containing the kanamycin-resistant marker gene (NPT1), in which the HindIII site had been destroyed (Y.-G.L., unpublished results). The P1 lytic replicon obtained from the P1 vector as a 1.7-kb AseI fragment then was inserted into the AatII site of pYL1 to yield pYL2. A T-DNA cassette was constructed in pBluescript II (Stratagene), which consists of components from pGA carrying the octopine-type left and right borders and the “overdrive” enhancer sequence (13) and pGDW32 carrying the hygromycin phosphotransferase gene (HPT) (14). This cassette was cloned into the BamHI site of pYL2 to produce pYL3. The cloning-selection marker gene sacB isolated from pAd10sacBII was inserted between the nopaline synthase gene (nos) 3′ region and the left border to produce pYL4. An 8.1-kb BamHI fragment containing the pRiA4 replicon (15, 16) was inserted into the BstEII site of pYL4. Finally, a synthetic, double-stranded oligonucleotide containing rare-cutter sites and cloning sites (Fig. 1) was introduced between the E. coli promoter and the sacB gene. To create a unique HindIII cloning site in the vector, the HindIII sites of the P1 plasmid replicon (two sites), the P1 lytic replicon (one site), and the pRiA4 replicon (one site) were destroyed sequentially after each component being cloned into the precursor TAC vector by HindIII digestion, end fill-in, and subsequent religation. The two HindIII sites of the sacB structural gene were destroyed by PCR site-directed mutagenesis. The complete sequence of the vector is available at the DDBJ/EMBL/GenBank accession number AB020028.
Figure 1.
Physical map of pYLTAC7. (A) The map shows the location of some sites for endonucleases that cleave the molecular once or twice. LB and RB, left and right borders, respectively; OD, overdrive sequence; Pnos, promoter of the nopaline synthase gene; HPT, coding region of the hygromycin phosphotransferase gene; nos 3′, polyadenylation signals of the nopaline synthase gene; KanR, kanamycin-resistance gene (NPT1). The complete sequence of the vector is available in the GenBank database (accession no. AB020028). (B) Sequence of the cloning-site region upstream of the sacB gene. The primer sets (R1, R2, R3, L1, L2, and L3) are designed for isolation of end fragments of the inserted DNA by thermal asymmetric interlaced PCR (TAIL-PCR) (22).
Construction of a TAC Library of A. thaliana.
Using a nuclei-based method of Liu and Whittier (17), very high molecular weight DNA (>2.5 Mb) was isolated from A. thaliana (Columbia ecotype). The DNA was digested partially with HindIII and size-fractionated in the 75- to 100-kb size range as described (10). The partially digested and size-selected DNA fragments were ligated with HindIII-digested pYLTAC7 and then used for transformation of E. coli DH10B by electroporation. Transformants carrying inserts were selected on LB agar plates containing 25 μg/ml kanamycin and 5% sucrose (4). Details of the library will be published elsewhere.
Plant Transformation.
TAC clones were selected randomly for plant transformation from a genomic DNA library of A. thaliana ecotype Columbia (unpublished results). TAC clones covering the SGR1 locus (18, 19) were isolated from the library by using two restriction fragment length polymorphism markers, CDC2B and KSAP3. E. coli cells carrying these clones were cultured at 37°C in LB containing 25 μg/ml kanamycin. When the cell density reached an OD600 of 0.4–0.8, isopropyl β-d-thiogalactoside was added to a concentration of 0.2 mM and the cells were cultured for an additional 5–12 hr. TAC plasmids were isolated by the alkaline lysis method. The TAC plasmids were introduced into A. tumefaciens strains C58C1(MP90), C58C1(GV2260), and EHA105 by electroporation using Gene Pulser (Bio-Rad) with parameters of 100 or 200 ohms and 2.5 kV/0.2 cm. A. tumefaciens colonies were selected on LB-agar plates containing 20 μg/ml kanamycin for EHA105, 20 μg/ml of kanamycin and 15 μg/ml gentamycin for MP90, or 25 μg/ml carbenicillin for GV2260. These bacteria were used for transformation of A. thaliana plants (3–4 weeks old) of ecotype Wassilewskija (WS) or the sgr1 mutant plants by the vacuum infiltration method (20) with minor modifications. Transformants (T1 generation) were selected on B5 medium containing 1% sucrose, 15 μg/ml hygromycin, and 250 μg/ml claforan (Hoechst–Roussel). Experiments were carried out by using the progeny of the transformants (T2 or T3 plants).
Genomic DNA Analysis.
For PCR analysis, plant genomic DNA was prepared on a small scale as described (21). Two primers, CATTACCCTGTTATCCCTA-3′ (sce) and AGGTTTGCAGAACCGGACC-3′ (sac), were used for amplification of the sacB gene (see Fig. 1). For Southern analysis of high-molecular-weight genomic DNA, megabase nuclear DNA was prepared from A. thaliana plants as described (17). The DNA in low-melting agarose plugs was digested with meganuclease I-SceI (Boehringer Mannheim) and separated on 0.8% agarose gels by field-inversion gel electrophoresis (FIGE) using a PPI-200 power inverter (MJ Research, Cambridge, MA) with programs 3 and 4. The DNA in the gels was irradiated by using UV Stratalinker 2400 (Stratagene) for 5 × 105 J and transferred to Hybond N+ membranes (Amersham) by alkaline transfer. Southern hybridization was done as described (20) by using the HPT gene sequence as a probe.
RESULTS
Design and Construction of the Transformation-Competent Vector.
We designed a TAC vector, pYLTAC7 (Fig. 1), to meet the following requirements: (i) efficient cloning of large DNA fragments, (ii) stable maintenance of inserted fragments in both E. coli and A. tumefaciens, and (iii) competence for transferring inserted DNA into plant genomes via Agrobacterium.
It is known that large, foreign DNA fragments are maintained stably in single-copy plasmids such as P1 or BAC (5, 23). We, therefore, used the P1 bacteriophage replicon (23) and the pRiA4 replicon of the Ri plasmid (15), which render the copy number of the plasmid single in E. coli and Agrobacterium, respectively. In fact, the structural stability of the genomic clones of Arabidopsis in P1 (10) and TAC (this study) vectors in E. coli has been shown during the genome-sequencing project conducted by Kazusa DNA Research Institute (Chiba, Japan) through analysis of a large number of the clones that cover, in total, 94% of chromosome V of A. thaliana (24). The low-yield disadvantage of single-copy plasmids for DNA preparation or library screening is overcome by amplifying the plasmid by releasing the suppresser of the P1 lytic replicon with isopropyl β-d-thiogalactoside (23).
Considering that electroporation is a conventional and efficient technique for transferring large plasmids into Agrobacterium, we did not introduce the pRK2 oriT sequence into our TAC vector, which is necessary for delivering plasmids from E. coli to Agrobacterium by the triparental-mating method (25).
We placed the plant-selectable marker gene (HPT), which is driven by the nopaline synthase gene promoter (Pnos), adjacent to the right border rather than to the left border in the vector. Because T-DNA transfer is initiated from the right border (25), transformants selected by the HPT gene could carry either the entire or truncated T-DNA. Most transgenes analyzed were not truncated (see below).
Because HindIII cohesive ends ligate efficiently, we created a unique HindIII-cloning site in the vector. The vector also has a unique BamHI-cloning site that is suitable for preparing libraries with small (especially <30-kb) Sau3AI/MboI fragments. The HindIII and BamHI sites were inserted between the sacB gene and its promoter. The production of levansucrase encoded by the sacB gene in E. coli is lethal in the presence of 5% sucrose in agar medium (26). Thus, insert-bearing clones can be selected on sucrose-containing agar plates, leading to a low level of “empty vector” transformants in libraries.
Adjacent to the HindIII and BamHI sites, five rare-cutter sites (AscI, SfiI, SrfI, FseI, and NotI) were created, which can be used for preparing nested deletion clones from a large genomic DNA fragment inserted in the vector. Two I-SceI sites flanking the cloning sites and the Pnos sequence were engineered in the vector. With an 18-bp recognition sequence, I-SceI should occur only once in 6.9 × 1010 bp for perfectly random sequence. This design enabled a size check of the entire transferred DNA segment in transgenic plants by probing Southern blots with the Pnos/HPT selection marker sequence.
Stability of TAC Clones.
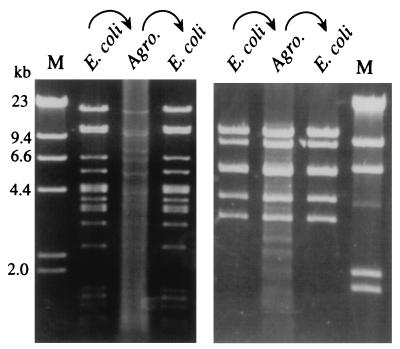
By ligating genomic DNA fragments (ca. 60–100 kb) of Arabidopsis ecotype Columbia into the HindIII site of the vector and subsequently transforming into E. coli DH10B, we constructed a TAC library consisting of ca. 10,000 clones (unpublished results). To investigate structural stability of TAC clones in E. coli and A. tumefaciens strains, 35 E. coli clones were selected randomly from the library and plasmids were isolated. The plasmid DNAs were electroporated into A. tumefaciens strain C58C1(MP90). Thus passaged, the plasmid DNAs were re-isolated from the Agrobacterium transformants and transferred back to E. coli. Restriction analysis of these plasmids indicated that 34 clones were maintained completely intact (Fig. 2). One clone in our experiments was found to be unstable in A. tumefaciens. Therefore, we recommend checking the stability of each TAC clone in A. tumefaciens before plant transformation.
Figure 2.
Stability of two TAC clones in E. coli and A. tumefaciens. TAC plasmid DNA of two independent clones were isolated from E. coli (DH10B) and were used for transformation of A. tumefaciens C58C1(MP90). The plasmid DNA in the Agrobacterium host was transferred back to the E. coli host. Digestion of plasmid DNA in each step is shown in this figure. (Left) A TAC clone digested by HindIII. (Right) Another TAC clone digested by PstI. M, molecular markers.
Transformation of A. thaliana with TAC Clones.
To assess the transformation efficiency with respect to T-DNA sizes, we conducted plant transformation by using TAC clones with or without a large genomic DNA insert. TAC clone 20D10 (the same clone shown in Fig. 4A) carries a ca. 80-kb insert of A. thaliana genomic DNA. This clone and the vector alone, which contains a T-DNA of 4.4 kb between the right and left borders, were introduced into A. tumefaciens strain C58C1(MP90). These plasmids were maintained stably in the host cells. We transformed Arabidopsis ecotype WS by using these bacteria by the vacuum-infiltration protocol. From three independent experiments, we obtained 482 hygromycin-resistant plants from 142 plants treated with the 20D10 bacterium and 688 hygromycin-resistant plants from 127 plants treated with the bacterium carrying the vector alone (Table 1). These results indicate that the transformation efficiency is not affected substantially by the sizes of introduced T-DNA within this range. This efficiency is comparable to those we obtained by using other binary vectors such as pBI121. Transformants also were obtained from the Columbia ecotype with other TAC clones (see Table 2 and data not shown). Our experiments showed that A. tumefaciens strains C58C1(GV2260) and EHA105 also were able to transform Arabidopsis with lower but sufficient efficiencies (data not shown).
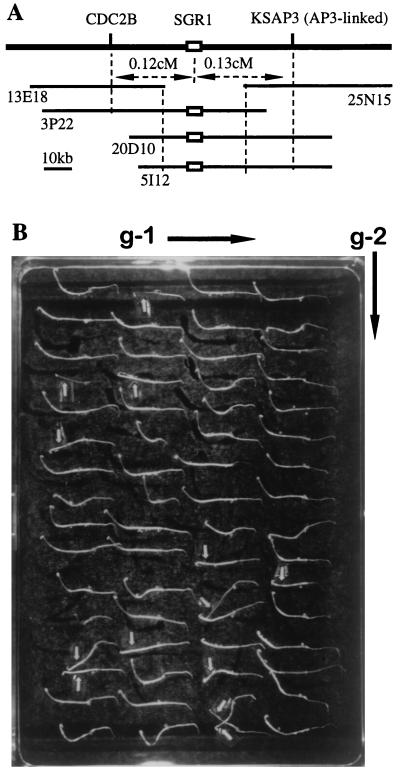
Figure 4.
Complementation of the sgr1 mutation with large TAC clones of wild type. (A) The SGR1 locus was covered contiguously by TAC clones (13E18, 3P22, 20D10, 5I12, and 25N15) carrying large (ca. 80-kb) genomic DNA fragments of A. thaliana Columbia ecotype that were isolated by using two DNA markers, CDC2B and KSAP3. (B) Segregation of the T2 family seedlings of the transformed line E for gravitropic responses. Seedlings were grown in darkness for 3 days after germination with the plate setting as the direction of gravity indicated by g-1. The plate then was turned by 90° as indicated by g-2, and the seedlings were grown in darkness for 24 hr. About 75% of seedlings showed distinct negative gravitropic curvature in hypocotyls as wild type, whereas the remaining (marked by arrows) did not show gravitropic curvature at all (sgr1 mutation phenotype).
Table 1.
Transformation efficiency of A. thaliana with TAC clones
| Exp. | Construct | Transformants/ treated plants | Seeds screened/ transformant |
|---|---|---|---|
| 1 | 1 | 115/46 (2.5) | 770 |
| 2 | 265/43 (6.2) | 404 | |
| 2 | 1 | 268/48 (5.6) | 328 |
| 2 | 325/36 (9.0) | 246 | |
| 3 | 1 | 99/48 (2.0) | 970 |
| 2 | 98/48 (2.0) | 876 | |
| Sum (1 + 2 + 3) | 1 | 482/142 (3.4) | 565 |
| 2 | 688/127 (5.4) | 397 |
A TAC clone having an insert of 80 kb (construct 1) and the TAC vector without an insert (construct 2) were used for transformation of A. thaliana ecotype Wassilewskija. Numbers of parentheses are transformation efficiency. The results of three independent experiments and their sum are shown.
Table 2.
Complementation of the A. thaliana sgr1 mutant with TAC clones carrying the SGR1 gene
| T1 line | Clone | Complementation | Segregation in T2 family for hygromycin resistance (R:S) | χ2 (3:1) |
|---|---|---|---|---|
| A | 5I12 | Yes | 139:50 | 0.213 |
| B | 5I12 | No | 94:29 | 0.133 |
| C | 5I12 | Yes | 166:63 | 0.770 |
| D | 5I12 | Yes | 169:44 | 2.142 |
| E | 5I12 | Yes | 215:64 | 0.632 |
| F | 5I12 | Yes | 221:94 | 3.938∗ |
| G | 5I12 | Yes | 199:25 | 22.88∗∗ |
| H | 20D10 | Yes | 201:81 | 2.085 |
| I | 20D10 | Yes | 208:76 | 0.469 |
,∗∗, Significant at the 5% and 1% levels, respectively.
Analysis of Transgenic Plants.
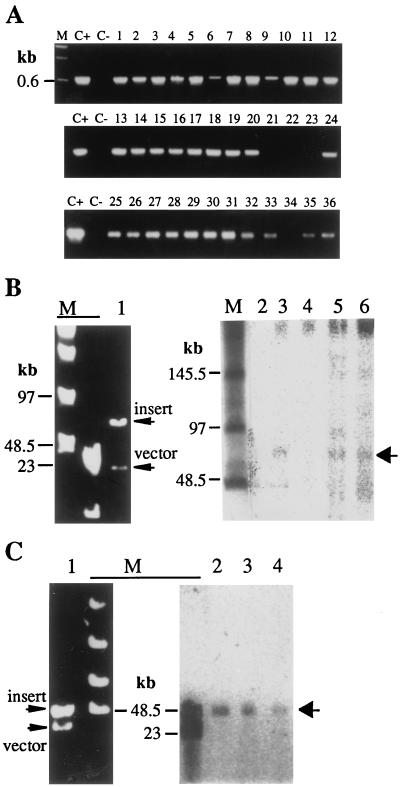
To examine whether large T-DNAs were integrated in their entirety into the plant genome, we analyzed the transferred segments by PCR and genomic Southern blotting. The sacB gene sequence, located near the left border, was amplified from 32 of 36 hygromycin-resistant plants obtained by using four distinct TAC clones carrying 40- to 80-kb inserts (Fig. 3A). This result suggests that in most transformants the entire inserts were transferred to the Arabidopsis genome.
Figure 3.
Transgenes in Arabidopsis plants transformed with TAC clones. (A) A. thaliana ecotype WS was transformed with TAC clones carrying either 40-kb (lanes 1–20) or 80-kb (lanes 21–36) genomic DNA fragments of ecotype Columbia. The sacB gene of 36 transgenic plants (hygromycin-resistant plants) was checked by PCR (Fig. 1). C+ and C−, positive (a TAC clone) and negative (untransformed plant) controls, respectively. (B) Transgenic lines transformed with a 75-kb TAC clone was self-crossed, and then the resulting T2 plants were analyzed by genomic Southern experiments. Genomic DNAs of transgenic and untransformed (negative control) plants were digested with I-SceI and hybridized with a HPT gene probe. The hybridized bands (lanes 3, 5, and 6) are shown by the arrow on the right. No hybridization band corresponding to the I-SceI fragment is seen in lanes 2 and 4. Lane 1, plasmid DNA digested with I-SceI; lanes 2–6, genomic Southern blotting of DNAs from untransformed plants (lane 2) and T2 lines (lanes 3–6). (C) The progenies of a transgenic line transformed with a 45-kb TAC clone were analyzed. Genomic DNAs of a T2 line (lane 2) and its T3 progenies (lanes 3 and 4) were digested with I-SceI and hybridized with the HPT gene probe. Lane 1, plasmid DNA digested with I-SceI; lanes 2–4, genomic Southern blotting of DNA digested with I-SceI.
The genomic DNAs of transformants that were created by using a TAC clone, 5I12, carrying a 75-kb insert (see below) were digested in agar plugs by I-SceI and subjected to Southern blotting experiments by using the HPT gene as a probe. The results showed that the sizes of the I-SceI fragments of three transformants were identical to those of the original plasmids (Fig. 3B, lanes 3, 5, and 6), indicating perfect, intact transfer of the 75-kb insert. However, one transgenic line (Fig. 3B, lane 4) did not show the I-SceI fragment, indicating the loss of at least one of the I-SceI sites.
The I-SceI fragment of 45 kb detected in a T2-generation transgenic line was inherited faithfully in the T3 generation (Fig. 3C).
Physical Mapping and Complementation of the Gravitropic Mutant sgr1.
We tested the usefulness of the TAC system for positional cloning with the SGR1 gene as a model. The Arabidopsis mutant sgr1 is deficient in the gravitropic response of its hypocotyl and stem (17). The SGR1 locus has been mapped on chromosome III with two adjacent DNA markers, CDC2B and KSAP3 (18). A TAC contig covering the locus was constructed by screening the TAC library using these DNA markers (Fig. 4A). While this study was underway, we noted that the SGR1 was allelic to the SCARECROW gene, which had been isolated as the gene for the radial organization of the Arabidopsis root (27). Therefore, we tested our contig by PCR with primers specific to the SCARECROW gene and identified three positive TAC clones. Two of these clones, 5I12 (ca. 75 kb) and 20D10 (ca. 80 kb), were introduced into the mutant plant sgr1 (ecotype Columbia background) by vacuum infiltration using A. tumefaciens strain C58C1(MP90). Of nine transgenic T1 plants, eight plants recovered the normal gravitropic response (Table 2). Line B appears to have a deletion in the transferred insert (Fig. 3B, lane 4). In the T2 generation of line E grown on medium without hygromycin, a 3:1 ratio of wild type to mutant was observed (Fig. 4B), as expected if the T1 generation carried the complementing DNA as a single-locus, hemizygous insertion. In fact, all hygromycin-resistant T2 plants in these eight lines exhibited wild-type gravitropism in shoot growth. These results indicate that the transgene SGR1 is expressed normally in the transgenic progeny plants, complementing the sgr1 mutation.
The results also demonstrate a single locus insertion of the transgene in all but one (line G) of the T2 families (Table 2) because segregation of the hygromycin-resistant phenotype of the transgenic lines was consistent with 3:1 segregation.
DISCUSSION
A major drawback of positional cloning is its dependence on detailed genetic analysis involving many progeny to achieve sufficiently fine mapping. Therefore, genetic mapping is a rate-limiting step in positional cloning. To accelerate this process for plants, we developed a TAC vector, pYLTAC7. This vector is suitable for stable maintenance of large genomic DNA fragments in both E. coli and A. tumefaciens and is competent for transfer of insert DNA into a plant genome by Agrobacterium-mediated transformation. To demonstrate this system’s practical utility in physical mapping and complementation, we complemented a sgr1 mutant of A. thaliana with large insert (ca. 80 kb) TAC clones carrying the wild-type allele. Recently, positional-cloning approaches have come to be preferred over DNA tagging for isolation of Arabidopsis genes identified through mutation (1). Although T-DNA and transposon tagging have been used for isolation of many plant genes, this approach has severe limitations because of the null function of most tagged alleles. It is difficult to dissect pathway interactions or processes vital for cell maintenance, especially any processes required after meiotic cell division. In contrast, chemical mutagens such as ethyl methanesulfonate can generate not only null mutants, but also mutants with partial or conditional gene function such as temperature-sensitive mutants. Analyses of these mutants and isolation of the genes defined by these mutations become desirable for understanding complex biological processes. In fact, the filamentous flower gene that is involved in flower development of A. thaliana was isolated successfully by using the TAC cloning system (S. Sawa and K. Okada, personal communication). A cell-wall synthesis gene that is defined by a temperature-sensitive mutation also was isolated by using this system (S. Sato, T. Kato, and D.S., unpublished results).
The TAC system is especially useful for positional cloning of genes when the position itself is imprecisely known. Mutations may exhibit incomplete penetrance whereby the mutant phenotype is subtle or depends on additional factors such as the external environment or other genetic loci. In this situation, perfect scoring of mapping crosses may not be achieved (e.g., cer9) (10). For the same reason, the map positions determined for quantitative trait loci (QTLs) are also approximate. Because QTLs control such important agronomic properties as yield, disease resistance, and stress tolerance, cloning of these elusive genes is a high economic priority. Genes located in recombination cold-spot regions (7) are also hard to be isolated by positional cloning. Thus, either scoring uncertainty or a scarcity of informative recombinational events can limit researchers’ ability to narrow a gene’s chromosomal position. TAC-based contigs can cover relatively wide chromosome regions with just a few clones. Thus, less mapping precision is required to make complementation testing feasible. Once a complementing clone is identified, the five rare-cutter restriction sites adjacent to the cloning site of TAC vector pYLTAC7 facilitate creation of nested deletion clones for further narrowing the complementing gene.
Elucidating the molecular genetics of Arabidopsis will be accelerated by using the TAC clones. At present, the genome mapping and sequencing project of A. thaliana at Kazusa DNA Research Institute (Chiba, Japan) has been using the TAC library for preparing contigs and sequencing parts of chromosomes V and III (24, 28, 29). The combination of TAC clone-based physical maps and their genome sequence data will greatly facilitate assignment and confirmation of gene function in Arabidopsis. The complete Arabidopsis genome sequence will be determined within a few years (2). With this in mind, sequencing of both ends of more than 2,000 TAC clones would suffice to identify clones covering nearly the entire genome of Arabidopsis. The TAC library of Arabidopsis will be distributed to academic researchers through the Arabidopsis Biological Resource Center at The Ohio State University.
Our results demonstrated high transformation efficiency with TAC clones; more than 1,000 transformants were obtained from three small-scale transformation experiments (Table 1). The virG- and virE-carrying helper plasmids used in the binary-BAC system for enhancing large T-DNA transfer (11) are not necessary for efficient transfer of 80-kb (this study) or larger TAC inserts. Among the transformants tested, most carry the entire T-DNA as confirmed by PCR and Southern analysis (Fig. 3B).
Acknowledgments
We thank Dr. Robert F. Whittier for helpful discussions and a critical reading of the manuscript.
ABBREVIATIONS
- TAC
transformation-competent artificial chromosome
- BAC
bacteria artificial chromosome
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB020028).
References
- 1.Somerville S, Somerville C. Plant Cell. 1996;8:1917–1932. doi: 10.1105/tpc.8.11.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meinke D W, Cherry J M, Dean C, Ronsley S D, Koorneef M. Science. 1998;282:662–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- 3.Burke D T, Carle G F, Olson M V. Science. 1987;236:806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- 4.Pierce J C, Sauer B, Sternberg N. Proc Natl Acad Sci USA. 1992;89:2056–2060. doi: 10.1073/pnas.89.6.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki T. Proc Natl Acad Sci USA. 1998;95:2027–2028. doi: 10.1073/pnas.95.5.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt R, West J, Love K, Lenehan Z, Lister C, Thompson H, Bouchez D, Dean C. Science. 1995;270:480–483. doi: 10.1126/science.270.5235.480. [DOI] [PubMed] [Google Scholar]
- 8.Olszewki E, Martin F B, Ausubel F M. Nucleic Acids Res. 1988;16:10765–10782. doi: 10.1093/nar/16.22.10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuse T, Kodama H, Hayashida N, Shinozaki K, Nishimura M, Iba K. Plant J. 1995;7:849–856. doi: 10.1046/j.1365-313x.1995.07050849.x. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y-G, Mitsukawa N, Vazquez-Tello A, Whittier R F. Plant J. 1995;7:351–358. [Google Scholar]
- 11.Hamilton C M, Frary A, Lewis C, Tanksley S D. Proc Natl Acad Sci USA. 1996;93:9975–9979. doi: 10.1073/pnas.93.18.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton C M. Gene. 1997;200:107–116. doi: 10.1016/s0378-1119(97)00388-0. [DOI] [PubMed] [Google Scholar]
- 13.An G, Watson B D, Chiang C C. Plant Physiol. 1986;81:301–305. doi: 10.1104/pp.81.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wing D, Koncz C, Schell J. Mol Gen Genet. 1989;219:9–16. doi: 10.1007/BF00261151. [DOI] [PubMed] [Google Scholar]
- 15.Jouanin L, Tourneur J, Tourneur C, Casse-Delbart F. Plasmid. 1985;16:124–134. doi: 10.1016/0147-619x(86)90071-5. [DOI] [PubMed] [Google Scholar]
- 16.Bouchez D, Camilleri C, Caboche M. C R Acad Sci. 1993;316:1188–1193. [Google Scholar]
- 17.Liu Y-G, Whittier R F. Nucleic Acids Res. 1994;22:2168–2169. doi: 10.1093/nar/22.11.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukaki H, Fujisawa H, Tasaka M. Plant Physiol. 1996;110:945–955. doi: 10.1104/pp.110.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey P N, Tasaka M. Plant J. 1998;14:425–430. doi: 10.1046/j.1365-313x.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- 20.Bechtold N, Ellis J, Pelletier G. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 21.Liu Y-G, Mitsukawa N, Oosumi T, Whittier R F. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y-G, Whittier R F. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 23.Sternberg N. Proc Natl Acad Sci USA. 1990;87:103–107. doi: 10.1073/pnas.87.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotani H, Sato S, Fukami M, Hosouchi T, Nakazaki N, Okumura S, Wada T, Liu Y-G, Shibata D, Tabata S. DNA Res. 1997;4:371–378. doi: 10.1093/dnares/4.6.371. [DOI] [PubMed] [Google Scholar]
- 25.Koncz C, Schell J. In: Methods in Arabidopsis Research. Koncz C, Chua N-H, Schell J, editors. Singapore: World Scientific; 1992. pp. 224–273. [Google Scholar]
- 26.Gay P, Le Coq D, Steinmetz T, Berkelman T, Kado C. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helariutta Y, Freshour G, Hahn M G, Feldmann K A, Benfey P N. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura Y, Sato S, Kaneko T, Kotani H, Asamisu E, Miyajima N, Tabata S. DNA Res. 1997;4:401–404. doi: 10.1093/dnares/4.6.401. [DOI] [PubMed] [Google Scholar]
- 29.Sato S, Kotani H, Hayashi R, Liu Y-G, Shibata D, Tabata S. DNA Res. 1998;5:163–168. doi: 10.1093/dnares/5.3.163. [DOI] [PubMed] [Google Scholar]