Summary
Background
In response to DNA damage, cells either undergo cell cycle arrest or apoptosis, depending on the extent of damage and the cell’s capacity for DNA repair. Cell cycle arrest induced by double-stranded DNA breaks depends on activation of the ataxia-telangiectasia (ATM) protein kinase, which phosphorylates cell cycle effectors such as Chk2 and p53 to inhibit cell cycle progression. ATM is recruited to double stranded DNA breaks by a complex of sensor proteins including Mre11/Rad50/Nbs1, resulting in autophosphorylation, monomerization, and activation of ATM kinase.
Results
In characterizing Aven protein, a previously reported apoptotic inhibitor, we have found that Aven can function as an ATM activator to inhibit G2/M progression. Aven bound to ATM and Aven overexpression in cycling Xenopus egg extracts prevented mitotic entry and induced phosphorylation of ATM and its substrates. Immunodepletion of endogenous Aven allowed mitotic entry even in the presence of damaged DNA, and RNAi-mediated knock-down of Aven in human cells prevented autophosphorylation of ATM at an activating site (S1981) in response to DNA damage. Interestingly, Aven is also a substrate of the ATM kinase. Mutation of ATM-mediated phosphorylation sites on Aven reduced its ability to activate ATM, suggesting that Aven activation of ATM following DNA damage is enhanced by ATM-mediated Aven phosphorylation.
Conclusions
These results identify Aven as a new ATM activator and describe a positive feedback loop operating between Aven and ATM. In aggregate, these findings place Aven, a known apoptotic inhibitor, as a critical transducer of the DNA damage signal.
Introduction
Entry into mitosis, driven by Cdc2/Cyclin B, can be inhibited following DNA damage by DNA-responsive checkpoints [1, 2]. These signaling pathways employ DNA damage sensors and transducers to inhibit cell cycle effectors [3]. To prevent mitotic entry, checkpoint pathways target the critical Cdc2 regulators, Wee1 and Cdc25 [4–7], which inhibit and activate Cdc2/Cyclin B, respectively [1, 2, 8]. When checkpoints are operative, Chk1/Chk2 kinases phosphorylate Wee1 and Cdc25, inducing activation and inhibition of the downstream molecules, respectively [6, 9]. Cdc25 is phosphorylated and inhibited by Chk1/Chk2–mediated phosphorylation at S287 (Xenopus numbering) which promotes docking of 14-3-3 [10–14]. Cdc25 activation depends upon active 14-3-3 removal and subsequent PP1-mediated S287 dephosphorylation [15–17].
Activation of Chk1/Chk2 requires DNA damage-mediated activation of upstream members of the phosphoinositide-3-kinase-related kinases (PIKKs), ATM and ATR [11, 18–21]. In the case of double stranded DNA breaks, a complex of proteins including Mre11, Rad50, and Nbs1 (the MRN complex) accumulates at DNA damage sites to form foci. ATM is then recruited to the MRN complex where ATM autophosphorylates at S1981 and is converted from an inactive dimer into an active monomer [22–26]. It is likely that there are other, as yet unidentified factors that participate in ATM regulation.
A variety of cellular processes have been reconstituted in extracts prepared from Xenopus eggs [27–29]. Cycling egg extracts [30], which oscillate between S and M phases of the cell cycle, can recapitulate DNA damage-responsive checkpoints in vitro; addition of annealed oligonucleotides mimicking damaged DNA prevents M phase entry, arresting extracts with inactive Cyclin B/Cdc2 complexes [18, 31, 32]. Prolonged incubation of egg extracts on the bench for 4–6 hours results in cytochrome c release from mitochondria and caspase activation [33–35]. These cycling and apoptotic properties of the Xenopus egg extracts make them well-suited to analyze factors implicated in cell cycle progression or apoptosis.
We wished to use egg extracts to analyze the function of a previously reported apoptotic inhibitor, Aven. Human Aven was originally identified as an interactor of the anti-apoptotic protein Bcl-xL. Aven mRNA was detected in all adult tissues, but was most abundant in heart, skeletal muscle, kidney, liver, pancreas, and testis. Although the predicted molecular weight of Aven is 38.6 kDa, the apparent molecular weight of Aven derived from HeLa cells, where it can be found in both cytoplasmic and nuclear fractions, is 55kD. Aven overexpression is reported to be associated with poor prognosis in childhood acute lymphoblastic leukemia[36–39]. In addition to interacting with Bcl-xL, Aven was reported to interact with and inhibit Apaf-1 oligomerization, required to activate caspase-9 [40, 41]. The molecular mechanism underlying the ability of Aven to impede Apaf-1 oligomerization and the significance of its association with Bcl-xL are not clear. Moreover, the possibility that Aven might have additional functions has not been explored.
We have now found that Aven protein is a potent activator of ATM, critical for its DNA damage-induced activation. Aven overexpression in cycling egg extracts induced phosphorylation of ATM substrates independent of DNA damage and depletion of Aven abrogated G2/M arrest in response to DNA damage. Similarly, RNAi knock-down of Aven in mammalian cells dampened DNA damage-induced ATM activation. Phosphorylation of Aven by ATM in a positive feedback loop appears to be required for its full potency as an ATM activator. These data implicate the apoptotic regulator Aven as an integral component of the DNA damage-responsive checkpoint pathway.
Results
Aven inhibits entry into mitosis
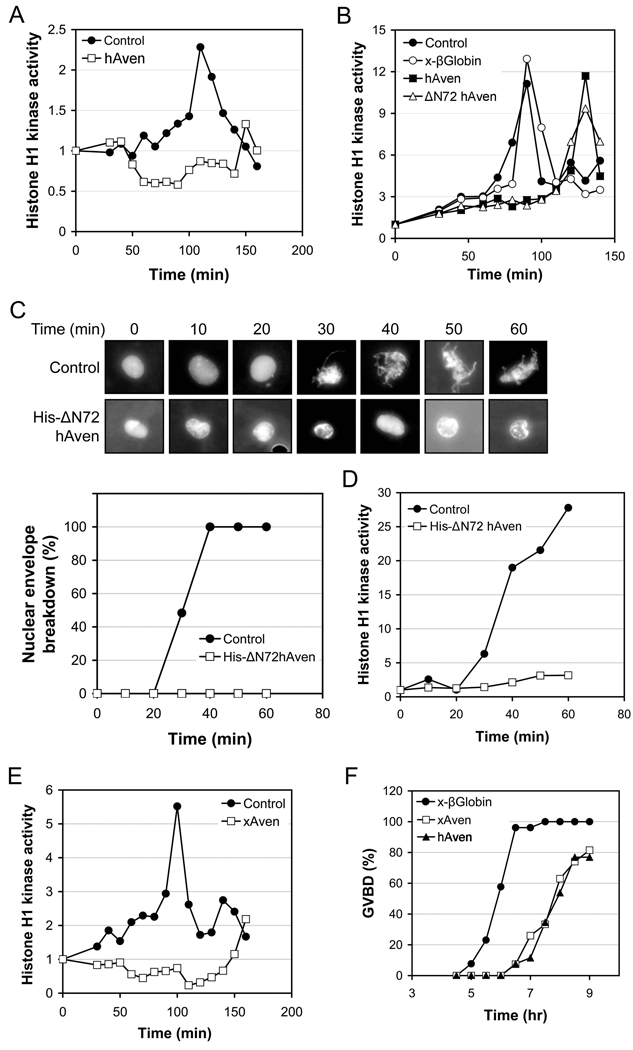
To further analyze the apoptotic function of Aven, we wished to overexpress Aven protein and determine its effects on caspase activation in Xenopus egg extracts. However, when we added excess mRNA encoding human Aven protein (approximately doubling the endogenous Aven concentration of 80nM) to cycling egg extracts, we found that Aven had a profound effect on cell cycle progression. The extracts expressing excess Aven either failed to enter mitosis or were significantly delayed in mitotic entry, as evidenced by a dampening of histone H1 kinase activation and inhibition of nuclear envelope breakdown/chromatin condensation (Fig. 1A and B and data not shown). To confirm that the protein product of Aven mRNA was responsible for cell cycle inhibition, recombinant His-tagged human Aven protein lacking its Gly-rich N-terminus (His- ΔN72), which posed difficulty for protein purification, was added to interphase egg extracts treated with recombinant Cyclin B to drive mitotic entry. His-ΔN72 Aven protein prevented nuclear envelope breakdown, chromatin condensation and activation of Cdc2/Cyclin B (Fig. 1C and D). Similar results were obtained with addition of Aven mRNA lacking codons 2–72 to cycling extracts (Fig. 1B), indicating that the Gly-rich N-terminal region was not required for the cell cycle arrest. To demonstrate that Xenopus Aven, which shares 42% amino acid sequence identity with human Aven could also inhibit mitotic entry, we cloned the Xenopus Aven cDNA (supplemental Fig. 1) and repeated our experiments analyzing the effects of full-length Xenopous Aven overexpression on M phase entry. As shown in Fig. 1E, Xenopus Aven acted similarly to human Aven in retarding mitotic entry. To assay the effects of Aven in an intact cell, we tested whether Aven overexpression would impede progesterone-induced oocyte maturation (progression through G2/M). Expression of either Xenopus or human Aven mRNAs, but not control Xenopus β-globin mRNA, markedly delayed M phase entry in oocytes (Fig. 1F).
Figure 1. Aven overexpression delays mitotic entry.
(A) Cycling Xenopus egg extracts were supplemented with mRNA encoding full-length human Aven (hAven), or an equal volume buffer (control). Samples were withdrawn at the indicated times and assayed for their ability to phosphorylate histone H1 in the presence of [γ-32P] ATP.
(B) mRNA encoding hAven, ΔN72hAven or Xenopus β-globin (x- βGlobin), or buffer control were added to cycling Xenopus egg extracts. Aliquots of extracts were withdrawn at the indicated times and assayed as (A).
(C and D) His-ΔN72hAven protein or GST control was added to interphase extracts and incubated for 30 minutes. Following the addition of non-degradable CyclinB1, aliquots were taken at the indicated times (C) to visualize nuclear envelope breakdown and chromatin condensation by fluorescence microscopy (Hoechst DNA stain), and (D) to assay histone H1 phosphorylation. (C) Bottom: represents the average of three replicates.
(E) Cycling extracts were incubated with mRNA encoding Xenopus Aven (xAven) or buffer, and aliquots were taken at the indicated times and assayed as (A).
(F) 40 stage VI oocytes were injected with mRNAs encoding x-βGlobin, xAven, or hAven. After 12 hr, oocytes were treated with progesterone and scored for germinal vesicle breakdown (GVBD).
Data shown in this and all subsequent figures are the result of at least three repetitions.
Aven inhibits activation of Cdc2/Cyclin B
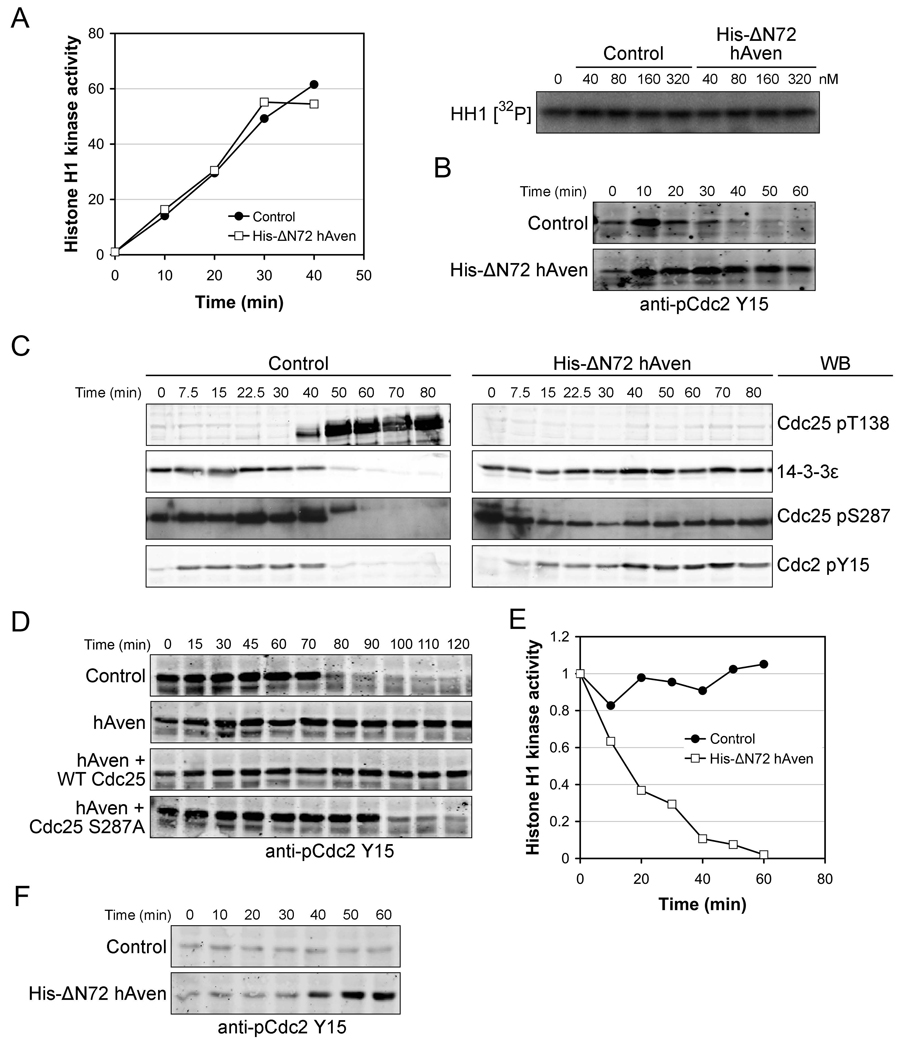
Aven’s ability to inhibit histone H1-directed kinase activity suggested that Aven might inhibit Cdc2 directly. However, as shown in Fig. 2A, addition of Aven protein to purified Cdc2/Cyclin B had no effect on Cdc2 kinase activity, even at protein levels as high as 320nM Aven. Cdc2 activity is controlled by Cyclin levels [42, 43] and through inhibitory Y15 phosphorylation of Cdc2 [1, 2]. Cyclin B protein synthesis was unaffected by Aven (data not shown). In contrast, Cdc2 dephosphorylation on Y15, which occurred on schedule in control extracts, was absent in the Aven-supplemented extracts, consistent with a role for Aven as a mitotic inhibitor (Fig. 2B).
Figure 2. Aven indirectly inhibits Cdc2/CyclinB kinase activity by modulating Cdc2 phosphorylation status.
(A) Left panel: 1 µl (4 unit) of purified Cdc2/Cyclin B kinase was incubated with 80 nm His-ΔN72hAven protein or GST control, radiolabeled ATP and Histone H1 substrate. Samples were taken at the indicated time points and assayed for Histone H1-directed kinase activity and quantified. Right panel: 1 µl (4 unit) of purified Cdc2/Cyclin B kinase was incubated with His-ΔN72hAven protein or His-GST control at different concentrations for 30 min, and assayed for Histone H1-directed kinase activity.
(B) Interphase extracts were incubated with His-ΔN72hAven protein or His-GST control for 30 minutes. Non-degradable cyclinB1 was then added to the extracts and aliquots were taken at the indicated times for immunoblotting with anti- pCdc2Y15.
(C) His-ΔN72hAven protein together with GST-Cdc25 coupled to glutathione beads was incubated in interphase egg extract for 30 minutes. Following the addition of non-degradable CyclinB1, GST-Cdc25 beads were retrieved and washed at the indicated times and immunoblotted with anti-14-3-3ε antibody. In addition, aliquots were taken at the same time points and immunoblotting was performed with anti-pCdc25T138, anti-pCdc25S287, or anti-pCdc2Y15 antibody.
(D) Cycling extracts were incubated with mRNA encoding hAven, or hAven together with wild type Cdc25 or Cdc25S287A. Samples were taken at the indicated times and immunoblotted with anti-pCdc2Y15 antibody.
(E and F) His-ΔN72hAven protein or GST control protein was added to mitotic extracts, aliquots were taken at the indicated times and assayed for their ability to phosphorylate histone H1 or immunoblotted with anti-pCdc2Y15 antibody.
Maintenance of Cdc2 Y15 phosphorylation could result from sustained activation of Y15-directed kinases (Wee1 and Myt1), from inhibition of the Y15-directed phosphatase, Cdc25, or both [1]. Activation of Cdc25 at mitotic entry requires T138 phosphorylation, which promotes removal of bound 14-3-3 and allows the reversal of inhibitory S287 phosphorylation on Cdc25 [15]. To determine if Aven could modulate Cdc25, we monitored Cdc25 modifications in the presence and absence of increased Aven protein. As shown in Fig. 2C, in the presence of Aven protein, T138 phosphorylation was not observed, Cdc25 remained 14-3-3-bound, and S287 remained phosphorylated, consistent with a role for Aven in controlling upstream factors regulating Cdc25 (Fig. 2C). Moreover, expression of an S287A mutant Cdc25 in cycling extracts, but not WT Cdc25, could override the effects of Aven addition, allowing Cdc2 Y15 dephosphorylation despite Aven overexpression (Fig. 2D).
Although Aven impinged on Cdc25 activation, this did not preclude a role for Aven in activation of Wee1 (or Myt1). In unperturbed extracts, Wee1/Myt1 activity is high during interphase and drops at mitotic entry to allow Cdc2 activation [44]. Remarkably, addition of recombinant Aven to extracts already in mitosis allowed suppression of Cdc2 activity and rephosphorylation of Y15 (Fig. 2E and 2F). As Wee1 and Myt1 are normally turned off in mitotic extracts and Cdc2 was fully dephosphorylated prior to Aven addition, these results strongly suggest that Aven can modulate both the Cdc25 and Wee1/Myt1 arms of the Cdc2-regulatory pathway.
Aven interacts with and activates ATM
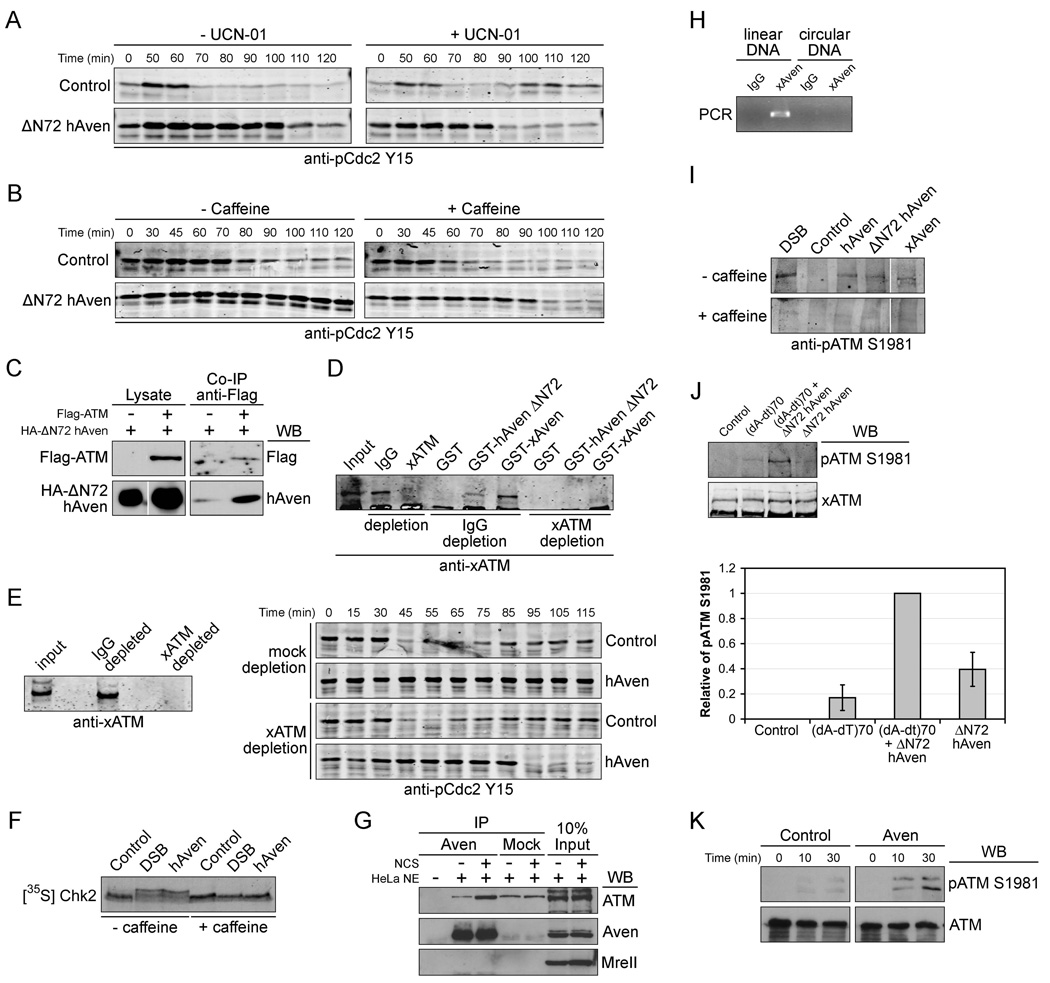
Because Aven appeared to control Cdc25 at known points of DNA-responsive checkpoint regulation (T138 and S287) and because both Wee1 and Cdc25 might be modulated downstream of Aven, we speculated that Aven might be an upstream regulator of factors that normally participate in DNA-responsive checkpoint signaling. In support of this, we found that both the inhibitor of Chk kinases, UCN-01, and the ATM/ATR inhibitor, caffeine, could override the ability of Aven to induce cell cycle arrest prior to mitotic entry (Fig. 3A and B).
Figure 3. DNA damage checkpoint pathway components are involved in Aven-induced delay of mitotic entry.
(A and B) mRNA encoding ΔN72hAven or control buffer was added to cycling extracts +/− UCN-01(5ng/ul) or caffeine (5mM). Aliquots were taken at the indicated times and immunoblotted with anti-pCdc2Y15 antibody.
(C) ΔN72hAven was transfected into 293T cells with or without Flag -tagged full-length ATM. Cell lysates and anti-Flag immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies.
(D) Glutathione beads coupled to GST, GST-ΔN72hAven or GST-xAven were incubated in interphase extracts depleted with xATM antibody or control IgG for 1 hr at room temperature, then retrieved, washed, and immunoblotted with anti-xATM antibody.
(E) Left panel: cycling extracts were immunodepleted with anti-xATM or control IgG and aliquots were immunoblotted with anti-xATM antibody. Right panel: hAven mRNA was incubated in cycling extracts immunodepleted of ATM or mock-depleted with IgG. Samples were taken at the indicated times and immunoblotted with anti-pCdc2Y15 antibody.
(F) Cycling extracts were supplemented with either buffer, hAven mRNA or DNA bearing double stranded breaks (DSB) in the presence of 35S-labeled Chk2 +/− caffeine (5mM) for 1 hr. Aliquots were resolved by SDS-PAGE and Chk2 electrophoretic mobility was examined by phosphoimager.
(G) Hela cells treated with or without NCS (200 ng/ml) for 1hr were harvested to make nuclear fractions. Rabbit anti-Aven antibody or control IgG immunoprecipitates were immunoblotted with indicated antibodies.
(H) Circular pGEX-KG-Emi2 or Not-I cut linear pGEX-KG-Emi2 plasmid was incubated in the interphase extracts for 30min. Anti-Xenopus Aven antibody or control IgG was used for immunoprecipitation and washed immunoprecipitates were used as PCR templates to detect DNA binding to Xenopus Aven.
(I) Cycling extracts were supplemented with buffer control, DSB DNA, or mRNA encoding full-length hAven, ΔN72hAven, or full-length xAven +/− caffeine (5mM) for 1hr. Samples were taken and immunoblotted with anti-pATM S1981 antibody.
(J) Top panel: buffer control, 10 ng of (dA-dT)70, 10 ng of (dA-dT)70 with mRNA encoding ΔN72hAven, or mRNA encoding ΔN72hAven was added to cycling extracts. After 1hr, samples were immunoblotted with the indicated antibodies. Bottom panel: the amount of pATM S1981 was quantified, normalized, and plotted. Error bars represent the standard deviation of three replicate experiments.
(K) Hela cell transfected with vector control or Aven were treated with NCS (200ng/ml) for 10 or 30 min. Cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Using Aven residues 1–289 as bait in a yeast two-hybrid screen of a cDNA library from 3-week-old rat brain (provided by Paul Worley; note that the C-terminal domain was eliminated because it had the intrinsic ability to activate transcription), we identified the C-terminal kinase domain (∼residues 2500–3000) of ATM kinase as a potential Aven interactor. Aven binding to both the kinase domain and full-length ATM was confirmed by co-immunoprecipitation experiments in transfected mammalian cells. Furthermore, both full-length and ΔN72-Aven interacted with ATM (data not shown and Fig. 3C). Moreover, GST-Aven coupled to glutathione beads retrieved ATM from Xenopus interphase extracts, and this co-immunoprecipitating band disappeared upon immunodepletion of ATM (Fig. 3D). Given these findings, we examined if Aven requires ATM to inhibit the cell cycle. Aven-induced arrest was markedly abrogated when ATM had been immunodepleted from egg extracts, suggesting that Aven acts, at least in part, through ATM activation (Fig. 3E). Moreover, as shown in Fig. 3F, expression of human Aven in egg extracts promoted an electrophoretic shift in Chk2 (indicative of activation) commensurate with the shift promoted by DNA bearing double-stranded breaks (DSB). The addition of either human Aven or DSB DNA to extracts also promoted commensurate phosphorylation of p53 at a known site of ATM-catalyzed phosphorylation (S15) (supplemental Fig. 2). We also found that the ability of endogenous Aven to co-immunoprecipitate with ATM from HeLa cell nuclear extracts was markedly increased by pre-treatment of cells with the radiomimetic drug, neocarzinostatin (NCS) (Fig. 3G; although some ATM signal is seen in mock immunoprecipitates, this signal is NCS-insenstive and substantially less than in the Aven precipitate from NCS- treated cells). Moreover, Aven could bind to plasmid DNA only after it had been linearized, suggesting that Aven might have affinity for DSB DNA (Fig. 3H).
Although Aven could promote phosphorylation of ATM substrates and induce cell cycle arrest, it was not clear whether Aven acted upstream of ATM to promote its activation, rather than at the level of the substrate(s). To address this, we examined phosphorylation of ATM itself at S1981, a site that is auto (or trans)-phosphorylated in response to DNA damage. Because it was possible that Aven addition promoted DNA damage to trigger ATM activation indirectly, we examined S1981 phosphorylation in extracts lacking DNA, but supplemented with Aven. As shown in Fig. 3I, Aven addition promoted endogenous ATM S1981 phosphorylation as seen following addition of DSB DNA. Moreover, this S1981 phosphorylation signal was overridden by caffeine and reduced by ATM depletion, consistent with Aven promoting ATM autophosphorylation (Fig. 3I and supplemental Fig. 3). Virtually identical results were obtained when a recombinant fragment that includes the ATM-S1981 site (GST-ATM aa1905–2100) was added to extracts as a trans substrate of endogenous ATM (supplemental Fig. 4). These data demonstrate that Aven addition to egg extracts triggers ATM activation to promote G2/M arrest. Furthermore, Aven overexpression acted synergistically with DSB DNA in Xenopus egg extracts or the radiomimetic drug neocarzinostatin (NCS) to induce ATM S1981 phosphorylation (Fig. 3J and K), consistent with the notion that Aven contributes to ATM activation in response to DNA damage.
Aven is required for DNA damage-induced inhibition of mitotic entry
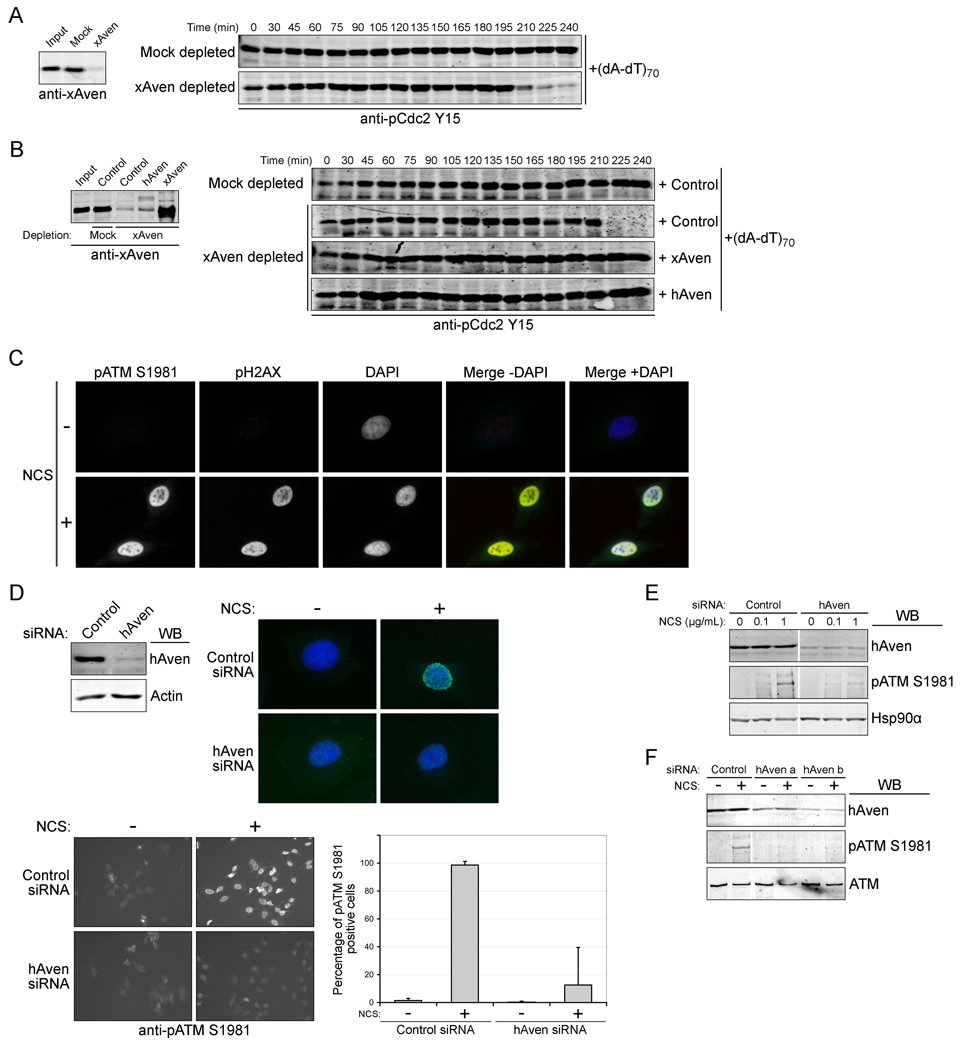
Although exogenous Aven appeared to promote ATM activation in the absence of DNA damage, we wished to determine if Aven was required for activation of ATM by endogenous DNA-responsive checkpoint pathways. Addition of (dA-dT)70 promotes robust activation of DNA-responsive checkpoints in egg extracts [32], resulting in stabilization of inhibitory Y15 phosphorylation of Cdc2. In contrast, immunodepletion of endogenous Aven protein allowed entry (albeit delayed) into mitosis even in the face of checkpoint activation (Fig. 4A). To rule out the possibility that removal of Aven co-depleted an associated protein that allowed DNA damage checkpoint override, Aven was added back to extracts following immunodepletion. When mRNA for either human or Xenopus Aven was added back to Aven-depleted extracts, (dA-dT)70-induced checkpoint activation was restored (Fig. 4B).
Figure 4. Aven depletion overrides a DNA damage checkpoint.
(A) Left panel: Anti-xAven immunoblotting of cycling extracts before and after depletion of Xenopus Aven by three sequential incubations with IgG or anti-xAven antibody at 4°C. Right panel: immunoblot of Aven- or mock-depleted extracts with anti-pCdc2Y15 antibody following addition of 10ng/µl (dA-dT)70; aliquots were removed at the indicated times.
(B) Left panel: immunoblot for xAven in depleted extracts (as in A) with subsequent addition of buffer control, mRNA for human or Xenopus Aven. Note that anti-xAven antibody weakly recognizes hAven in cycling extracts. Right Panel: immunoblotting of the same extracts with anti-pCdc2Y15 antibody at the indicated times following addition of 10ng/µl (dA-dT)70.
(C) Hela cells treated with NCS (200 ng/ml) for 30 minutes were stained with anti-pATM S1981(green), anti-pH2AX (red) or with DAPI (blue) to stain DNA. Yellow coloring indicates co-localization of pATM S1981 and pH2AX.
(D) Top left panel: immunoblotting of human Aven in HeLa cells transfected with control siRNA or pooled hAven siRNAs with anti-hAven antibody. Actin serves as a loading control. Top right and bottom left panels: control or hAven siRNA-transfected Hela cells were incubated with NCS (200 ng/ml) for 30 minutes and stained with anti-pATM S1981 (green) or with DAPI to stain DNA (blue). Top right panel shows a single high magnification nucleus; pATM S1981 staining of a larger field of cells is shown on the bottom left. Bottom right: Columns represent the average of pATM S1981 positive cells from three replicate experiments and error bars represent the standard deviation.
(E) anti-pATM S1981 immunoblots of HeLa cells transfected with control siRNA, or pooled hAven siRNAs and treated +/− 100 or 1000 ng/ml of NCS. Hsp90α was used as a loading control.
(F) Immunoblots of HeLa cells transfected with single hAven siRNAs (a or b) or control siRNA and treated +/− NCS (200 ng/ml) with the indicated antibodies.
In response to DNA damage, ATM is phosphorylated at S1981 and is localized in foci, that can be detected as punctate staining by immunofluorescence [25, 45]. To examine the role of Aven in checkpoint-induced ATM activation in a mammalian system, we utilized RNAi interference (pooled oligonucleotides) to knock-down the expression of endogenous human Aven in HeLa cells and monitored the activation status of ATM upon addition of the radiomimetic drug neocarzinostatin (NCS). As shown in Fig. 4C, NCS-treated HeLa cells displayed discrete foci of phospho-ATM that co-localized with phospho-H2AX. Remarkably, the phospho-ATM signal was absent from cells ablated for Aven by RNA interference (Fig. 4D). These data strongly suggest that ATM could not be activated or that the activation could not be sustained in the absence of Aven. These findings were confirmed by immunoblotting of pATM S1981, as depletion of Aven abolished detectable autophosphorylation on S1981 of ATM in response to DNA damage (Fig. 4E). To further validate these findings, individual siRNAs from the pool were used to knock-down Aven; two different oligonucleotides that decreased Aven levels dampened the appearance of S1981 phosphorylation in response to NCS (Fig. 4F; Note that the Aven knock-down in these experiments is not complete). These data reinforce the conclusion that Aven is a required for linking DNA damage to ATM activation.
ATM phosphorylates and activates Aven in a positive feedback loop
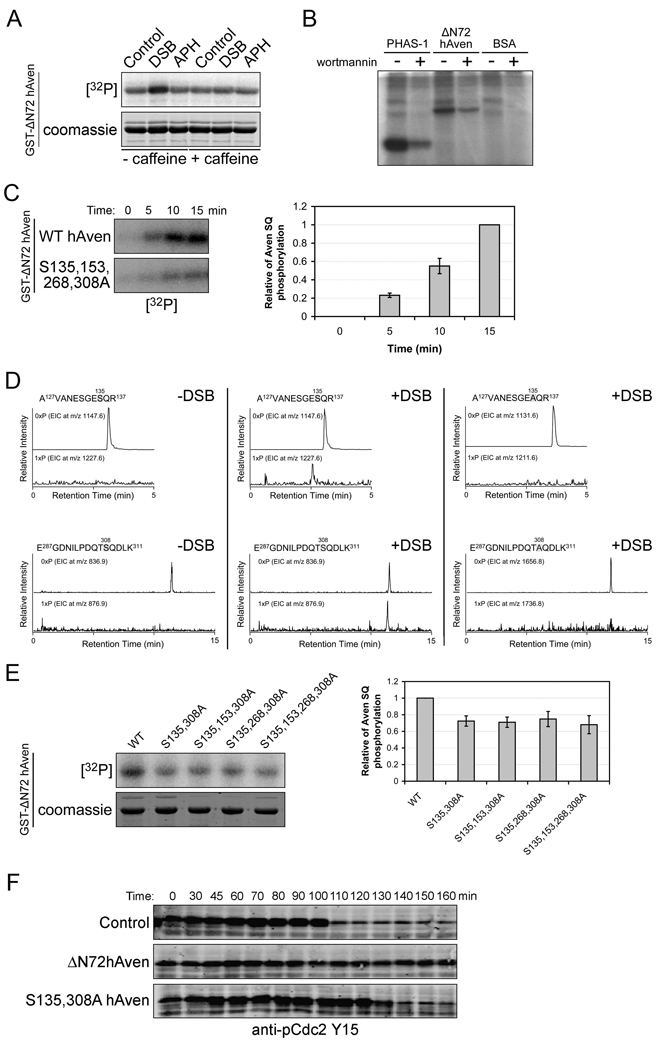
Positive and negative feedback loops are common in cell cycle regulatory networks [1, 2, 16, 46, 47]. Hence, we sought to determine if Aven might also be a substrate of ATM, in addition to its role as a modulator of the ATM pathway. Indeed, treatment of egg extracts with DSB DNA (but not aphidicolin-treated nuclei to activate the replication checkpoint) enhanced phosphorylation of Aven, an effect that could be abrogated by addition of caffeine (Fig. 5A), addition of KU55933, an ATM specific inhibitor (supplemental Fig. 5), or immunodepletion of ATM (Fig. 6). In addition, active ATM immunoprecipitated from 293T cells could phosphorylate Aven in vitro and this phosphorylation was largely inhibited by treatment with an inhibitor of PI3K family kinases, wortmannin (Fig. 5B). Moreover, when ATM was immunodepleted from extracts, Aven phosphorylation by ATM immunoprecipitates was largely abrogated (supplemental Fig. 7).
Figure 5. Positive feedback activation of Aven by phosphorylation.
(A) GST-ΔN72hAven coupled to glutathione beads were dipped into interphase egg extracts pre-treated with DSB or sperm chromatin and Aphidicolin (APH) in the presence of [γ-32P] ATP +/− caffeine (5mM) for 15 min, then retrieved, washed, and resolved on 10% SDS-PAGE. Phosphorylated hAven was detected using a phosphoimager.
(B) ATM kinase was immunoprecipitated from 293T cells treated with NCS +/− wortmannin using anti-ATM antibody coupled to protein A sepharose beads. Purified His-ΔN72hAven protein was then incubated with the immunoprecipitates in kinase buffer for 30 min and resolved by SDS-PAGE. PHAS-1, a known ATM substrate, and BSA were included as positive and negative controls, respectively. Phosphorylation was detected by phosphoimager.
(C) Left panel: GST-ΔN72hAven wild type (WT) or S135,153,268,308A mutant protein conjugated to glutathione beads were dipped into interphase extracts in the presence of [γ-32P] ATP. Extracts were then treated with DSB and aliquots were taken at the indicated times and assayed as (A). Right panel: the amount of Aven SQ phosphorylation was quantified, normalized, and plotted. Error bars represent standard deviation of three replicate experiments.
(D) GST-ΔN72hAven wild type or S135, 153, 268, 308A protein was incubated in interphase extracts - (left) or + (middle and right) DSB DNA. After incubation, the protein was retrieved on glutathione Sepharose beads and resolved by SDS-PAGE, and the relevant bands were excised from the gel, trypsin digested, and analyzed by LC/MS mass spectrometry. LC/MS-extracted ion chromatograms (EIC) of unphosphorylated (0×P) and presumed monophosphorylated (1×P) tryptic peptides were examined, and EIC peaks (middle; at 2.6 and 11.5 min in the 1×P traces) indicate the phosphorylation of the tryptic peptides upon DSB DNA treatment. Such peaks were not detected in the tryptic peptides derived from the GST-AN72hAven S135, 153, 268, 308A protein (right), indicating a lack of phosphorylation.
(E) Left panel: xATM kinase was immunoprecipitated from interphase extracts pre-treatment with DSB. Purified GST-ΔN72hAven WT or AQ mutant proteins were incubated with the immunoprecipitates in the kinase buffer for 30 min and assayed as (A). Right panel: the amount of Aven SQ phosphorylation was quantified, normalized, and plotted. Error bars represent standard deviation of three replicate experiments (T test, P<0.05).
(F) mRNA-encoding ΔN72hAven or ΔN72hAvenS135, 308A was added to the cycling extracts, and aliquots were taken at the indicated times and immunoblotted with anti-pCdc2Y15 antibody.
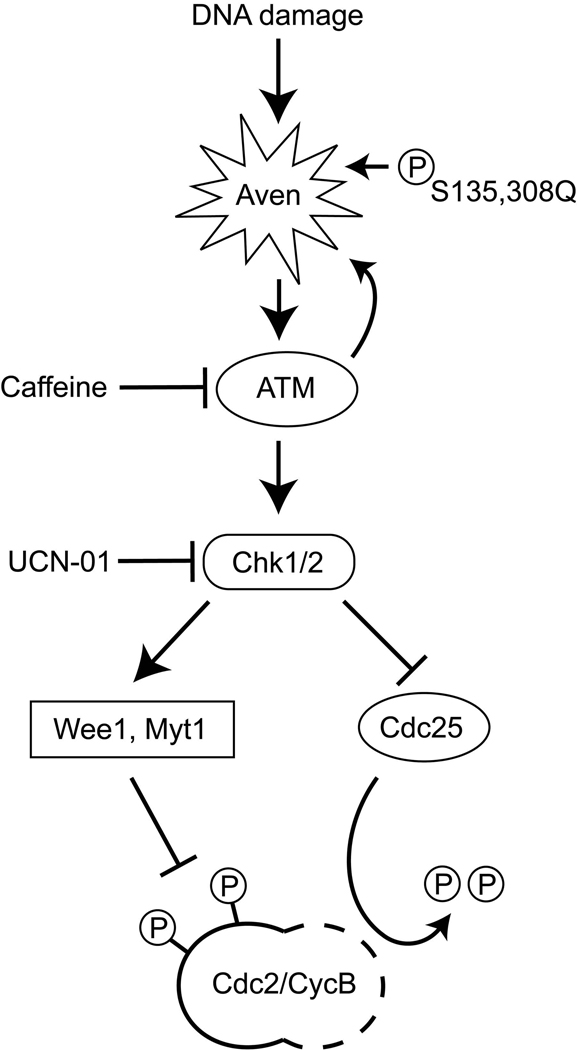
Figure 6. Aven’s role in DNA damage-induced ATM activation and G2/M arrest.
During DNA damage, Aven serves as a signal transducer in the ATM activation pathway. Activated ATM in turn phosphorylates Aven on S135 and S308, and causes full activation of Aven. Fully activated Aven then further enhances ATM activation, leading to activation of downstream pathway components to inhibit Cdc25 activation and enhance Wee1/Myt kinase activity, leading to Cdc2/CyclinB inactivation and inhibition of mitotic entry.
ATM typically phosphorylates serine residues followed by glutamine (SQ sites) [48], and analysis of the amino acid sequence of human Aven revealed four such motifs located at residues S135, 153, 268, and 308. To study the potential effect of ATM-mediated phosphorylation on Aven function, we mutated these four SQ sites to AQ and analyzed the phosphorylation status of the resulting mutant. As shown in Fig. 5C and supplemental Fig. 5, DSB DNA enhanced Aven phosphorylation over time and mutating all four putative ATM phosphorylation sites largely eliminated the DSB DNA enhanced phosphorylation of Aven (though basal phosphorylation remained). We performed Mass Spectrometry on recombinant Aven protein that had been dipped into egg extracts treated with or without DSB DNA and identified S135 and S308 as conserved sites of phosphorylation (Fig. 5D and supplemental Fig. S9). To confirm the significance of these two phospho-SQ sites, we mutated these to AQ and performed an in vitro ATM kinase assay using ATM immunoprecipitated from Xenopus egg extracts. We found that mutating S135 and S308 sites to Alanine largely dampened Aven’s phosphorylation by ATM (though some phosphorylation remained, due to either a contaminating kinase or an unidentified ATM phosphorylation site). However, it is noteworthy that we did not observe any additional decrease in phosphorylation when we changed S153 or S268 to Alanine (Fig. 5E and supplemental Fig. S10), consistent with our Mass Spec identification of phospho-S135 and S308 following ATM activation. To analyze the effects of Aven phosphorylation, we added mRNA encoding either WT or non-phosphorylatable (S135, 308A) mutant Aven to cycling egg extracts and monitored mitotic entry. Mutant Aven protein was partially active but failed to maintain the cell cycle arrest observed with similar levels of WT Aven (Fig. 5F). These data suggest that Aven overexpression can activate ATM and that Aven phosphorylation in a positive feedback loop enforces Aven activity, making it a more potent ATM activator.
Discussion
The results described here implicate Aven as a key regulator of the G2/M DNA damage checkpoint. Aven’s ability to prevent M phase entry stems from its ability to activate ATM to promote phosphorylation and activation of cell cycle inhibitors (e.g., Chk2) controlling Cdc2/Cyclin B (see model, Fig. 6). Precisely how Aven is engaged following DNA damage remains to be determined, but it is clear that the absence of Aven impairs checkpoint-mediated ATM function.
Aven-mediated ATM activation
Because Aven interacted with ATM and induced ATM S1981 phosphorylation, Aven protein may initiate activation of ATM (i.e., recruitment to sites of DNA damage, monomerization and autophosphorylation). Although we did not detect Aven interaction with MRN complex, we have noticed that Aven protein binds efficiently to restriction enzyme-digested plasmids, but not to intact plasmids added to egg extracts. In addition, the endogenous Aven-ATM interaction was increased following DNA damage, suggesting that Aven might be involved in ATM recruitment to sites of double stranded DNA breaks or that double stranded breaks provide a platform for Aven-mediated ATM activation. Alternatively, Aven may inhibit ATM S1981 dephosphorylation, thereby interfering with maintenance of ATM activity. Both PP2A and Wip1 dephosphorylate ATM S1981 [49, 50]. However, we have not observed any direct interaction of Aven with PP2A or Wip1, nor have we seen alterations in overall PP2A activity in egg extracts supplemented with Aven (data not shown). It has also been reported that protein phosphatase 5 (PP5) is required for ATM activation, so it is plausible that Aven could activate PP5 or inactivate a PP5 inhibitor [51].
A positive feedback loop linking ATM and Aven
Depletion of Aven in both mammalian and Xenopus systems compromised DNA damage-induced ATM activation, and overexpression of Aven prevented mitotic entry in egg extracts. We propose that overexpression of Aven mimics a signaling pathway that would normally be engaged following DNA damage. Following ATM activation by DSB or Aven overproduction, ATM may then phosphorylate Aven, enhancing Aven-mediated ATM activation. Following DNA damage, it may be that low level ATM activation promotes Aven phosphorylation, leading to higher ATM activation. The impaired ability of the SQ mutant Aven to promote cell cycle arrest and ATM activation is consistent with a role for these phosphorylation sites in controlling Aven function.
Although Aven could be phosphorylated by ATM, basal phosphorylation of Aven was also evident when we incubated GST-Aven protein with egg extracts. Mass Spectrometric analysis of Aven dipped into egg extracts demonstrated basal phosphorylation on Ser and Thr sites in addition to the SQ sites mentioned here (JYG, MZW and SK, unpublished data). Thus, other kinases/pathways may contribute to Aven phosphorylation, potentially providing additional inputs to Aven regulation.
A dual regulator of cell cycle and apoptotic progression
Although Aven was reported as an apoptotic regulator, we do not know whether its cell cycle and apoptotic modulatory properties stem from the same underlying biochemical activity. It would be interesting to determine whether ATM is required for Aven’s modulation of the apoptosome. Conversely, it will be of interest to determine whether Aven mutants that cannot modulate apoptosis can influence cell cycle progression. Aven may require entirely different domains/functions to regulate cell division and apoptosis. In this regard, it is of interest that Apaf-1 was recently reported to induce S-phase arrest following DNA damage [52]. Although Aven has been shown to be an apoptosome regulator and Apaf-1 interactor (Chau et al 2000, PMI and JMH, unpublished), Apaf-1 was reported not to be required for ATM S1981 phosphorylation and activation, while Aven binds and promotes ATM activation. Thus Aven and Apaf-1 may act at different points in checkpoint control. Indeed, it may be that Apaf-1 is required for the intra-S phase, but not the G2/M checkpoint. It would be of significant interest to determine whether extensive DNA damage alleviates Aven’s apoptotic inhibitory functions. Regardless of its precise mechanism of action, Aven is ideally situated as mediator of both cell cycle progression and cell death to participate in the cellular decision to live or die following DNA damage.
Supplementary Material
Acknowledgments
We are grateful to Dr. James Edwin Hall (UNC< Chapel Hill) for mass spectrometry resources. We thank Jen Perry and Carrie Johnson for reading and discussing the manuscript. This work was supported by RO1 GM67225 and RO1 CA102707 to SK, RO1 NS34704 to JMH and RO1 AI54952 to PI.
References
- 1.Lew DJ, Kornbluth S. Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Current opinion in cell biology. 1996;8:795–804. doi: 10.1016/s0955-0674(96)80080-9. [DOI] [PubMed] [Google Scholar]
- 2.Coleman TR, Dunphy WG. Cdc2 regulatory factors. Current opinion in cell biology. 1994;6:877–882. doi: 10.1016/0955-0674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 3.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & development. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 4.Raleigh JM, O'Connell MJ. The G(2) DNA damage checkpoint targets both Wee1 and Cdc25. Journal of cell science. 2000;113(Pt 10):1727–1736. doi: 10.1242/jcs.113.10.1727. [DOI] [PubMed] [Google Scholar]
- 5.Furnari B, Blasina A, Boddy MN, McGowan CH, Russell P. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Molecular biology of the cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanford JS, Ruderman JV. Changes in regulatory phosphorylation of Cdc25C Ser287 and Wee1 Ser549 during normal cell cycle progression and checkpoint arrests. Molecular biology of the cell. 2005;16:5749–5760. doi: 10.1091/mbc.E05-06-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Kumagai A, Dunphy WG. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Molecular biology of the cell. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman TR, Tang Z, Dunphy WG. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 1993;72:919–929. doi: 10.1016/0092-8674(93)90580-j. [DOI] [PubMed] [Google Scholar]
- 9.Perry JA, Kornbluth S. Cdc25 and Wee1: analogous opposites? Cell division. 2007;2:12. doi: 10.1186/1747-1028-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. The Journal of cell biology. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaturvedi P, Eng WK, Zhu Y, Mattern MR, Mishra R, Hurle MR, Zhang X, Annan RS, Lu Q, Faucette LF, et al. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–4054. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- 12.Furnari B, Rhind N, Russell P. Science. Vol. 277. New York, N.Y: 1997. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase; pp. 1495–1497. [DOI] [PubMed] [Google Scholar]
- 13.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Science. Vol. 277. New York, N.Y: 1997. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216; pp. 1501–1505. [DOI] [PubMed] [Google Scholar]
- 14.Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 15.Margolis SS, Perry JA, Forester CM, Nutt LK, Guo Y, Jardim MJ, Thomenius MJ, Freel CD, Darbandi R, Ahn JH, et al. Role for the PP2A/B56delta phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell. 2006;127:759–773. doi: 10.1016/j.cell.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margolis SS, Perry JA, Weitzel DH, Freel CD, Yoshida M, Haystead TA, Kornbluth S. A role for PP1 in the Cdc2/Cyclin B-mediated positive feedback activation of Cdc25. Molecular biology of the cell. 2006;17:1779–1789. doi: 10.1091/mbc.E05-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolis SS, Walsh S, Weiser DC, Yoshida M, Shenolikar S, Kornbluth S. PP1 control of M phase entry exerted through 14-3-3-regulated Cdc25 dephosphorylation. The EMBO journal. 2003;22:5734–5745. doi: 10.1093/emboj/cdg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Z, Dunphy WG. Response of Xenopus Cds1 in cell-free extracts to DNA templates with double-stranded ends. Molecular biology of the cell. 2000;11:1535–1546. doi: 10.1091/mbc.11.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer research. 2000;60:5934–5936. [PubMed] [Google Scholar]
- 20.Melchionna R, Chen XB, Blasina A, McGowan CH. Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nature cell biology. 2000;2:762–765. doi: 10.1038/35036406. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & development. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 22.Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 23.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annual review of genetics. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 24.Lavin MF, Kozlov S. Cell cycle. Vol. 6. Georgetown, Tex: 2007. ATM activation and DNA damage response; pp. 931–942. [DOI] [PubMed] [Google Scholar]
- 25.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 26.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. The EMBO journal. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutchison CJ, Cox R, Drepaul RS, Gomperts M, Ford CC. Periodic DNA synthesis in cell-free extracts of Xenopus eggs. The EMBO journal. 1987;6:2003–2010. doi: 10.1002/j.1460-2075.1987.tb02464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchison CJ, Cox R, Ford CC. Development. Vol. 103. Cambridge, England: 1988. The control of DNA replication in a cell-free extract that recapitulates a basic cell cycle in vitro; pp. 553–566. [DOI] [PubMed] [Google Scholar]
- 29.Smythe C, Newport JW. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods in cell biology. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- 30.Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 31.Yoo HY, Shevchenko A, Shevchenko A, Dunphy WG. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. The Journal of biological chemistry. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- 32.Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Molecular cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 33.Newmeyer DD, Farschon DM, Reed JC. Cell-free apoptosis in Xenopus egg extracts: inhibition by Bcl-2 and requirement for an organelle fraction enriched in mitochondria. Cell. 1994;79:353–364. doi: 10.1016/0092-8674(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 34.Kornbluth S. Apoptosis in Xenopus egg extracts. Methods in enzymology. 1997;283:600–614. doi: 10.1016/s0076-6879(97)83047-9. [DOI] [PubMed] [Google Scholar]
- 35.Deming P, Kornbluth S. Methods in molecular biology. Vol. 322. Clifton, N.J: 2006. Study of apoptosis in vitro using the Xenopus egg extract reconstitution system; pp. 379–393. [DOI] [PubMed] [Google Scholar]
- 36.Chau BN, Cheng EH, Kerr DA, Hardwick JM. Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Molecular cell. 2000;6:31–40. [PubMed] [Google Scholar]
- 37.Paydas S, Tanriverdi K, Yavuz S, Disel U, Sahin B, Burgut R. Survivin and aven: two distinct antiapoptotic signals in acute leukemias. Ann Oncol. 2003;14:1045–1050. doi: 10.1093/annonc/mdg277. [DOI] [PubMed] [Google Scholar]
- 38.Figueroa B, Jr, Chen S, Oyler GA, Hardwick JM, Betenbaugh MJ. Aven and Bcl-xL enhance protection against apoptosis for mammalian cells exposed to various culture conditions. Biotechnology and bioengineering. 2004;85:589–600. doi: 10.1002/bit.10913. [DOI] [PubMed] [Google Scholar]
- 39.Choi J, Hwang YK, Sung KW, Kim DH, Yoo KH, Jung HL, Koo HH. Aven overexpression: association with poor prognosis in childhood acute lymphoblastic leukemia. Leukemia research. 2006;30:1019–1025. doi: 10.1016/j.leukres.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Bratton SB, Walker G, Srinivasula SM, Sun XM, Butterworth M, Alnemri ES, Cohen GM. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. The EMBO journal. 2001;20:998–1009. doi: 10.1093/emboj/20.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y. Mechanical aspects of apoptosome assembly. Current opinion in cell biology. 2006;18:677–684. doi: 10.1016/j.ceb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Minshull J, Blow JJ, Hunt T. Translation of cyclin mRNA is necessary for extracts of activated xenopus eggs to enter mitosis. Cell. 1989;56:947–956. doi: 10.1016/0092-8674(89)90628-4. [DOI] [PubMed] [Google Scholar]
- 43.Hunt T. Maturation promoting factor, cyclin and the control of M-phase. Current opinion in cell biology. 1989;1:268–274. doi: 10.1016/0955-0674(89)90099-9. [DOI] [PubMed] [Google Scholar]
- 44.Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1-like kinase. Molecular biology of the cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartkova J, Bakkenist CJ, Rajpert-De Meyts E, Skakkebaek NE, Sehested M, Lukas J, Kastan MB, Bartek J. Cell cycle. Vol. 4. Georgetown, Tex: 2005. ATM activation in normal human tissues and testicular cancer; pp. 838–845. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2--cyclin B and its involvement in the self-amplification of MPF at mitosis. The EMBO journal. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izumi T, Maller JL. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Molecular biology of the cell. 1993;4:1337–1350. doi: 10.1091/mbc.4.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Traven A, Heierhorst J. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays. 2005;27:397–407. doi: 10.1002/bies.20204. [DOI] [PubMed] [Google Scholar]
- 49.Petersen P, Chou DM, You Z, Hunter T, Walter JC, Walter G. Protein phosphatase 2A antagonizes ATM and ATR in a Cdk2-and Cdc7-independent DNA damage checkpoint. Molecular and cellular biology. 2006;26:1997–2011. doi: 10.1128/MCB.26.5.1997-2011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Molecular cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 51.Ali A, Zhang J, Bao S, Liu I, Otterness D, Dean NM, Abraham RT, Wang XF. Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes & development. 2004;18:249–254. doi: 10.1101/gad.1176004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zermati Y, Mouhamad S, Stergiou L, Besse B, Galluzzi L, Boehrer S, Pauleau AL, Rosselli F, D'Amelio M, Amendola R, et al. Nonapoptotic role for apaf-1 in the DNA damage checkpoint. Molecular cell. 2007;28:624–637. doi: 10.1016/j.molcel.2007.09.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.