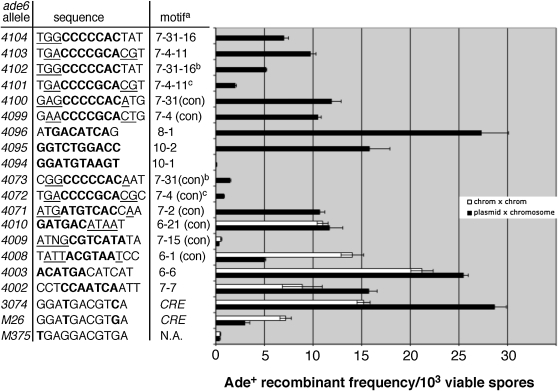
Figure 3.—
Reconstructed sequence motifs produce hotspots. High-frequency sequence motifs (Table 2 and Figure S1) were reconstructed in the ade6 gene and tested for hotspot activity in homothallic crosses (plasmid pWS35 × chromosome; solid bars) or heterothallic crosses (chromosome × chromosome; open bars). Heterothallic strains were crossed with WS315 (ade6-469; Table 1). Each bar represents the average of at least three crosses ±1 SEM. Sequences show the original motif (Table 2; boldface type) with some flanking nucleotides (regular type). Consensus sequence nucleotides (Figure S1) are underlined when applicable. In this table, only ade6-4009 and ade6-4094 are not significantly more active than ade6-M375 (P > 0.01, Student's t-test). Notes: aThe motif (Table 2) on which the allele is based. Con, consensus sequence. NA, not applicable. Motifs with three numbers indicate reconstruction of a particular allele from a subscreen (Figure S1), e.g., 7-31-16 = sequence number 16 from subscreen of motif 7-31. bThese motifs are inverted relative to ade6-4100 and -4104. cThese motifs are located 36 bp downstream relative to ade6-4099 and -4103.