Abstract
The role of sex chromosomes in sex determination has been well studied in diverse groups of organisms. However, the role of the genes on the sex chromosomes in conferring sexual dimorphism is still being experimentally evaluated. An unequal complement of sex chromosomes between two sexes makes them amenable to sex-specific evolutionary forces. Sex-linked genes preferentially expressed in one sex over the other offer a potential means of addressing the role of sex chromosomes in sexual dimorphism. We examined the testis transcriptome of the silkworm, Bombyx mori, which has a ZW chromosome constitution in the female and ZZ in the male, and show that the Z chromosome harbors a significantly higher number of genes expressed preferentially in testis compared to the autosomes. We hypothesize that sexual antagonism and absence of dosage compensation have possibly led to the accumulation of many male-specific genes on the Z chromosome. Further, our analysis of testis-specific paralogous genes suggests that the accumulation on the Z chromosome of genes advantageous to males has occurred primarily by translocation or tandem duplication.
IN the silkworm, Bombyx mori, as in other organisms, differentiation of the testis and formation of sperm are under the control of a large number of tissue- and stage-specific genes. In previous studies, a few testis-specific genes involved in silkworm spermatogenesis, such as the genes coding for BmAHA1 (Miyagawa et al. 2005), BmDmc1 (Kusakabe et al. 2001), and testis-specific tektin (Ota et al. 2002), have been reported. However, no comprehensive data on the testis transcriptomes are available for B. mori. Among insects, the testis transcriptome has been studied in detail only in Drosophila melanogaster (Andrews et al. 2000; Parisi et al. 2003; Ranz et al. 2003; Mikhaylova et al. 2008).
Recent high-throughput genomics projects have focused on the construction, annotation, and analysis of cell-specific and tissue-specific transcriptomes, providing fundamental insights into biological processes (Fizames et al. 2004; Urushihara et al. 2006). Data gathered from expressed sequence tags (ESTs) and microarray- based gene expression profiles are important resources for discovery of novel genes expressed in a tissue-specific manner. In this context, we analyzed the B. mori testis transcriptome to identify testis-specific genes and their functions. Further, we annotated the testis-specific ESTs by using protein homology data and Gene Ontology and deduced gene structures of a few testis-specific genes using whole-genome shotgun (WGS) sequence data and ESTs.
Another rewarding exercise of analyzing the testis transcriptome is to examine whether there is any correlation between the sex-specific expression of the genes and their chromosomal location. In eukaryotes, an unequal complement of sex-determining chromosomes exists between the two sexes. Unlike mammals and dipteran insects, which have an XX/XY system of sex chromosome composition, lepidopteran insects have a ZZ/ZW or a ZZ/ZO system of sex chromosome composition. Two current hypotheses propose a contradictory fate for the genes that reside on the sex chromosomes and are preferentially or exclusively expressed in one sex. Rice's hypothesis (Rice 1984) proposes that genes with sex-biased or sex-specific expression are overrepresented on the X chromosome and has been tested in human (Saifi and Chandra 1999; Vallender and Lahn 2004) and mouse (Wang et al. 2001). An alternative hypothesis suggests that the X becomes feminized because the X chromosome is present in females 50% more frequently than in males, providing evolution with more opportunity to act on genes that benefit females (Reinke et al. 2000); thus, genes with female-biased expression should reside preferentially on the X chromosome. So which hypothesis is correct? Or are the two hypotheses mutually compatible? Studies by Wang et al. (2001) on mouse male germ cells and by Saifi and Chandra (1999) and Lercher et al. (2003) on the human X chromosome concluded that genes expressed specifically in males are predominantly X-linked, far in excess of male-specific genes on the autosomes, thus corroborating the Rice hypothesis. In contrast, in a study of sex-biased expression in mouse, Khil et al. (2004) showed that genes involved in spermatogenesis are relatively underrepresented on the X chromosome whereas female-biased genes are overrepresented on it. Further, they showed that meiotic sex chromosome inactivation (MSCI), in which sex chromosomes become heterochromatic and transcriptionally inactive, accounts for the depletion of testis-expressed genes on the X chromosome. This was inferred because the genes expressed before MSCI are overrepresented on the X chromosome. In sharp contrast, male-biased genes in Drosophila (Parisi et al. 2003; Ranz et al. 2003) and genes expressed in spermatogenic and oogenic cells in Caenorhabditis elegans (Reinke et al. 2004) are underrepresented on the X chromosome.
In this study we tested this phenomenon in B. mori, a lepidopteran model system. In silkworm, the female is the heterogametic sex and has one Z and one W chromosome, whereas the male has two Z chromosomes. We analyzed the distribution of 1104 testis-specific genes, identified by microarray analysis in an earlier study (Xia et al. 2007), and 1984 full-length testis-specific cDNAs and ESTs, which were mapped onto their specific locations on B. mori chromosomes. Our results showed that the Z chromosome (linkage group 1) harbors a significantly higher number of testis-specific genes than the autosomes. We suggest that lack of dosage compensation and sexual antagonism could have led to the accumulation of male advantageous genes on the Z chromosome. In the course of evolution, proteins that are required in higher amounts in males than in females would have been favored on the Z chromosome to facilitate phenotypic sexual dimorphism. Finally, initial analysis of testis-specific paralogs suggests that male-advantageous genes are accumulated on the Z chromosome by translocation from autosomes or tandem duplication.
MATERIALS AND METHODS
Sequence source:
More than 100,000 ESTs are available for B. mori in the NCBI dbEST (Mita et al. 2003; Xia et al. 2004). We downloaded 9614 testis ESTs and a total of 96,051 ESTs derived from tissues other than testis (supporting information, Table S1).
Sequence analysis:
Since many ESTs may be derived from the same gene, the sequences were assembled into clusters with the TGICL program (Pertea et al. 2003). A cluster is defined as a unique set of sequences that have sequence similarity. A cluster that has only one sequence is termed a singleton.
On the basis of Gene Ontology (GO) annotation of closely related homologs, ESTs were assigned with molecular functions, biological process, and cellular component from the GO database (Ashburner et al. 2000) (see File S1, Figure S1, Figure S2, and Figure S3 for details).
To identify putative homologies to known proteins, 3622 clusters were subjected to BLASTx searches against the nonredundant (nr) database of the NCBI, using a cutoff E-value of 1e-05 for parsing output files (Altschul et al. 1990). An E-value of 1e-20 was used as an upper limit to assign significant homology. In cases where no significant homology was found, an E-value limit of 1e-05 was used to assign weak homology. We found this additional category useful for data mining as many clusters do not represent full-length sequences, making it possible that only a highly divergent region of a gene sequence is available in our collection. The category of weak homology allowed us to find potential homologs in such situations.
Digital differential display:
We carried out in silico differential display to identify genes expressed specifically in the testis. This was done by carrying out MegaBLAST with a cutoff score ≥50, percentage of similarity ≥94, and E-value ≤1, against the nonredundant EST set obtained by clustering and assembly of B. mori ESTs from tissues other than testis and using nonredundant testis ESTs as queries. Testis ESTs that did not show any similarity with ESTs from other tissues were considered as putative testis-specific genes.
Experimental validation of a few evolutionarily conserved testis-specific genes:
To confirm the testis specificity of genes predicted by digital differential display, semiquantitative RT–PCR was carried out for a select set of 15 transcripts (Table S2) using total RNA from six different tissues: midgut, fatbody, head, silk gland, epidermis, and gonads of fifth instar larvae as templates. Sex-limited silkworm strains carrying translocations of a region of chromosome 2 that harbors the gene for larval markings to the W chromosome were utilized for this purpose. Male and female larvae of this silkworm stock can be distinguished from the third instar onward by the presence of markings on the thoracic segment in females but not in males. Total RNA was isolated separately from all tissues using Trizol (Invitrogen, Carlsbad, CA), followed by DNase (Invitrogen) treatment to remove genomic DNA.
PCR was carried out using a thermal cycler (Eppendorf, Germany) under the following conditions: initial denaturation at 94° for 2 min; 30 cycles of 94° for 30 sec, 58° for 30 sec, and 72° for 2 min; and a final elongation step at 72° for 10 min. Silkworm cytoplasmic β-actin cDNA was amplified as an endogenous control. The PCR reaction components included 1× buffer, 100 μm dNTPs, 1.5 mm MgCl2, 0.5 unit Taq polymerase (MBI), and 0.5 μm primers.
Physical mapping of testis-specific genes and analysis of testis-specific gene paralogs:
In general, tissue-specific gene expression is considered as an indicator or a predictor of a tissue-specific function. Using the criterion that the intensity of expression of a gene in a particular tissue exceeds twice that in other tissues, a total of 1104 genes were identified as being testis specific in B. mori (Xia et al. 2007).
Testis-specific genes validated in this manner by microarray results were assigned chromosomal positions on the B. mori genome to examine their distribution on different chromosomes. Mapping was carried out using the B. mori physical chromosome map implemented in KAIKOBLAST and the University of Tokyo Genome Browser (UTGB). In brief, all of the gene sequences were queried against the physical map database using the BLAST program. The results were then manually parsed to determine the exact location of genes. The total number of genes on each chromosome and the number of genes per megabase of each chromosome were calculated using the map data. We also mapped 465 genes from other somatic tissues identified in Xia et al. (2007) through microarray analysis onto chromosomes to compare their distribution with that of testis-specific genes.
From the testis-specific genes identified through microarray analysis, paralogs were selected by combining BLAST parameters and homology to published annotated genes. Translated BLAST (tBLASTx) was performed for each of the testis-specific genes against the complete set of testis-specific genes. The BLAST result was parsed to obtain information on hits with a score >100. The parsed data were manually checked to group the paralogs and assigned chromosomal locations. Each group was then analyzed by carrying out BLAST against the NCBI protein nr database. Only groups that had the same functional annotation for the genes included in a group were regarded as genuine paralogs.
To strengthen our findings, we clustered 20,000 full-length cDNAs (fl-cDNAS) generated from testis (K. Mita, unpublished data) and 9614 testis ESTs (Mita et al. 2003; Xia et al. 2004) to obtain a nonredundant set of sequences. From these sequences we identified 2559 testis-specific genes by removing the sequences that were present in EST libraries derived from other tissues. Further, after excluding the microarray-validated testis-specific genes, 1857 testis-specific genes were identified as new ones and were mapped onto B. mori chromosomes to study their distribution on the Z chromosome and the autosomes.
RESULTS AND DISCUSSION
In the study reported here in silico analysis of 9614 ESTs derived from B. mori fifth instar larval testis revealed that several testis-specific genes are evolutionarily conserved from insects to mammals. These findings are consistent with the idea that the testis expresses a complex set of transcripts, as observed in Drosophila (Andrews et al. 2000). The present study also identified several families of testis-specific genes, such as those that encode tektins, dyneins, kinases, and tubulins, involved in silkworm spermatogenesis.
High gene diversity in the testis transcriptome:
The availability of 9× coverage of the B. mori genome sequence and abundant EST resources for this insect facilitated the identification of a subset of testis-specific genes. Clustering and assembly of 9614 testis ESTs resulted in a total of 3622 unique clusters containing 1112 contigs and 2510 singletons. We also obtained 24,857 unique clusters (8819 contigs and 16,038 singletons) by clustering and assembly of the ESTs derived from other tissues.
To identify putative homologs and orthologs, we subjected the clusters to BLASTx searches against the nr database of the NCBI. A total of 2385 (66%) unique sequences showed homology with known proteins and could be assigned a putative identity. Of the 3622 clusters, 2.6% matched proteins with an E-value of ≤1e-99 and were considered to be genuine orthologs. Thirty percent of the clusters found a hit with an E-value between 1e-20 and 1e-99 and were assigned significant homology. Finally, 16% of these clusters had a first hit with an E-value between 1e-19 and 1e-05 and were assigned weak homology to a protein from the nr database.
In silico differential display of B.mori testis ESTs:
Several interesting features about the testis transcriptome were revealed by in silico subtraction of testis-derived nonredundant transcripts from the total transcripts from other somatic tissues. More than 900 of the 3622 unique clusters of testis originally identified to be of testis origin were found to be expressed only in the testis. These were regarded as putative testis-specific genes. Similar results were reported by Xia and co-workers (Xia et al. 2007), who found 1104 testis-specific genes using microarray analysis.
Several testis-specific genes are conserved across phyla:
Using the EST database, we identified four β-tubulin and three α-tubulin genes that could be classified into at least three distinct subfamilies: ubiquitously expressed, developmentally regulated, and testis specific. Among them we identified a previously unknown α-tubulin gene, which we have named bmtua4 (accession no. Bmo.6186). This gene has a single exon, similar to the three genes reported previously (Kawasaki et al. 2003). bmtua4 showed ∼76% similarity to bmtua1 and bmtua2 and ∼71% similarity to bmtua3 genes. Whereas the β-tubulin (bmtub4) (Mita et al. 1995) and α-tubulin (bmtua3) genes reported earlier (Kawasaki et al. 2003) were testis specific, bmtua4 showed enhanced expression in testis. In this respect silkworm spermatogenesis has the same distinctive properties as other species in that many gene families include paralogs such as tubulin, dynein light chain genes that are expressed solely in spermatogenic cells (Eddy and O'Brien 1998), and others are expressed in somatic and germinal cells.
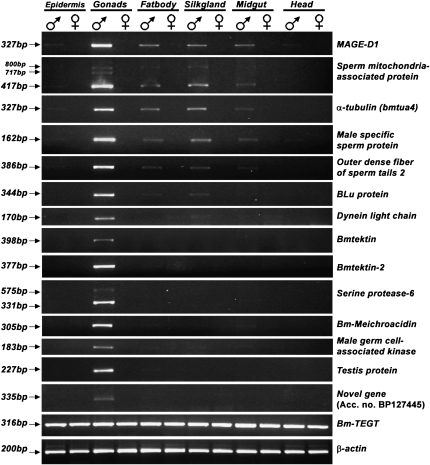
Through BLAST analysis we found 34 testis-specific genes (Table S3) that are known to have specialized functions in testis. All these genes have testis-specific homologs in species as diverse as mammals (Homo sapiens, Mus musculus, Rattus norvegicus, Canis familiaris, Bos taurus), birds (Gallus gallus), amphibia (Xenopus laevis), fishes (Tetraodon nigroviridis, Danio rerio), ascidians (Ciona intestinalis), and sea urchins (Strongylocentrotus purpuratus). To confirm their testis specificity, RT–PCR expression analysis of 15 of these evolutionarily conserved genes was carried out. Of these, 9, including a 717-bp testis-specific splice product of Sperm mitochondria associated protein, were found to be expressed exclusively in the testis; the remaining 6 genes showed male specificity with enhanced expression in the testis. Only 1 gene, a homolog of TEGT (Testis Enhanced Gene Transcript), showed equal expression in all the tissues examined (Figure 1). Expression analysis using RT–PCR was consistent with the tissue distribution of their ESTs as revealed by in silico analysis. Expression analysis also revealed that most of the testis-enhanced genes were male specific. Conservation of several testis-specific genes from insects to mammals suggests that the proteins they encode are probably indispensable for spermatogenesis.
Figure 1.—
RT–PCR analysis of expression of 15 predicted testis-specific genes. β-Actin was used as an internal control. Details of accession numbers and tissue specificity as predicted in silico are given in Table S3. Primer sequences are given in Table S2.
Expression analysis confirmed the presence of predicted alternative splice forms of Sperm mitochondria associated protein and Serine protease genes. The Sperm mitochondria associated protein gene showed three transcripts with amplicons of 800, 717, and 417 bp, whereas the Serine protease gene showed two products of 575 and 331 bp. We found one testis-specific transcript (accession no. BP127445), having no apparent similarity to any protein from the nr database of the NCBI, and it was confirmed to be testis specific through RT–PCR (Figure 1).
Physical mapping reveals abundance of testis-specific genes on the Z chromosome:
An increasing body of evidence suggests that the evolutionary significance of sex chromosomes probably involves not only the mechanism of sex determination but also the evolution of sexually dimorphic traits (Charlesworth 1996; Rice 1996; Reinhold et al. 1998; Gotter et al. 1999; Hurst and Randerson 1999; Gibson et al. 2002).
Analyzing the distribution of sex-specific genes between sex chromosomes and autosomes could uncover the underlying molecular mechanisms of sexual dimorphism since the factors governing the molecular evolution of sex-linked genes differ in several ways compared to those affecting the evolution of autosomal genes. In humans, the X chromosome has more than its fair share of genes involved in sex and reproduction (Saifi and Chandra 1999; Vallender and Lahn 2004) compared to Drosophila (Parisi et al. 2003; Ranz et al. 2003) and C. elegans (Reinke et al. 2004), where the X chromosome is deficient in male-biased genes.
The biased representation of sex-specific genes on sex chromosomes is attributed to two main reasons, sexual antagonism and dosage compensation. According to the hypothesis of sexual antagonism (Rice 1984; Hurst and Randerson 1999), an unusual homogametic sex chromosome gene content reflects a nonrandom accumulation of sexually antagonistic mutations on this chromosome. The other hypothesis concerns the epigenetic modifications of the sex chromosomes associated with meiotic sex chromosome inactivation and dosage compensation (Parisi et al. 2003; Rogers et al. 2003; Reinke et al. 2004; Khil et al. 2005). However, the exact role of these mechanisms still remains elusive, as all the studies have been carried out in male heterogametic sex chromosome systems (XX/XY system).
Alternatively, the mechanisms responsible for the nonrandom representation of sex-biased and sex-specific genes on the homogametic sex chromosome may be clarified by analyzing the gene content of the Z chromosome, a homogametic sex chromosome in heterogametic female organisms. Unlike the X chromosome, the Z chromosome spends more time in male individuals. If sexually antagonistic selection were the primary mechanism affecting the sex chromosome gene content, the opposite trend would be expected in the representation of sex-biased and sex-specific genes on the X and Z chromosomes (Storchova and Divina 2006). In a recent study in chicken, which is a female heterogametic system like B. mori, the Z chromosome was found to be significantly enriched in male-biased genes expressed in brain compared to female-biased genes. However, the study did not find any bias in distribution of testis-specific genes on the Z chromosome (Storchova and Divina 2006).
Unlike in Drosophila, the recent findings show that there is no dosage compensation in B. mori (Koike et al. 2003) and other lepidopterans (Johnson et al. 1979; Charlesworth 1996; Rice 1996; Reinhold et al. 1998; Gotter et al. 1999; Hurst and Randerson 1999; Gibson et al. 2002).
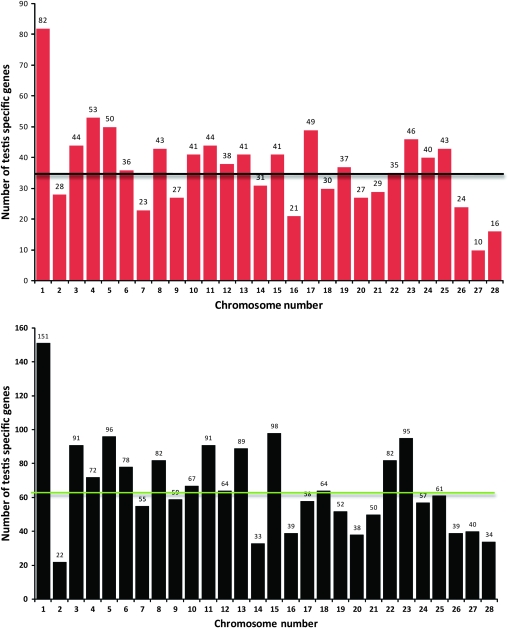
To address the question whether or not the sex-specific genes are distributed in a biased manner on the Z chromosome, we investigated the genomic distribution of B. mori genes that are preferentially expressed only in testis. For this purpose, we assayed two data sets, one set comprising 1104 microarray-validated testis-specific genes (Xia et al. 2007) and the second comprising 1984 testis-specific genes derived from fl-cDNAs and ESTs. These were assigned chromosomal positions using the B. mori physical chromosome map implemented in KAIKOBLAST and UTGB. We were able to map 1029 genes and 1857 genes in the first and second data sets, respectively, onto their respective chromosomal locations (Figure 2). Physical mapping revealed many interesting features about the frequency and chromosomal distribution of testis-specific genes. Surprisingly, the Z chromosome harbored the highest number of testis-specific genes (82 and 151 from the first and second data sets, respectively) as compared to the average number of testis-specific genes on autosomes (35 ± 2 and 63 ± 4; Figure 2). A Student's t-test revealed that the Z chromosome harbored a significantly higher (P < 0.001) number of genes compared to that on autosomes. These results suggest that the locus on the Z chromosome is more than two times more likely to have testis-specific expression than an autosomal locus.
Figure 2.—
Distribution of testis-specific genes identified through microarray analysis (top panel) and through EST and fl-DNA sequence analysis (bottom panel) on different chromosomes of B. mori. There is a significant difference (P < 0.001) between the number of testis-specific genes present on the Z chromosome and on autosomes in both the cases. Among 1104 microarray-validated testis-specific genes, 1029 were successfully mapped onto B. mori chromosomes. The average number of testis-specific genes on autosomes was calculated to be 35 ± 2, which is indicated by a black horizontal line on the histogram. Of 1984 testis-specific genes identified through analysis of testis-derived fl-cDNAs and ESTs, 1857 genes could be mapped onto B. mori chromosomes. The average number of genes on autosomes was calculated to be 63 ± 4 (green horizontal line).
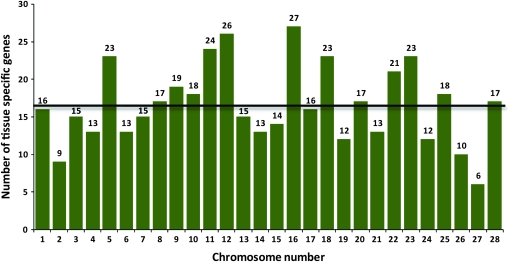
We also assessed the allocation of genes expressed exclusively in a single tissue excluding testis and ovary, between autosomes and the Z chromosome. For this purpose, we mapped 465 additional tissue-specific genes (Xia et al. 2007), which showed no apparent chromosome bias in their distribution (Figure 3). These results indicate that testis-specific genes are more often located on the Z chromosome than is expected by chance.
Figure 3.—
Distribution of tissue-specific genes (excluding testis and ovary) on different chromosomes of B. mori. Of 501 other tissue-specific genes, 465 were successfully mapped. The average number of other tissue-specific genes on autosomes was calculated to be 16, which is indicated by the black horizontal line on the histogram.
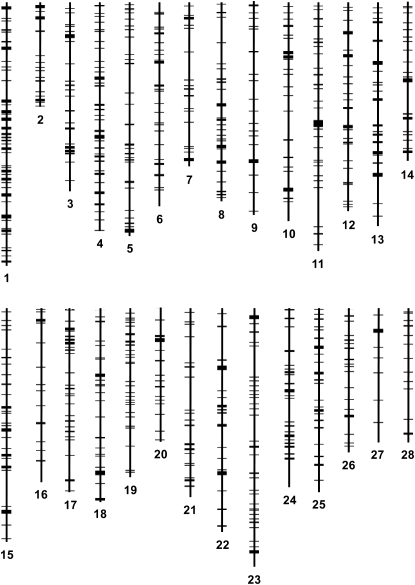
To study the density of testis-specific genes on different chromosomes, we calculated the number of genes per megabase for each chromosome. The Z showed a significantly higher frequency of testis-specific genes (Student's t-test, P < 0.001), with 11.45 genes per megabase of chromosome compared to 6.54 ± 0.18 genes on autosomes (Table 1). These results provide further evidence that overrepresentation of testis-specific genes on the Z is not due to its sheer size or to a higher frequency of genes on the Z chromosome. To determine the presence of any clusters of testes-specific genes, we constructed a physical map using Karyoview (http://www.ensembl.org/index.html). The map showed the spatial distribution of genes on all B. mori chromosomes (Figure 4). It was evident from the map that genes are quite evenly and densely distributed on the Z chromosome. However, the analysis confirmed the presence of several clusters of testis-specific genes on different autosomes, suggesting an apparent bias in their distribution compared to the Z. Information on the exact chromosomal location of each gene can be found in Table S4.
TABLE 1.
Number and frequency of testis-specific genes on different chromosomes of the silkworm, B. mori
| Chromosome | Size (Mb) | No. of testis-specific genes (full-length cDNA and ESTs, and microarray-validated genes) | No. of testis-specific genes/Mb |
|---|---|---|---|
| Chromosome 1 (Z chromosome) | 20.35 | 233 | 11.45 |
| Chromosome 2 | 7.94 | 50 | 6.30 |
| Chromosome 3 | 14.68 | 135 | 9.20 |
| Chromosome 4 | 17.87 | 125 | 6.99 |
| Chromosome 5 | 18.37 | 146 | 7.95 |
| Chromosome 6 | 15.91 | 114 | 7.17 |
| Chromosome 7 | 12.75 | 78 | 6.12 |
| Chromosome 8 | 15.36 | 125 | 8.14 |
| Chromosome 9 | 16.47 | 86 | 5.23 |
| Chromosome 10 | 17.22 | 108 | 6.27 |
| Chromosome 11 | 19.58 | 135 | 6.89 |
| Chromosome 12 | 16.47 | 102 | 6.19 |
| Chromosome 13 | 17.45 | 130 | 7.45 |
| Chromosome 14 | 12.60 | 64 | 5.08 |
| Chromosome 15 | 17.97 | 139 | 7.74 |
| Chromosome 16 | 13.70 | 60 | 4.38 |
| Chromosome 17 | 14.34 | 107 | 7.46 |
| Chromosome 18 | 15.16 | 94 | 6.20 |
| Chromosome 19 | 13.19 | 89 | 6.75 |
| Chromosome 20 | 10.61 | 65 | 6.12 |
| Chromosome 21 | 14.92 | 79 | 5.29 |
| Chromosome 22 | 17.36 | 117 | 6.74 |
| Chromosome 23 | 20.08 | 141 | 7.01 |
| Chromosome 24 | 13.96 | 97 | 6.95 |
| Chromosome 25 | 14.14 | 104 | 7.36 |
| Chromosome 26 | 10.76 | 63 | 5.86 |
| Chromosome 27 | 10.44 | 50 | 4.79 |
| Chromosome 28 | 10.33 | 50 | 4.84 |
| Total | 419.98 | 2886 | |
| Autosomal average ± SE | 14.80 ± 0.47 | 98 ± 6 | 6.54 ± 0.18 |
Figure 4.—
Physical map showing the distribution of 1029 microarray-validated testis-specific genes on B. mori chromosomes.
Why is a certain category of genes abundant on the Z chromosome? Previous studies have shown that in butterflies a disproportionate number of genes related to sexuality, reproduction, and speciation are located on the Z chromosome, which forms approximately one-sixtieth of the genome in females. Female mate-selection behavior, male courtship signals, female limitation of color polymorphism, and mimicry are thought to result largely from interactions between autosomal genes and uncompensated Z-linked regulatory genes (Stehr 1959; Sheppard 1961; Cook 1964; Grula and Taylor 1980; Sperling 1994). In B. mori, four muscle protein genes, Bmkettin, Bmtitin1, Bmtitin2, and Bmprojectin, and another gene involved in locomotor behavior, Bmhig, are Z-linked and are not dosage compensated (Suzuki et al. 1998, 1999; Koike et al. 2003). These genes are functionally conserved between B. mori and Drosophila. In silkworm, although adult moths have lost flight during the course of domestication, male moths flap their wings more vigorously than females to approach sedentary females. In this context, Z-linkage of uncompensated muscle proteins encoding genes ensures a higher quantity of these proteins in males. It is speculated that the Z chromosome has evolved through a process of genome shuffling to accumulate genes whose products are required at higher levels in males (Koike et al. 2003). Considering these results together, we argue that the absence of dosage compensation and nonrandom distribution of male-biased genes on the Z chromosome have ensured accumulation of male advantageous genes in silkworm.
Our results are consistent with the Rice thesis (Rice 1984), which suggests that sex-linked genes are likely to evolve sex-related functions. The beneficial mutations will be selected naturally if they are located on sex chromosomes rather than on autosomes. The evolutionary dynamics of male-benefit mutations were considered when they first appear as rare alleles on X chromosomes in contrast with autosomes. When male-benefit mutations are rare, autosomal recessive alleles offer no advantage to (heterozygous) males and thus would be unlikely to spread in the population. By contrast, X-linked recessive alleles would confer a benefit to hemizygous males, greatly favoring the alleles' likelihood of permeating the population. Eventually, as an allele's frequency increased in the population, female fitness would be diminished by the detrimental effects of homozygosity. This would generate adaptive pressure to limit the gene's expression to males through additional mutations. Therefore, it was postulated that X chromosomes should evolve to harbor a disproportionate share of male-specific genes functioning in male differentiation (Rice 1984; Wang et al. 2001).
By contrast, Z linkage can facilitate the increase of traits that also favor the homogametic sex. Dominant mutations favoring the homogametic sex have a greater chance to be fixed on the Z chromosome where they are exposed to selection, because two-thirds of the Z chromosomes reside in homogametic individuals compared to only one-third in heterogametic individuals and due to absence of dosage compensation. In the Z chromosome, dominant sexually antagonistic genes that favor the homogametic sex are expressed at a higher rate in the sex where they are positively selected. Our findings are consistent with this prediction.
Possible translocation of male advantageous genes onto Z chromosomes from autosomes:
Duplicated genes may acquire novel functions and altered expression patterns (Ohno et al. 1968) and thus contribute to diversification of tissues during development (Mikhaylova et al. 2008). Analysis of such paralogs provides a unique opportunity to study genome evolution as it sheds light on the history of gene duplication and gene trafficking between chromosomes through translocation events. In Drosophila, a number of testis-specific genes have been reported to be generated by gene duplications, where the duplicated gene is specifically expressed in the male reproductive system while the parental gene is ubiquitous (Nurminsky et al. 1998; Betran and Long 2003). In the present study we have used the data on paralogous genes that express in B. mori testis to investigate the evolutionary causes that lead to the enrichment of testis-specific genes on the Z chromosome. Through BLAST analysis we obtained 30 groups of paralogs (comprising 74 of 1104 testis-specific genes), and among them only 12 were found in clusters on different chromosomes. In contrast, the remaining 18 did not show any clustering, and although some paralogs were mapped onto the same chromosome, they were located far apart from each other. Duplications were more frequent among genes coding for dynein proteins (3 of 30 paralogous groups), probably because of their requirement in large amounts in sperm for motility (Moss et al. 1992). None of the clustered coexpressed testis-specific genes showed similarity to transposable elements, in contrast with nonclustered paralogous groups, where 6 paralogous groups were found to code for transposable elements (e.g., reverse transcriptase, transposon polyprotein, and transposase).
One or more genes in the six nonclustered paralogous groups were located on the Z chromosome (Table S5), corresponding to ∼30% of the nonclustered paralogs, the highest for any chromosome. Our speculation is that during the course of evolution male advantageous genes were translocated onto the Z chromosome and selected positively. This may explain the preponderance of one of the copies of testis-specific nonclustered paralogs on the Z chromosome. Also, 3 (25%) of the 12 coexpressed clusters were present on the Z chromosome, again the highest number for any chromosome. Analysis of expression patterns of paralogous genes in testes would provide valuable information to clarify the functional relationships between these genes (Mikhaylova et al. 2008). On the basis of these results we surmise that testis-specific paralogs are more concentrated on Z chromosomes either by clustering or by the location of one of the copies of nonclustered paralogs. Such translocations followed by fixation may be responsible for the enrichment of testis-specific genes on the Z chromosome. This argument is in line with the proposal by Charlesworth and Charlesworth (1980) that translocations from the autosomes to the X chromosome would be favored if the translocated region harbored sexually antagonistic genes.
Conclusions:
In silico analyses of the B. mori transcriptome provide a foundation for further in-depth analysis of activities in male germ cells. Information produced by the sequencing of B. mori cDNA libraries and identification of testis-specific genes in the present study provide new insights into the structure, diversity, and molecular evolution of genes involved in spermatogenesis in lepidopterans in particular and insects in general. Our study of the distribution of testis-specific genes on B. mori chromosomes has shown that testis-specific genes are accumulated on the Z chromosomes either by translocation from other chromosomes or by tandem duplication on the Z chromosome.
Acknowledgments
We thank two anonymous reviewers and Marian R. Goldsmith for their comments on the manuscript. This work was supported by a Centre of Excellence grant from the Department of Biotechnology, Government of India to J.N. K.P.A. is a recipient of a fellowship from the Council of Scientific and Industrial Research, India.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.108.099994/DC1.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Andrews, J., G. G. Bouffard, C. Cheadle, J. Lu, K. G. Becker et al., 2000. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler et al., 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran, E., and M. Long, 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1996. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6 149–162. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., and B. Charlesworth, 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet. Res. 35 205–214. [DOI] [PubMed] [Google Scholar]
- Cook, A. G., 1964. Dosage compensation and sex-chromatin in non-mammals. Genet. Res. 5 354–365. [Google Scholar]
- Eddy, E. M., and D. A. O'Brien, 1998. Gene expression during mammalian meiosis. Curr. Top. Dev. Biol. 37 141–200. [PubMed] [Google Scholar]
- Fizames, C., S. Munos, C. Cazettes, P. Nacry, J. Boucherez et al., 2004. The Arabidopsis root transcriptome by serial analysis of gene expression. Gene identification using the genome sequence. Plant Physiol. 134 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, J. R., A. K. Chippindale and W. R. Rice, 2002. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc. Biol. Sci. 269 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter, A. L., J. D. Levine and S. M. Reppert, 1999. Sex-linked period genes in the silkmoth, Antheraea pernyi: implications for circadian clock regulation and the evolution of sex chromosomes. Neuron 24 953–965. [DOI] [PubMed] [Google Scholar]
- Grula, J. W., and O. R. Taylor, Jr., 1980. The effect of X–chromosome inheritance on mate-selection behavior in the sulfur butterflies, Colias eurytheme and C. philodice. Evolution 34 688–695. [DOI] [PubMed] [Google Scholar]
- Hurst, L. D., and J. P. Randerson, 1999. An eXceptional chromosome. Trends Genet. 15 383–385. [DOI] [PubMed] [Google Scholar]
- Johnson, L. D., M. Binder and R. A. Lazzarini, 1979. A defective interfering vesicular stomatitis virus particle that directs the synthesis of functional proteins in the absence of helper virus. Virology 99 203–206. [DOI] [PubMed] [Google Scholar]
- Kawasaki, H., K. Sugaya, G. X. Quan, J. Nohata and K. Mita, 2003. Analysis of alpha- and beta-tubulin genes of Bombyx mori using an EST database. Insect Biochem. Mol. Biol. 33 131–137. [DOI] [PubMed] [Google Scholar]
- Khil, P. P., N. A. Smirnova, P. J. Romanienko and R. D. Camerini-Otero, 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 36 642–646. [DOI] [PubMed] [Google Scholar]
- Khil, P. P., B. Oliver and R. D. Camerini-Otero, 2005. X for intersection: retrotransposition both on and off the X chromosome is more frequent. Trends Genet. 21 3–7. [DOI] [PubMed] [Google Scholar]
- Koike, Y., K. Mita, M. G. Suzuki, S. Maeda, H. Abe et al., 2003. Genomic sequence of a 320-kb segment of the Z chromosome of Bombyx mori containing a kettin ortholog. Mol. Genet. Genomics 269 137–149. [DOI] [PubMed] [Google Scholar]
- Kusakabe, T., Y. Kawaguchi, T. Maeda and K. Koga, 2001. Role of interaction between two silkworm RecA homologs in homologous DNA pairing. Arch. Biochem. Biophys. 388 39–44. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., A. O. Urrutia and L. D. Hurst, 2003. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol. Biol. Evol. 20 1113–1116. [DOI] [PubMed] [Google Scholar]
- Mikhaylova, L. M., K. Nguyen and D. I. Nurminsky, 2008. Analysis of the Drosophila melanogaster testes transcriptome reveals coordinate regulation of paralogous genes. Genetics 179 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita, K., M. Nenoi, M. Morimyo, H. Tsuji, S. Ichimura et al., 1995. Expression of the Bombyx mori beta-tubulin-encoding gene in testis. Gene 162 329–330. [DOI] [PubMed] [Google Scholar]
- Mita, K., M. Morimyo, K. Okano, Y. Koike, J. Nohata et al., 2003. The construction of an EST database for Bombyx mori and its application. Proc. Natl. Acad. Sci. USA 100 14121–14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa, Y., J. M. Lee, T. Maeda, K. Koga, Y. Kawaguchi et al., 2005. Differential expression of a Bombyx mori AHA1 homologue during spermatogenesis. Insect Mol. Biol. 14 245–253. [DOI] [PubMed] [Google Scholar]
- Moss, A. G., W. S. Sale, L. A. Fox and G. B. Witman, 1992. The alpha subunit of sea urchin sperm outer arm dynein mediates structural and rigor binding to microtubules. J. Cell Biol. 118 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminsky, D. I., M. V. Nurminskaya, D. De Aguiar and D. L. Hartl, 1998. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 396 572–575. [DOI] [PubMed] [Google Scholar]
- Ohno, S., U. Wolf and N. B. Atkin, 1968. Evolution from fish to mammals by gene duplication. Hereditas 59 169–187. [DOI] [PubMed] [Google Scholar]
- Ota, A., T. Kusakabe, Y. Sugimoto, M. Takahashi, Y. Nakajima et al., 2002. Cloning and characterization of testis-specific tektin in Bombyx mori. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 133 371–382. [DOI] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, G., X. Huang, F. Liang, V. Antonescu, R. Sultana et al., 2003. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19 651–652. [DOI] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300 1742–1745. [DOI] [PubMed] [Google Scholar]
- Reinhold, K., M. D. Greenfield, Y. Jang and A. Broce, 1998. Energetic cost of sexual attractiveness: ultrasonic advertisement in wax moths. Anim. Behav. 55 905–913. [DOI] [PubMed] [Google Scholar]
- Reinke, V., H. E. Smith, J. Nance, J. Wang, C. Van Doren et al., 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6 605–616. [DOI] [PubMed] [Google Scholar]
- Reinke, V., I. S. Gil, S. Ward and K. Kazmer, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311–323. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38 735–742. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 1996. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381 232–234. [DOI] [PubMed] [Google Scholar]
- Rogers, D. W., M. Carr and A. Pomiankowski, 2003. Male genes: X-pelled or X-cluded? BioEssays 25 739–741. [DOI] [PubMed] [Google Scholar]
- Saifi, G. M., and H. S. Chandra, 1999. An apparent excess of sex- and reproduction-related genes on the human X chromosome. Proc. Biol. Sci. 266 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, P. M., 1961. Some contributions to population genetics resulting from the study of the Lepidoptera. Adv. Genet. 10 165–216. [DOI] [PubMed] [Google Scholar]
- Sperling, F. A. H., 1994. Sex-linked genes and species differences in Lepidoptera. Can. Entomol. 126 807–818. [Google Scholar]
- Stehr, G., 1959. Hemolymph polymorphism in a moth and the nature of sex-controlled inheritance. Evolution 13 537–560. [Google Scholar]
- Storchova, R., and P. Divina, 2006. Nonrandom representation of sex-biased genes on chicken Z chromosome. J. Mol. Evol. 63 676–681. [DOI] [PubMed] [Google Scholar]
- Suzuki, M. G., T. Shimada and M. Kobayashi, 1998. Absence of dosage compensation at the transcription level of a sex-linked gene in a female heterogametic insect, Bombyx mori. Heredity 81(Pt. 3): 275–283. [DOI] [PubMed] [Google Scholar]
- Suzuki, M. G., T. Shimada and M. Kobayashi, 1999. Bm kettin, homologue of the Drosophila kettin gene, is located on the Z chromosome in Bombyx mori and is not dosage compensated. Heredity 82(Pt. 2): 170–179. [DOI] [PubMed] [Google Scholar]
- Urushihara, H., T. Morio and Y. Tanaka, 2006. The cDNA sequencing project. Methods Mol. Biol. 346 31–49. [DOI] [PubMed] [Google Scholar]
- Vallender, E. J., and B. T. Lahn, 2004. How mammalian sex chromosomes acquired their peculiar gene content. BioEssays 26 159–169. [DOI] [PubMed] [Google Scholar]
- Wang, P. J., J. R. McCarrey, F. Yang and D. C. Page, 2001. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27 422–426. [DOI] [PubMed] [Google Scholar]
- Xia, Q., Z. Zhou, C. Lu, D. Cheng, F. Dai et al., 2004. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306 1937–1940. [DOI] [PubMed] [Google Scholar]
- Xia, Q., D. Cheng, J. Duan, G. Wang, T. Cheng et al., 2007. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 8 R162. [DOI] [PMC free article] [PubMed] [Google Scholar]