Abstract
Polyploidy is an important force in the evolution of flowering plants. Genomic merger and doubling induce an extensive array of genomic effects, including immediate and long-term alterations in the expression of duplicate genes (“homeologs”). Here we employed a novel high-resolution, genome-specific, mass-spectrometry technology and a well-established phylogenetic framework to investigate relative expression levels of each homeolog for 63 gene pairs in 24 tissues in naturally occurring allopolyploid cotton (Gossypium L.), a synthetic allopolyploid of the same genomic composition, and models of the diploid progenitor species. Results from a total of 2177 successful expression assays permitted us to determine the extent of expression evolution accompanying genomic merger of divergent diploid parents, genome doubling, and genomic coevolution in a common nucleus subsequent to polyploid formation. We demonstrate that 40% of homeologs are transcriptionally biased in at least one stage of cotton development, that genome merger per se has a large effect on relative expression of homeologs, and that the majority of these alterations are caused by cis-regulatory divergence between the diploid progenitors. We describe the scope of transcriptional subfunctionalization and 15 cases of probable neofunctionalization among 8 tissues. To our knowledge, this study represents the first characterization of transcriptional neofunctionalization in an allopolyploid. These results provide a novel temporal perspective on expression evolution of duplicate genomes and add to our understanding of the importance of polyploidy in plants.
DUPLICATE genes are widespread in genomes of almost all eukaryotes. Among flowering plants, polyploidy (whole-genome duplication) is a primary source of duplicate genes (Soltis and Soltis 1999; Wendel 2000; Bowers et al. 2003; Lockton and Gaut 2005). All flowering plants either are contemporary polyploids or harbor the evolutionary signature of paleopolyploidy (ancient polyploidy) in their genomes. Polyploidy may have influenced flowering plant diversification, as it provides raw material for the evolution of novelty by relaxing purifying selection on duplicate genes (Stephens 1951; Ohno 1970; Lynch and Conery 2000; Wendel 2000). Through genic redundancy, polyploids may be subject to an array of evolutionary processes, including subfunctionalization (evolution of partitioned ancestral functions among duplicate genes) and neofunctionalization (evolution of novel functions among duplicate genes).
Subfunctionalization and neofunctionalization have been demonstrated in several species (Force et al. 1999; Adams et al. 2003; Duarte et al. 2006; Cusack and Wolfe 2007; Liu and Adams 2007; Teshima and Innan 2008). From an evolutionary perspective, both processes can lead to the preservation of the two members of a duplicate gene pair (Ohno 1970; Lynch and Force 2000). Because duplicate genes tend to be lost rapidly through mutational processes (Lynch and Conery 2000), subfunctionalization is thought to be most important shortly after gene duplication. As the age of the duplicate pair increases, neofunctionalization becomes increasingly likely (Stephens 1951; Ohno 1970). Further linking these two processes, it has been suggested that subfunctionalization could serve as a preservational transition state leading to neofunctionalization (Rastogi and Liberles 2005). Thus following polyploidy, both subfunctionalization and neofunctionalization may make significant contributions to duplicate gene retention and functional diversification.
In addition to subfunctionalization and neofunctionalization, allopolyploid plants also generate diversity through rapid genomic changes at various levels, including chromosomal lesions and intergenomic exchanges, as in wheat (Shaked et al. 2001), Brassica (Song et al. 1995; Pires et al. 2004; Udall et al. 2004), and Arabidopsis (Madlung et al. 2002), epigenetic modifications (Lee and Chen 2001; Madlung et al. 2002; Wang et al. 2004; Salmon et al. 2005; Gaeta et al. 2007), and gene expression changes (Adams et al. 2003; Wang et al. 2004; Bottley et al. 2006; Adams 2007; Flagel et al. 2008). It is thought that these changes result from “genomic shock” caused by the joint effects of genome merger and genome doubling during allopolyploid formation (Adams et al. 2004; Hegarty et al. 2006; Flagel et al. 2008). Additionally, allopolyploidy entails combining homeologous regulatory variation and may lead to expression variation through interacting cis- and trans-regulatory factors, as has been shown for allelic variation (Wittkopp et al. 2004; Stupar and Springer 2006; Swanson-Wagner et al. 2006). Collectively, these results demonstrate that both genomic and genic evolutionary processes play a role in allopolyploid evolution.
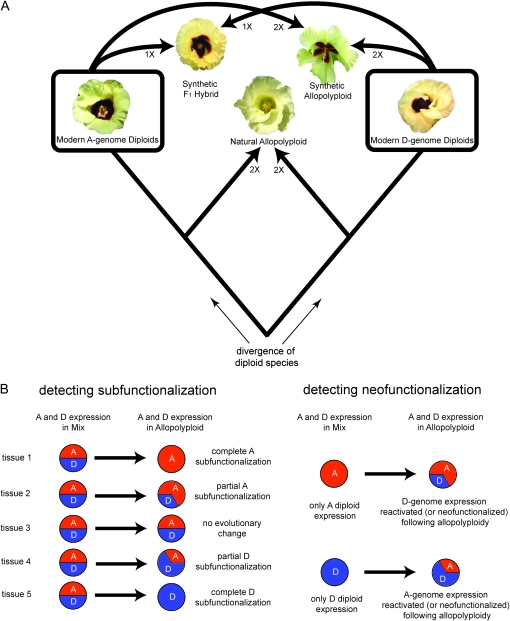
The cotton genus (Gossypium) is a useful system to study the extent of genomic changes that accompany genome merger and allopolyploidization (Wendel and Cronn 2003). Allotetraploid cottons were formed by the merger of two diploid species originating, respectively, from the cotton A- and D-genome groups. This event took place 1–2 million years ago (Percy and Wendel 1990; Wendel and Albert 1992; Seelanan et al. 1997; Cronn et al. 2002; Senchina et al. 2003) (Figure 1A). The modern diploid species Gossypium arboreum (A genome) and G. raimondii (D genome) are extant diploids most similar to the ancestral A- and D-genome diploids involved in the formation of natural allotetraploids (Percy and Wendel 1990; Wendel and Albert 1992; Seelanan et al. 1997; Cronn et al. 2002; Senchina et al. 2003) (Figure 1A). Following formation, the allotetraploid lineage diverged into five extant species. Furthermore, F1 hybrids and allotetraploids synthetically derived from A- and D-genome species mergers are also available (Figure 1A and Table 1). These synthetic accessions have proved particularly useful in teasing apart the effects of genome merger and genome doubling during the formation of the natural allopolyploid (Adams et al. 2004; Adams and Wendel 2005a; Flagel et al. 2008). Although these studies and others (Comai et al. 2000; Adams et al. 2004; Soltis et al. 2004; Hegarty et al. 2006; Tate et al. 2006; Chen 2007) have provided insights into the formation and immediate genetic consequences of polyploidy, there is still much to be learned about stabilization and evolution of polyploid genomes following formation.
Figure 1.—
Phylogenetic framework and detection of subfunctionalization and neofunctionalization among homeologs. (A) Phylogenetic history of diploid and allopolyploid cotton (Gossypium). Allopolyploidy occurred ∼1–2 MYA by hybridization between A- and D-genome diploid species, most similar to the modern species G. arboreum and G. raimondii. The modern F1 hybrid and synthetic allopolyploid, both derived from A- and D-genome diploid species, mimic the stages of genome merger and genome duplication during allopolyploid formation. (B) After genome merger regulatory changes may cause allelic/homeologous gene expression patterns to diverge. These patterns can result in subfunctionalization, the partitioning of ancestral expression, or neofunctionalization, operationally defined here as the development of novel expression patterns relative to that of the ancestor. The latter was detected by comparing ancestral expression (1:1 mix) to the expression found in the F1, synthetic, and Maxxa.
TABLE 1.
Details of plant materials used
| Taxon | Genome designation | Accession | Ploidy level | Location of origin |
|---|---|---|---|---|
| G. arboreum | A2 | AKA-8401 | Diploid | Africa |
| G. raimondii | D5 | Jfw | Diploid | Peru |
| G. arboreum × G. raimondii | (A2 × D5) | NA | Diploid | Laboratory |
| G. hirsutum | AD1 | cv. Maxxa | Tetraploid | Mexico/Central America |
| G. arboreum × G. davidsonii | 2(A2 × D3) | NA | Tetraploid | Laboratory |
The natural allotetraploid (G. hirsutum) was derived from hybridization, 1–2 MYA, between diploid A- and D-genome species most similar to the modern species G. arboreum and G. raimondii. The cytoplasmic donor of G. hirsutum is its A-genome parent and thus the F1 cross was created in the same direction, with A2 as the maternal parent. The synthetic allotetraploid was created by crossing A2 and D3 diploid parents followed by genome doubling through colchicine treatment.
In this study we employ a genome-specific, mass-spectrometry technology to study relative levels of allelic and homeologous (gene pairs duplicated by polyploidy) gene expression in diploid and allopolyploid cotton. By contrasting allelic and homeologous gene expression in cotton species within an appropriate phylogenetic framework (Figure 1A), we have detected expression patterns consistent with subfunctionalization and neofunctionalization (Figure 1B). Because the cotton accessions selected for this study represent three successive stages in allopolyploid evolution, i.e., genomic merger of divergent parents, genome doubling, and finally genomic coevolution in a common nucleus, we were able to determine the extent of expression evolution accompanying each stage.
MATERIALS AND METHODS
Maintenance of cotton germplasm and tissue collection:
Seedling tissues:
Seeds of two diploid cottons, G. arboreum (A2) and G. raimondii (D5), and a natural (G. hirsutum L. cv. Maxxa) and synthetic [2(A2 × D3)] allotetraploid cotton (Table 1), were sown and grown in steamed potting mix in the Pohl Conservatory at Iowa State University at 24° day/20° night with a photoperiod of 16 hr light/8 hr dark. The synthetic allotetraploid cotton was formed by colchicine doubling the hybrid resulting from a cross between A2 and the D-genome species G. davidsonii (D3). Three biological replicates were planted for each species and seedling-stage tissues were sampled at 10 days postemergence. Additionally, a sterile F1 hybrid (A2 × D5) population has been maintain through vegetative propagation, and was also sampled for some tissues. The above accessions include representatives of both diploid progenitor genomes (A and D genomes), their synthetic F1 hybrid and synthetic allotetraploid, and a natural allopolyploid cotton (Wendel and Cronn 2003) (Figure 1A). All seedlings were sampled between 9 am to 10 am to minimize circadian effects, and tissues were flash frozen in liquid nitrogen and stored at −80° prior to RNA isolation.
Vegetative and floral tissues:
Seedlings were grown for 3–5 weeks before transfer to larger pots and maintained at 32° and a photoperiod of 16 hr light/8 hr dark. After the emergence of the fifth leaf, the first, third, and fifth leaves were harvested from all five taxa on the same day and flash frozen immediately and stored. Petioles were sampled from the fifth leaf of each biological replicate, and midrib and lamina tissues were harvested from young and newly emerged leaves at the same time. After 3–4 months, flowers from all species, except D5, were harvested on 0 days postanthesis (dpa) (0 dpa is the day the flower opened). Juvenile plants from D5 were grown separately under a shade regime for ∼1 month, a treatment necessary to induce flowering. Fully opened flowers were collected between 9 am and 11 am to mitigate circadian effects. All flower tissues were manually excised and immediately flash frozen in liquid nitrogen.
Fiber:
Plants from all five taxa were grown in three replicates in the horticulture greenhouse at Iowa State University and flowers were harvested for four different stages of fiber development (5, 10, 20, and 25 dpa). For each replicate and developmental time point, ovules were excised and immediately frozen in liquid nitrogen. Ovules were visually inspected for cell damage and fibers were inspected for contaminating tissue.
Isolation of total RNA and sample platform preparation:
RNA isolation:
All 24 tissues (Table 2) from the five taxa and three biological replicates were collected in 1.7-ml microfuge tubes. RNAs were extracted from all seedling, vegetative, and floral tissues using a modified QIAGEN RNA extraction protocol according to the manufacturer's instructions (QIAGEN, Valencia, CA) with modifications as follows: Tissues were ground in fresh XT buffer (Wan and Wilkins 1994) in microfuge tubes with plastic pestles and incubated at 42° for 1.5 hr. Then 2 m KCl was added and the sample was incubated on ice for 1 hr. After incubation, the samples were transferred to Qiashredder columns supplied with the QIAGEN Plant RNeasy kit and all subsequent steps followed this kit's protocol.
TABLE 2.
Number of successful genome-specific assays calculated for all genes in all tissues
| No. of genes successfully assayed | ||||
|---|---|---|---|---|
| Tissues | Mix | F1 hybrid | Synthetic | Maxxa |
| Seedling stage | ||||
| Primary root | 41 | NA | 33 | 33 |
| Hypocotyl | NA | NA | 7 | 7 |
| Cotyledon | 45 | NA | 38 | 38 |
| Vegetative stage | ||||
| First leaf | NA | NA | 31 | 36 |
| Third leaf | NA | NA | 26 | 26 |
| Fifth leaf | NA | NA | 22 | 36 |
| Petiole | 50 | 50 | 42 | 42 |
| Apical shoot | NA | 48 | 40 | 40 |
| Leaf midrib | NA | 42 | 37 | 38 |
| Leaf lamina | 40 | 34 | 28 | 26 |
| Floral stage | ||||
| Pedicel | NA | 32 | 27 | 25 |
| Bract | NA | 30 | 24 | 21 |
| Calyx | NA | 32 | 22 | 22 |
| Petal | NA | 35 | 26 | 24 |
| Anther | NA | 40 | 24 | 29 |
| Stamina tube | NA | 39 | 27 | 28 |
| Pollen | NA | NA | NA | 11 |
| Style and stigma | NA | 41 | 30 | 27 |
| Ovary wall (0 dpa) | 30 | 32 | 25 | 21 |
| Ovule (0 dpa) | 37 | 38 | 26 | 26 |
| Fiber | ||||
| Fiber (5 dpa) | 25 | NA | 21 | 19 |
| Fiber (10 dpa) | 32 | NA | 26 | 23 |
| Fiber (20 dpa) | 37 | NA | 30 | 30 |
| Fiber (25 dpa) | 41 | NA | 34 | 32 |
| Total | 378 | 493 | 646 | 660 |
NA, tissue not available.
RNAs were extracted from fibers at each developmental time point using a liquid nitrogen/glass bead shearing approach following a lithium chloride hot borate protocol (Hovav et al. 2007; Taliercio and Boykin 2007). Purified RNA samples were quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and assayed for degradation using a BioAnalyzer (Agilent, Palo Alto, CA).
cDNA preparation:
A total of 307 tissue samples were used for RNA isolations, each yielding ∼5 μg of total RNA. All RNA samples were treated with DNase following the manufacturer's protocol [New England Biolabs (DNase I, M0303S)], and assayed for genomic DNA contamination by PCR amplification with primers flanking intron eight of a Gossypium RNA helicase with high similarity to the gene At4G00660 in Arabidopsis (GenBank accession NM_179204). Following DNase treatment, cDNAs were synthesized using Superscript III reverse transcriptase, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). In tissues with surplus RNA yields, cDNAs were also synthesized from equal RNA mixes of A2 and D5 accessions. These mixes served as an in vitro model for mid-parent expression within the allopolyploid and hybrid accessions.
Probe selection for multiplex PCR:
MALDI-TOF mass-spectrometry assays for genome-specific expression were designed for the Sequenom (San Diego) MassARRAY platform. Genes for this platform were selected from 1231 cotton EST contigs (Udall et al. 2006), derived from A2, D5, and AD1 accessions. These contigs were inferred to represent homeologous relationships in AD1 on the basis of comparisons to orthologous sequences from the A- and D-genome diploids This led to the identification of genome-specific SNPs, which were processed using the Sequenom probe selection software. From these results we selected four multiplexes, each including 29 genes.
Genome-specific expression assays:
For each multiplex, forward and reverse primers from all 29 genes were pooled and used to amplify each cDNA sample using the manufacturer's specifications (Sequenom). Amplified cDNAs were visualized on agarose gels to confirm amplification and loaded on a 384-well plate in three technical replicates. Mass-spectrometry quantification of genome-specific expression ratios was performed at the University of Minnesota genotyping facility.
Data processing, filtering, and analysis:
Identification of diagnostic assays:
All expression data recovered from the MassARRAY process were first filtered on the basis of internal measures of assay quality, which included removing all assays flagged as “Bad Spectra,” or having a frequency of uncertainty >0.2 or an unused extension primer frequency >0.5. Next all genes were filtered on the basis of assays of A2 and D5 DNA samples, which were mixed in known ratios (4:1, 2:1, 1:1, 1:2, and 1:4) and used to standardize the genome-specific quantification procedure for each gene (Stupar and Springer 2006). All genes were required to display a strong correlation (R2 > 0.9) between the expected and observed A2:D5 DNA ratios. Additionally, DNAs from Maxxa, the synthetic polyploid, and the F1 hybrid were also assayed as controls for lineage-specific SNPs, which could potentially arise in these accessions, with the expectation that good assays would yield ∼1:1 A- to D-genome values. Maxxa, synthetic, and F1 hybrid assays were excluded if these DNA control values exceeded the expected 1:1 ratio by ±25%. Following filtering, a maximum of nine replicates (three biological × three technical) could potentially be recovered for each assay. Each of these replicate pools represents one gene by tissue and evaluates the proportion of A- and D-genome contribution to the transcriptome. These values were stored as the percentage of D-genome contribution to the transcriptome (% D) and outlier replicates were identified and removed if they deviated from the median of the replicate pool by ±50% D. Next, the mean % D and standard deviation of the remaining values were recorded and used for all subsequent analyses; this complete data set can be found as supporting information, File S2.
Statistical contrasts of genome-specific expression ratios:
Contrasts of A- and D-genome expression ratios were made using a t-test. P-values were then converted to q-values using the method of Storey and Tibshirani (2003), and individual contrasts were considered equivalent when q > 0.05.
RESULTS
Assessment of Sequenom MassARRAY performance:
Using a single nucleotide polymorphism (SNP)-based Sequenom MassARRAY technology, we simultaneously assayed the A- and D-genome contribution to the transcriptome for 63 gene pairs (Table S1). These A- and D-genome gene pairs are hereafter termed “homeologous” in the polyploid genotypes and “allelic” in the diploid F1 hybrid genotypes (note however, in the F1 hybrid chromosomes from the A- and D-genome chromosome pairing is limited; Endrizzi et al. 1985). The MassARRAY technology has previously been shown to be effective in determining the relative allelic transcript levels in hybrid maize (Stupar and Springer 2006). Assays of cotton A- and D-genome expression were made possible by the availability of A- and D-genome-specific SNPs obtained from cotton EST contig assemblies (Udall et al. 2006), which included transcripts from the diploid members of the A and D genome (A2 and D5) and an allotetraploid (AD1). The presence of a genome-specific SNP alters the molecular weight such that the MassARRAY platform can distinguish either variant from a mixed transcript pool and estimate relative abundance.
Expression assays were filtered using a rigorous quality-control protocol (see materials and methods), yielding the total number of successful assays summarized in Table 2. The percentage of successful assays varied among tissues from a maximum of ∼73% in petioles to a minimum of 5% in pollen and hypocotyl tissues (Table 2 and Figure S1). Among 63 genes and 24 tissue types examined, 660 and 646 gene-by-tissue combinations were successful in the natural (“Maxxa” hereafter) and synthetic (“synthetic” hereafter) allopolyploids. Due to limited tissue and sample availability in the F1 and 1:1 A- and D-genome mix, we examined 13 and 10 tissue types in these accessions resulting in 493 and 378 successful assays, respectively (Table 2).
Patterns of genome-specific gene expression in cotton tissues:
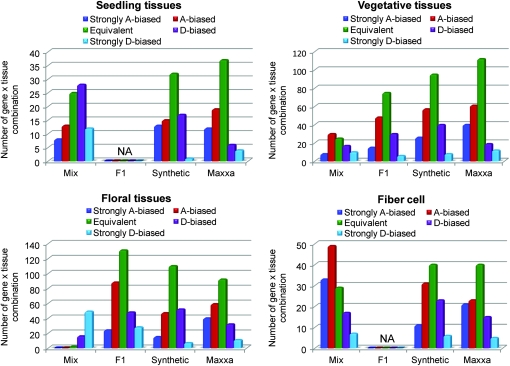
The primary goal of this study was to quantify genome-specific expression among a sampling of cotton tissues and developmental conditions in an evolutionary context. This was accomplished by assaying 24 tissues or developmental stages, which fit into four general categories: seedling, vegetative, and floral tissues, as well as developing fibers (Table 2). For each of these categories, genome-specific expression values were extracted for the mix, F1, and the synthetic and natural (Maxxa) allopolyploids and binned into five groups, using the percentage of D-genome expression as a metric (0–20% D, 20–40% D, 40–60% D, 60–80% D, and 80–100% D) (Figure 2). In the F1, synthetic, and Maxxa, biases indicate differential gene expression between the A- and D-genome transcripts within the same nucleus, whereas in the mix, which pools two biologically different species, a bias reflects differential gene expression between the A2 and D5 parents.
Figure 2.—
Distribution of genome-specific expression states among accessions and within different tissue categories. Each panel represents a tissue category and shows histograms for the mix F1, synthetic, and Maxxa. The expression categories correspond to the following values: strongly A biased (0–20% D expression); A biased (20–40% D expression); equivalent (40–60% D expression); D biased (60–80% D expression); strongly D biased (80–100% D expression). The y-axes indicate the number of gene-by-tissue combinations that fell under each category. NA, tissue type not available.
Overall, F1, synthetic, and Maxxa show an A-genome bias in seedling and vegetative tissues, but in floral tissues the mix shows a D bias whereas the F1 and Maxxa show A-genome biases and the synthetic is nearly equivalent (Figure 2). For “floral” samples the mix is represented by only ovary wall and ovule tissues, though both individually support a D bias. Fiber expression in the mix and Maxxa show a substantial level of A-genome bias, whereas the synthetic is less A-genome biased (Figure 2; note that the F1 hybrid between A2 and D5 is sterile and hence fibers could not be studied). These expression patterns are interesting, as they highlight previous observations (Adams et al. 2003, 2004; Adams and Wendel 2005a; Udall et al. 2006; Yang et al. 2006; Flagel et al. 2008; Hovav et al. 2008b) that neither the A nor D genome is globally dominant with regard to genome-specific expression.
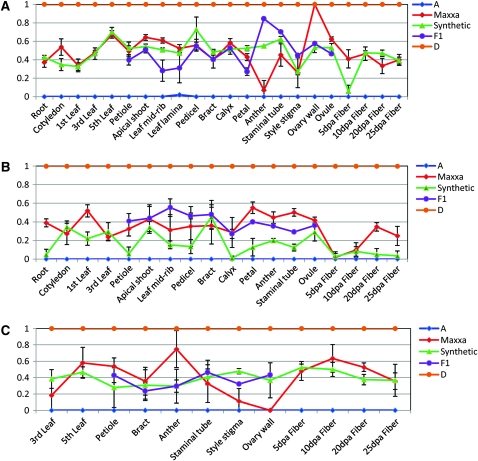
These general trends describe the overall patterns of expression states for this sampling of genes and tissues. At the individual gene level there is considerable variation. An interesting example is the gene CO131164 (a putative phytochrome-associated protein), which shows highly variable expression among tissues and accessions. In Maxxa this gene demonstrates nearly complete A-genome expression in anthers and complete D-genome expression in ovary wall (Figure 3A), indicative of developmentally regulated reciprocal silencing of alternative homeologs in different parts of the same flower (cf. Adams et al. 2003). Additionally, shortly after fiber initiation (5 dpa), CO131164 is strongly A-genome biased in the synthetic, though Maxxa shows approximately equivalent expression (Figure 3A). Another illustrative gene is CO130747 (a putative CBL-interacting protein kinase), which shows significant differences in tissue-specific homeolog expression between Maxxa and the synthetic during many developmental stages (Figure 3B). The synthetic is more A-genome biased in seedling, vegetative, and floral stages, including almost total A-genome expression in roots, petioles, the calyx, and all four developmental stages of fiber. In contrast, Maxxa is only strongly A-genome biased in 5 and 10 dpa fibers. A third example gene illustrated (Figure 3C) is DW008528 (similar to a putative protein with unknown function in Arabidopsis thaliana), for which we observed equivalent A- and D-genome homeolog expression in all tissues for the synthetic and the F1, but considerable expression variation for vegetative tissues in Maxxa. In all fiber stages studied, both Maxxa and the synthetic show nearly equal expression of homeologs.
Figure 3.—
Tissue-specific and genome-specific gene expression among three gene pairs in A2, D5, F1 hybrid, synthetic, and Maxxa. Tissues are arrayed along the x-axis while the proportion of D-genome expression is on the y-axis. The lines linking tissues do not imply a strict order of plant development; instead they serve as a viewing aid. (A) Gene CO131164 (a putative phytochrome-associated protein). (B) Gene CO130747 (a putative CBL-interacting protein kinase). (C) Gene DW008528 (a protein of unknown function).
Genome-specific expression biases during genome merger and doubling:
The accessions studied were selected to provide insight into the various stages involved in allopolyploid speciation, including diploid divergence, genome merger, genome doubling, and subsequent evolution and stabilization. To assess homeolog transcriptional alteration accompanying each of these stages, we identified all gene × tissue combinations shared by all four accessions (mix, F1, synthetic, and Maxxa), as well as those shared just by the F1, synthetic, and Maxxa, and finally just by the synthetic and Maxxa. For each of these groups, we assigned all gene × tissue relationships as either equivalent (“=”; q-value > 0.05) or nonequivalent (“≠”; q-value ≤ 0.05). Specific examples of several expression patterns and their biological interpretation can be found in Figure 4.
Figure 4.—
Examples of tissue-specific expression alteration arising from parental divergence, genomic merger, polyploidy, and polyploidy evolution. Shown are the proportions of D-genome (y-axis) homeolog expression, including the associated standard deviation. (A) Four representative genes from petioles, which exhibit statistically equivalent ratios in all accessions, indicating little expression evolution since divergence between the A- and D-genome parents. (B) Four representative genes from leaf lamina, each showing equivalent expression among the F1 hybrid, synthetic, and Maxxa, which is not equivalent to the mix, indicating an expression change resulting from genomic merger. (C) Four representative genes from petioles (genes CAO71171, CO131379, and CAO49511) and leaf lamina (gene CO131379) showing equal genome-specific expression values in the mix and F1 hybrid, which differ from the synthetic and Maxxa, suggesting that the change occurred as a result of genome doubling. (D) Four genes in petioles showing no change among mix, F1, and synthetic, but a new expression pattern in Maxxa, indicating a change in homeolog-specific expression during the evolution (∼1–2 million years) of allopolyploid cotton.
As summarized in Table 3, when comparing all four accessions, the category that induced the most expression alteration was genome merger (implicated in 23 + 16 + 9 + 9 = 57 gene × tissue events) followed by change due to polyploid evolution (implicated in 9 + 16 + 6 + 9 = 40 gene × tissue events). From these results, it is clear that genome merger and polyploidy evolution (subsequent to formation) have the greatest effect on homeologous gene expression, though diploid divergence and genome doubling are implicated in 11 and 30 gene × tissue events, respectively. For genes lacking data from the mix sample, more homeolog expression changes occurred due to polyploid evolution than polyploidy alone, corroborating the foregoing result. Alternatively, some of the above observations could be due to the divergence between the model diploid progenitors used in this study and the actual ancient parents of natural allopolyploid cotton. Similar findings have been reported in cotton and other polyploid systems regarding the relative importance of genome merger (Adams and Wendel 2005a; Wang et al. 2005; Hegarty et al. 2006; Flagel et al. 2008) and genome doubling (Stupar et al. 2007). However, to our knowledge, this is the first study wherein the specific effects of each of these four components (divergence, merger, polyploidy, and polyploidy evolution) have been disentangled.
TABLE 3.
Distribution of expression states among the mix, F1, synthetic, and Maxxa and their biological interpretation
| Genotype comparison | Gene × tissue combinations showing pattern (% of total) | Biological description |
|---|---|---|
| Mix = F1 = synthetic = Maxxa with equal A–D expression | 6 (6.4) | No change |
| Mix = F1 = synthetic = Maxxa with unequal A–D expression | 11 (11.7) | Change due to A–D divergence |
| Mix ≠ F1 = synthetic = Maxxa | 23 (24.4) | Change due to genome merger |
| Mix = F1 ≠ synthetic = Maxxa | 5 (5.3) | Change due to polyploidy alone |
| Mix = F1 = synthetic ≠ Maxxa | 9 (9.6) | Change due to polyploid evolution |
| Mix ≠ F1 ≠ synthetic ≠ Maxxa | 16 (17) | Change due to all sources |
| Mix = F1 ≠ synthetic ≠ Maxxa | 6 (6.4) | Change due to polyploidy and polyploid evolution |
| Mix ≠ F1 ≠ synthetic = Maxxa | 9 (9.6) | Change due to genome merger and polyploidy |
| Mix ≠ F1 = synthetic ≠ Maxxa | 9 (9.6) | Change due to genome merger and polyploid evolution |
| Total | 94 | |
| F1 = synthetic = Maxxa | 104 (34.9) | No change |
| F1 ≠ synthetic = Maxxa | 57 (19.1) | Change due to polyploidy alone |
| F1 = synthetic ≠ Maxxa | 67 (22.5) | Change due to polyploid evolution |
| F1 ≠ synthetic ≠ Maxxa | 70 (23.4) | Change due to all sources |
| Total | 298 | |
| Synthetic = Maxxa | 275 (50.6) | No change |
| Synthetic ≠ Maxxa | 269 (49.4) | Change due to polyploid evolution |
| Total | 544 |
The first nine rows compare the distribution of expression categories for all available gene × tissue combinations among all four taxa. The next four rows compare just F1, synthetic, and Maxxa, and the last two only synthetic and Maxxa.
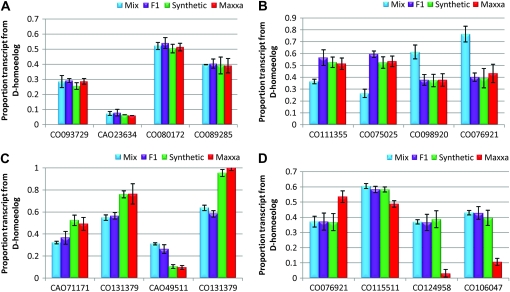
Tissue-specific subfunctionalization and gene silencing:
To address the prevalence of subfunctionalization between homeologous genomes, we searched for patterns of highly differential homeolog expression biases between tissues from the F1, synthetic, and Maxxa (see Figure 1B). We did not detect any cases of complete reciprocal homeolog silencing (here silencing is operationally defined as the absence of detectable transcript) among the 63 genes assayed, but the most subfunctionalized genes and their respective tissues are listed in Table 4. In Maxxa, the most striking example is the gene CO131164, where the A-genome homeolog has been silenced in the ovary wall, but the reverse is observed in anthers, where the A-genome homeolog accounts for 93% of homeologous expression. Other genes showed similar patterns of subfunctionalization in various tissues (Table 4). Interestingly, among those genes displaying the largest degree of expression subfunctionalization, it appears that reproductive tissues such as anthers, style/stigma, staminal tube, and ovary wall are often involved (these tissues comprise 12 of 18 tissues in Table 4). This observation mirrors similar findings from Adams et al. (2003).
TABLE 4.
Proportional transcript contribution of A and D homeologs in different tissues
| Genotype | GenBank accession | Strongly A-biased tissue | A-biased tissue expression (A, D) | Strongly D-biased tissue | D-biased tissue expression (A, D) |
|---|---|---|---|---|---|
| Maxxa | CO131164 | Anther | 93, 7 | Ovary wall | 0, 100 |
| CO080701 | Pollen | 94, 6 | 10 dpa fiber | 14, 86 | |
| CO077994 | 10 dpa fiber | 77, 23 | Ovary wall | 0, 100 | |
| DW008528 | Ovary wall | 100, 0 | Anther | 25, 75 | |
| CO082621 | Anther | 71, 29 | Ovary wall | 0, 100 | |
| Synthetic | CO124958 | First leaf | 100, 0 | Style/stigma | 25, 75 |
| CO098920 | Cotyledon | 91, 9 | Anther | 22, 78 | |
| F1 hybrid | CO121715 | Staminal tube | 100, 0 | Leaf lamina | 25, 75 |
| CAO23634 | Leaf lamina | 100, 0 | Staminal tube | 39, 61 |
Each gene (listed by GenBank accession) demonstrates nearly complete expression subfunctionalization among the two tissues shown.
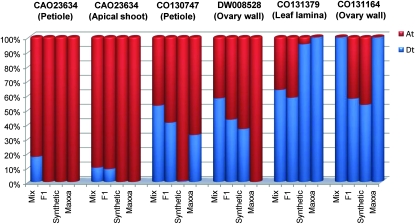
In addition to subfunctionalization, hybrid and polyploid plants also display genome-specific silencing biases. For each genotype, the percentage of completely silenced genes varied from a maximum of ∼6% D-homeolog silencing in Maxxa to a minimum ∼0.3% A-homeolog silencing in the synthetic (Table 5). In most cases, complete silencing remains in each subsequent stage along the pathway to allopolyploidy. For example, the gene CAO23634 (a putative S-formylglutathione hydrolase), is D silenced in petioles and apical shoot meristems in the F1, and this silencing remains in the synthetic and Maxxa polyploids (Figure 5). However, there are also counterexamples, such as the gene CO130747. This gene is D silenced in petioles of the synthetic but the D genome is once again expressed in Maxxa (Figure 5). Another example is gene CO131164, where there is silencing of the A2 diploid in ovary walls, but this gene is expressed in the F1 and synthetic, and then once again the A genome is silenced in Maxxa (Figure 5).
TABLE 5.
Distribution of tissue-specific homeologous gene silencing events
| Genotype | No. of gene × tissue assayed | Gene × tissue combinations with D silencing | Gene × tissue combinations with A silencing | % D silenced | % A silenced |
|---|---|---|---|---|---|
| F1 | 493 | 7 (4 genes) | 6 (3 genes) | 1.41 | 1.21 |
| Synthetic | 646 | 16 (5 genes) | 2 (2 genes) | 2.48 | 0.31 |
| Maxxa | 660 | 42 (11 genes) | 9 (5 genes) | 6.36 | 1.36 |
Figure 5.—
Examples of tissue-specific expression partitioning. The y-axis represents the proportional transcript contribution from A and D homeologs.
Tissue-specific transcriptional neofunctionalization:
Neofunctionalization may be detected in our framework by first indentifying all gene × tissue assays that lack expression of either the A- or D-genome ortholog in the mix (i.e., not expressed in the A2 or D5 parent) and which gain expression in the F1, synthetic, or Maxxa (Figure 1B). It is important to note that the pattern above can arise de novo, as a totally novel form of expression, or as a product of the reactivation of a lost ancestral expression regime, and our experiment cannot distinguish between these two forms of transcriptional neofunctionalization.
Using the criteria above, a total of 15 genes across eight different tissues exhibit transcriptional neofunctionalization. Additionally, by observing the range of expression values for the 1:1 parental mixtures in all available genes (Figure S2), it appears unlikely that these cases of neofunctionalization are a product of an inaccurate mix. Among the neofunctionalized genes, 10 showed substantial contributions from both genomes in the F1, synthetic, and Maxxa, reinforcing the presence of gene expression neofunctionalization (Table 6). Genes CO108066 (a putative glyceraldehyde-3-phosphate dehydrogenase) and CO076921 (a putative vacuolar ATP synthase catalytic subunit) show lack of expression of either the A or D orthologs, respectively, in leaf lamina, but both homeologs are expressed in the same tissue in the F1, synthetic, or Maxxa (Table 6). In addition, in those cases where neofunctionalization has occurred, it has been maintained in all genomically merged samples (F1, synthetic, and Maxxa; Figure S3). Overall, the nonfunctional alleles were usually from the diploid A genome (11 of 14 cases), indicative of the potential for a genome-of-origin bias for neofunctionalization in cotton, albeit for a relatively small sampling of genes.
TABLE 6.
Expression neofunctionalization
| Proportion genomic expression
|
|||||
|---|---|---|---|---|---|
| Gene | Tissue | Mix (A, D) | F1 (A, D) | Synthetic (A, D) | Maxxa (A, D) |
| CAO62858 | Root | 0, 100 | NA | 32, 68 | 41, 59 |
| Ovule | 0, 100 | 30, 70 | 25, 75 | 32, 68 | |
| CO111212 | Leaf petiole | 0, 100 | – | 27, 73 | 33, 67 |
| 5 dpa fiber | 0, 100 | NA | 34, 66 | 49, 51 | |
| 10 dpa fiber | 0, 100 | NA | 29, 71 | 42, 58 | |
| CO108066 | Leaf lamina | 0, 100 | 49, 51 | 66, 34 | 63, 37 |
| Ovule | 0, 100 | 59, 41 | 68, 32 | 34, 66 | |
| CO076921 | Leaf lamina | 100, 0 | 61, 39 | 93, 07 | 57, 43 |
| AAP41846 | Ovule | 0, 100 | 53, 47 | 50, 50 | 48, 52 |
| CO081422 | Ovule | 0, 100 | 48, 52 | 40, 60 | – |
| CAO71171 | Ovule | 0, 100 | 61, 59 | 40, 60 | 47, 53 |
| AAK69758 | Ovule | 0, 100 | 49, 51 | 22, 78 | 38, 62 |
| CO077994 | 20 dpa fiber | 100, 0 | NA | 63, 37 | 58, 42 |
| CO093729 | 25 dpa fiber | 100, 0 | NA | 72, 28 | 86, 14 |
Each gene exhibited differential expression between the diploids (shown by the 1:1 parental mix), but expression from both the A and D genomes in the F1, synthetic, and natural allopolyploid (Maxxa). NA, tissue not available. –, value could not be determined.
Evolution of cis- and trans-regulatory variations in cotton:
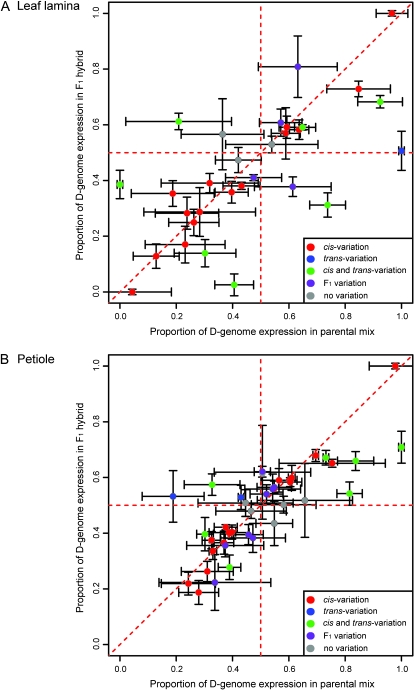
Expression variation can originate via either cis- or trans-regulatory evolution, or both. By comparing genome-specific expression between the mix and F1 it is possible to partition expression variation into cis- and trans-origins, using the procedures described by Wittkopp et al. (2004) and Stupar and Springer (2006). Our analysis of cis- and trans-acting regulation in cotton includes 30 genes in leaf lamina and 38 genes in the petiole (Figure 6). Among both leaf lamina and petiole tissues the most prevalent type of regulatory divergence is cis-regulatory evolution (50% and 39% in lamina and petiole, respectively) followed by a combination of cis- and trans-factors. This result is similar to other studies regarding the prevalence of these modes of regulatory evolution (Wittkopp et al. 2004; Stupar and Springer 2006; Springer and Stupar 2007a; Zhuang and Adams 2007). Additionally this result gives an indication that some of the expression changes attributed to genome merger (Table 3) are likely caused by cis-regulatory divergence between the A and D genomes.
Figure 6.—
Plots of A- and D-genome parental mix (mix) vs. F1 hybrid (F1) for leaf lamina (A) and petiole (B). In principle, genome-specific expression differences initiated by cis-regulatory divergence are expected to share this difference between both the mix and the F1 and will accordingly fall on a 1:1 diagonal when plotted against one another (red points), whereas trans-regulatory divergence will equilibrate genome-specific expression when coresident in the F1 nucleus and instead fall on equivalently expressed horizontal line for the F1 only (blue points). Genes that fall along neither of these lines are inferred to be regulated by a combination of cis- and trans-factors (Wittkopp et al. 2004; Stupar and Springer 2006) (green points). Finally, genes with divergence only in the F1 (purple points) or no expression divergence (gray points) offer no insight into cis- or trans-expression evolution.
DISCUSSION
Homeologous contributions to the transcriptome:
We used a mass-spectrometry-based SNP detection technique to measure allele- and homeolog-specific contributions to the transcriptome of diploid and allopolyploid cotton accessions that were selected to be informative with respect to the evolutionary stages involved in allopolyploid speciation and subsequent evolution (Figure 1A). Although the representative progenitor diploid species used in this study (A2, D3, and D5) are not the actual parents of natural allopolyploid cotton, which formed 1–2 million years ago, a substantial body of evidence indicates that they represent close approximations (reviewed in Wendel and Cronn 2003). Furthermore, to evaluate differences between D3 and D5 [and as a corollary species-specific biases associated with the 2(A2 × D3) synthetic allotetraploid] we compared expression between these species from 18 randomly selected genes in petiole tissues and 17 genes in leaf tissues. These comparisons were made relative to a common A2 reference sample and were conducted using the Sequenom platform following the procedures outlined in materials and methods. These experiments show that D3 and D5 are similar in their expression, having an average expression difference of 15.5% among the 35 comparisons (Figure S4). For comparison, the average variation between biological replicates within D3 and D5 was 12.5%, meaning that within-species variation was ∼81% of the level of the difference between D3 and D5. These results indicate that species-specific differences between D3 and D5 are small.
Contrasting genome-specific expression in these accessions allowed us to allocate expression alterations to the stages of genome merger, genome doubling, and subsequent evolution within the allopolyploid lineage, while revealing examples of subfunctionalization and neofunctionalization (Figure 1B).
To substantiate the MassARRAY-based interpretations, we validated these estimates of genome-specific expression through comparisons to expression data generated by a genome-specific microarray platform (Udall et al. 2006). These validations were conducted for both petals (Flagel et al. 2008) and fibers from several developmental stages (Hovav et al. 2008a), and demonstrate significantly positive correlations.
Allopolyploidy entails the merger of two diploid genomes, which may contribute either equally or disproportionately to the transcriptome. Data presented here demonstrate that genomically biased expression in cotton is a common phenomenon, occurring in vegetative and floral tissues, and also in single-celled fibers, consistent with previous studies using other genes and analytical methods (Adams et al. 2003, 2004; Adams and Wendel 2005a; Udall et al. 2006; Yang et al. 2006; Flagel et al. 2008; Hovav et al. 2008a). In this study, among 49 homeologous genes sampled in Maxxa, ∼40% exhibit biased expression toward the A or D homeolog, in all tissues examined (Figure 2). Furthermore, the extent of genome-specific bias varies substantially among tissues, from nearly equal expression to complete silencing (here again, silencing refers to an absence of detectable transcript). The accumulated results from this study and others noted above indicate that among hybrid and allopolyploid cotton both the A and D genome contribute unequally to the transcript pool, but that neither genome displays an overall expression preference. This result differs from natural and synthetic allotetraploids in Arabidopsis, which show a global downregulation of the A. thaliana genome in favor of the A. arenosa genome (Wang et al. 2006; Chen et al. 2008).
Although genomic preference was not detected at a global scale, relative transcript abundance from individual genes varied greatly. Genome-specific silencing was observed in 4 genes in the F1 hybrid, 5 genes in the synthetic, and 11 genes in Maxxa, noting that this differed widely among tissue types for many of those genes (Table 5). These results indicate that silencing is most prevalent in the natural allopolyploid, following 1–2 MY of allopolyploid evolution. Furthermore, in Maxxa, silencing is more prevalent among D-genome homeologs than among A-genome homeologs (Table 5). Both of these findings regarding the enhancement of silencing in Maxxa and a greater level of D-genome silencing mirror the findings of Flagel et al. (2008), though their study was limited to only petal tissues. Though the phenotypic effects of homeolog silencing in cotton are unknown, it is possible that tissue-specific homeolog silencing has had an impact on the evolution of allotetraploid cotton. For the AdhA gene in G. hirsutum, Liu and Adams (2007) have shown that homeologous expression biases can occur as a response to abiotic stress. These findings of altered homeologous expression patterns in response to genomic stress may hint at the adaptive potential of polyploidy. In this vein, our findings shed additional light on the extensive breadth and diversity of homeolog expression patterns in natural allotetraploid cotton.
Distinguishing the effects of genome merger, genome doubling, and polyploid evolution on gene expression:
By partitioning genome-specific expression changes within a selected framework of cotton accessions (Figure 1A), we were able to determine that genome merger has the largest impact on biased expression of homeologs along the pathway to polyploidy in cotton (Table 3). Allelic expression differences, detectable immediately in the F1 hybrid, likely arise as a result of the merger of the divergent regulatory machinery of the A and D genomes within cotton. As many expression biases are shared with ancient allopolyploid cotton, the early establishment of expression patterns may play a role in gene expression evolution during the formation and subsequent evolution of natural cotton allopolyploids (Adams 2007; Chen 2007). Similar results have been previously noted in cotton (Adams and Wendel 2005a; Flagel et al. 2008), as well as Senecio (Hegarty et al. 2006) and Brassica (Albertin et al. 2006). These authors all found that a considerable portion of gene expression alteration took place at the F1 hybrid stage when compared to resynthesized allopolyploids. In Senecio and Brassica the effect of genome merger was, in fact, found to contribute a majority of the observed expression changes. Hegarty et al. (2006) classified this result as an example of “genomic shock,” a phenomenon which has often been observed in plant hybrids, but remains poorly understood at the molecular level. Some insight may derive from estimating the relative roles of cis- and trans-regulation within the F1 (Figure 6), and in this respect our data indicate that cis-evolutionary factors (those arising from A- and D-genome cis-regulatory divergence), appear to be most prevalent. Taken together, these data indicate that reuniting divergent cis-regulatory domains may be a major component of genomic shock as it pertains to cotton hybrids and allopolyploids.
Following genomic merger, we found that allopolyploid evolution was the next most prevalent contributor to expression evolution (Table 3). This result is interesting as it implicates a significant role for the action of long-term evolutionary processes, such as sub- and neofunctionalization. Furthermore, changes that occur via allopolyploid evolution are more prevalent than those occurring via genome duplication alone (40 vs. 30 gene × tissue events; Table 3). This result indicates that genomic duplication alone may play a less significant role in altering homeologous gene expression states in cotton, possibly affecting only those homeologs with dosage-regulated expression (Osborn et al. 2003).
Mechanisms of functional divergence and retention of homeologs following allopolyploidy:
Tissue-specific and developmental expression variation between coresident genomes may occur via several mechanisms, including altered regulatory interactions, epigenetic modifications, and gene dosage changes (Comai et al. 2000; Birchler et al. 2003; Osborn et al. 2003; Riddle and Birchler 2003; Adams and Wendel 2005b). At present, we lack an explanation of the underlying mechanisms of allelic and homeologous gene expression biases, though our results indicate that both short- (genome merger) and long-term (duplicate gene evolution) evolutionary processes play a role in determining homeolog expression states in allopolyploid cotton. Recent work in allotetraploid Arabidopsis has shown that genome-specific methylation may play a crucial role in establishing homeolog expression patterns (Chen et al. 2008). Using RNAi to silence met1, a cytosine methyltransferase, Chen et al. (2008) demonstrated that many previously identified cases of genome-specific gene silencing were caused by or connected to methylation. Though these results may offer a promising mechanistic explanation of our findings of genome-specific biased expression and silencing, changes in methylation do not appear to accompany allopolyploidy in cotton (Liu et al. 2001). This difference between Arabidopsis and cotton indicates that there may be no single unifying factor that governs genome-specific expression biases in allopolyploid plant species; instead genome-specific expression evolution may occur via a unique and ad hoc mixture of genetic and epigenetic regulatory mechanisms within different species.
Following allopolyploid establishment, several mechanisms may affect the fate of homeologous genes (Leitch and Bennett 1997; Matzke et al. 1999; Wendel 2000; Levy and Feldman 2002; Liu and Wendel 2002; Soltis et al. 2004; Comai 2005; Chen and Ni 2006). One model of homeologous gene retention is subfunctionalization, which is the partitioning of ancestral function and/or expression domains between duplicated genes, such that both copies continue to be necessary (Ohno 1970; Force et al. 1999; Lynch and Force 2000). Various studies of subfunctionalization, including MADS-box genes in Arabidopsis (Duarte et al. 2006), germin genes in barley (Federico et al. 2006), ZMM1 and ZAG1 genes in maize (Mena et al. 1996), and the AdhA gene in cotton (Adams et al. 2003), have shown that expression subfunctionalization occurs in plants. Here we show that instantaneous expression subfunctionalization may occur immediately following genomic merger (Table 4). Because of this, the preservational forces of subfunctionalization may be immediately initiated for a significant number of genes within allopolyploid cotton, as previously suggested (Adams et al. 2003; Adams and Wendel 2005a; Flagel et al. 2008). Recent genomic analyses comparing homeologous regions in G. hirsutum lend support to this claim, as homeologous gene loss appears to be rare (Grover et al. 2004, 2007).
During allopolyploid evolution, duplicate genes not subject to subfunctionalization may still be retained if one copy evolves a novel function via neofunctionalization (Force et al. 1999; Lynch et al. 2001). Several studies have identified neofunctionalization among duplicate genes in diploid plants, including lectins in legumes (Van Damme et al. 2007), MADS-box genes in Physalis (He and Saedler 2005) and Arabidopsis (Duarte et al. 2006), LEAFY paralogs in Idahoa scapigera (Brassicaceae) (Sliwinski et al. 2007), and diterpene synthase paralogs in conifers (Keeling et al. 2008). Expression neofunctionalization was also detected in this study, which makes this the first example of neofunctionalization in an allopolyploid, as far as we are aware. We found 15 genes in eight different tissues where expression was undetectable in one of the parental diploids but appeared in the F1, synthetic, and Maxxa. This pattern, which indicates an expansion of ancestral expression domains, is consistent with expression neofunctionalization.
In addition to the processes described above, cis-and trans-regulatory changes provide insight into the evolution of regulatory networks in cotton. We observed that most variation in gene expression following genome merger is the result of cis-regulatory variation. This finding suggests a mechanism for additive expression patterns detected for many genes in a microarray study of the F1 hybrid (Flagel et al. 2008). Additionally, cis-regulatory variation has been found to be a prevalent mechanism for generating expression differences in F1 maize hybrids (Stupar and Springer 2006; Swanson-Wagner et al. 2006). While cis-regulatory evolution may be more common, it is also possible that trans-regulatory effects may affect gene expression, and even profoundly so. For example, reactivation of a silenced gene copy in a hybrid background, due to a trans-effect, may generate novel expression cascades that have evolutionary consequences. Mechanistic studies that determine the exact nature of important cis-changes would be of tremendous help in advancing our understanding of underpinnings of the observation of a prevalence of cis-regulatory in the divergence in hybrid and allopolyploid plants.
Evolutionary consequences of homeologous gene expression in cotton:
Recurrent polyploidization has played a significant role in adding genetic variation to the genomes of plant species. It has been demonstrated that most duplicate genes are lost quickly on evolutionary times scales (Lynch and Conery 2000; Kellis et al. 2004; Thomas et al. 2006). Despite these rapid losses some homeologous genes are retained, and various explanations have been put forth to explain this retention, including dosage sensitivity (Thomas et al. 2006) and gene function (Blanc and Wolfe 2004). For example, among the retained homeologs in A. thaliana, transcription factors and signal transduction genes have been preferentially retained, whereas genes performing enzymatic functions have not (Blanc and Wolfe 2004). It has also been suggested that alteration in duplicate gene expression patterns may enhance retention (Adams et al. 2003; Flagel et al. 2008). In cotton, this form of duplicate gene retention may be facilitated by expression subfunctionalization and neofunctionalization. These forms of divergence can occur rapidly after polyploidization; indeed we show here that many changes occur immediately in synthetic F1 hybrids and allopolyploids. From an evolutionary perspective, this immediate form of expression divergence can enhance expression variation and phenotypic diversification in the short-term with the long-term consequence of homeolog retention. Together these processes may add to genetic and phenotypic variation with a species, thus enhancing the future potential for natural selection to lead to adaptive evolution.
Acknowledgments
The authors thank two anonymous reviewers for their helpful comments. This project was supported by the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education and Extension Service, grant no. 2005-35301-15700. B.C. received financial assistance from the Department of Biotechnology, India. L.F. received financial assistance through a graduate fellowship from the Plant Sciences Institute at Iowa State University.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.102608/DC1.
References
- Adams, K. L., 2007. Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 98 136–141. [DOI] [PubMed] [Google Scholar]
- Adams, K. L., R. Cronn, R. Percifield and J. F. Wendel, 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K. L., R. Percifield and J. F. Wendel, 2004. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K. L., and J. F. Wendel, 2005. a Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics 171 2139–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K. L., and J. F. Wendel, 2005. b Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8 135–141. [DOI] [PubMed] [Google Scholar]
- Albertin, W., T. Balliau, P. Brabant, A.-M. Chevre, F. Eber et al., 2006. Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics 173 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., D. L. Auger and N. C. Riddle, 2003. In search of the molecular basis of heterosis. Plant Cell 15 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., and K. H. Wolfe, 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottley, A., G. M. Xia and R. M. D. Koebner, 2006. Homoeologous gene silencing in hexaploid wheat. Plant J. 47 897–906. [DOI] [PubMed] [Google Scholar]
- Bowers, J. E., B. A. Chapman, J. Rong and A. H. Paterson, 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422 433–438. [DOI] [PubMed] [Google Scholar]
- Chen, M., M. Ha, E. Lackey, J. Wang and Z. J. Chen, 2008. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics 178 1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. J., 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Ann. Rev. Plant Biol. 58 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. J., and Z. Ni, 2006. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays 28 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6 836–846. [DOI] [PubMed] [Google Scholar]
- Comai, L., A. P. Tyagi, K. Winter, R. Holmes-Davis, S. H. Reynolds et al., 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12 1551–1568.11006331 [Google Scholar]
- Cronn, R. C., R. L. Small, T. Haselkorn and J. F. Wendel, 2002. Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am. J. Bot. 89 707–725. [DOI] [PubMed] [Google Scholar]
- Cusack, B. P., and K. H. Wolfe, 2007. When gene marriages don't work out: divorce by subfunctionalization. Trends Genet. 23 270–272. [DOI] [PubMed] [Google Scholar]
- Duarte, J. M., L. Cui, P. K. Wall, Q. Zhang, X. Zhang et al., 2006. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 23 469–478. [DOI] [PubMed] [Google Scholar]
- Endrizzi, J. E., E. L. Turcotte and R. J. Kohel, 1985. Genetics, cytogenetics, and evolution of Gossypium. Adv. Genet. 23 271–375. [Google Scholar]
- Federico, M. L., F. L. Iniguez-Luy, R. W. Skadsen and H. F. Kaeppler, 2006. Spatial and temporal divergence of expression in duplicated Barley germin-like protein-encoding genes. Genetics 174 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel, L., J. A. Udall, D. Nettleton and J. F. Wendel, 2008. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y.-L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta, R. T., J. C. Pires, F. Iniguez-Luy, E. Leon and T. C. Osborn, 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover, C. E., H. Kim, R. A. Wing, A. H. Paterson and J. F. Wendel, 2004. Incongruent patterns of local and global genome size evolution in cotton. Genome Res. 14 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover, C. E., H. Kim, R. A. Wing, A. H. Paterson and J. F. Wendel, 2007. Microcolinearity and genome evolution in the AdhA region of diploid and polyploid cotton (Gossypium). Plant J. 50 995–1006. [DOI] [PubMed] [Google Scholar]
- He, C., and H. Saedler, 2005. Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae. Proc. Natl. Acad. Sci. USA 102 5779–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty, M. J., G. L. Barker, I. D. Wilson, R. J. Abbott, K. J. Edwards et al., 2006. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr. Biol. 16 1652–1659. [DOI] [PubMed] [Google Scholar]
- Hovav, R., J. A. Udall, B. Chaudhary, E. Hovav, L. Flagel et al., 2008. a The evolution of spinable cotton fiber entailed natural selection for prolonged development and a novel metabolism. PLoS Genet. 4(2): e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav, R., J. A. Udall, B. Chaudhary, R. Rapp, L. Flagel et al., 2008. b Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. USA 105 6191–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav, R., J. A. Udall, E. Hovav, R. A. Rapp, L. Flagel et al., 2007. A majority of genes are expressed in the single-celled seed trichome of cotton. Planta 227 319–329. [DOI] [PubMed] [Google Scholar]
- Keeling, C. I., S. Weisshaar, R. P. C. Lin and J. Bohlmann, 2008. Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc. Natl. Acad. Sci. USA 105 1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., B. W. Birren and E. S. Lander, 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428 617–624. [DOI] [PubMed] [Google Scholar]
- Lee, H.-S., and Z. J. Chen, 2001. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl. Acad. Sci. USA 98 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch, I. J., and M. D. Bennett, 1997. Polyploidy in angiosperms. Trends Plant Sci. 2 470–476. [Google Scholar]
- Levy, A. A., and M. Feldman, 2002. The impact of polyploidy on grass genome evolution. Plant Physiol. 130 1587–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., C. L. Brubaker, G. Mergeai, R. C. Cronn and J. F. Wendel, 2001. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 44 321–330. [PubMed] [Google Scholar]
- Liu, B., and J. Wendel, 2002. Non-Mendelian phenomenon in allopolyploid genome evolution. Curr. Genomics 3 489–505. [Google Scholar]
- Liu, Z., and K. L. Adams, 2007. Expression partitioning between genes duplicated by polyploidy under abiotic stress and during organ development. Curr. Biol. 17 1669–1674. [DOI] [PubMed] [Google Scholar]
- Lockton, S., and B. S. Gaut, 2005. Plant conserved non-coding sequences and paralogue evolution. Trends Genet. 21 60–65. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and A. Force, 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., M. O'Hely, B. Walsh and A. Force, 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung, A., R. W. Masuelli, B. Watson, S. H. Reynolds, J. Davison et al., 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M. A., S. O. Mittelsten and A. J. Matzke, 1999. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. BioEssays 21 761–767. [DOI] [PubMed] [Google Scholar]
- Mena, M., B. A. Ambrose, R. B. Meeley, S. P. Briggs, M. F. Yanofsky et al., 1996. Diversification of C-function activity in maize flower development. Science 274 1537–1540. [DOI] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, New York.
- Osborn, T. C., J. Chris Pires, J. A. Birchler, D. L. Auger, Z. Jeffery Chen et al., 2003. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19 141–147. [DOI] [PubMed] [Google Scholar]
- Percy, R. G., and J. F. Wendel, 1990. Allozyme evidence for the origin and diversification of Gossypium barbadense L. Theor. Appl. Genet. 79 529–542. [DOI] [PubMed] [Google Scholar]
- Pires, J. C., J. Zhao, M. E. Schranz, E. J. Leon, P. A. Quijada et al., 2004. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Bio. J. Linn. Soc. 82 675–688. [Google Scholar]
- Rastogi, S., and D. A. Liberles, 2005. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 5 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, N. C., and J. A. Birchler, 2003. Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 19 597–600. [DOI] [PubMed] [Google Scholar]
- Salmon, A., M. L. Ainouche and J. F. Wendel, 2005. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14 1163–1175. [DOI] [PubMed] [Google Scholar]
- Seelanan, T., A. Schnabel and J. F. Wendel, 1997. Congruence and consensus in the cotton tribe (Malvaceae). Syst. Bot. 22 259–290. [Google Scholar]
- Senchina, D. S., I. Alvarez, R. C. Cronn, B. Liu, J. Rong et al., 2003. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20 633–643. [DOI] [PubMed] [Google Scholar]
- Shaked, H., K. Kashkush, H. Ozkan, M. Feldman and A. A. Levy, 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwinski, M. K., J. A. Bosch, H.-S. Yoon, M. v. Balthazar and D. A. Baum, 2007. The role of two LEAFY paralogs from Idahoa scapigera (Brassicaceae) in the evolution of a derived plant architecture. Plant J. 51 211–219. [DOI] [PubMed] [Google Scholar]
- Soltis, D. E., and P. S. Soltis, 1999. Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 14 348–352. [DOI] [PubMed] [Google Scholar]
- Soltis, D. E., P. S. Soltis and J. A. Tate, 2004. Advances in the study of polyploidy since plant speciation. New Phytol. 161 173–191. [Google Scholar]
- Song, K., P. Lu, K. Tang and T. C. Osborn, 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, N. M., and R. M. Stupar, 2007. a Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19 2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, S. G., 1951. Possible significances of duplication in evolution. Adv. Genet. 4 247–265. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R. M., P. B. Bhaskar, B. S. Yandell, W. A. Rensink, A. L. Hart et al., 2007. Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 176 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R. M., and N. M. Springer, 2006. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner, R. A., Y. Jia, R. DeCook, L. A. Borsuk, D. Nettleton et al., 2006. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA 103 6805–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliercio, E. W., and D. Boykin, 2007. Analysis of gene expression in cotton fiber initials. BMC Plant Biol. 7 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate, J. A., Z. Ni, A. C. Scheen, J. Koh, C. A. Gilbert et al., 2006. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 173 1599–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima, K. M., and H. Innan, 2008. Neofunctionalization of duplicated genes under the pressure of gene conversion. Genetics 178 1385–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B. C., B. Pedersen and M. Freeling, 2006. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall, J. A., P. A. Quijada, H. Polewicz, R. Vogelzang and T. C. Osborn, 2004. Phenotypic effects of introgressing chinese winter and resynthesized Brassica napus L. germplasm into hybrid spring canola. Crop Sci. 44 1990–1996. [Google Scholar]
- Udall, J. A., J. M. Swanson, D. Nettleton, R. J. Percifield and J. F. Wendel, 2006. A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics 173 1823–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme, E. J. M., R. Culerrier, A. Barre, R. Alvarez, P. Rouge et al., 2007. A novel family of lectins evolutionarily related to class V chitinases: an example of neofunctionalization in legumes. Plant Physiol. 144 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, C., and T. Wilkins, 1994. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal. Biochem. 223 7–12. [DOI] [PubMed] [Google Scholar]
- Wang, J., L. Tian, A. Madlung, H.-S. Lee, M. Chen et al., 2004. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., L. Tian, H.-S. Lee, N. E. Wei, H. Jiang et al., 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.-M., Z.-Y. Dong, Z.-J. Zhang, X.-Y. Lin, Y. Shen et al., 2005. Extensive de novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.). Genetics 170 1945–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42 225–249. [PubMed] [Google Scholar]
- Wendel, J. F., and V. A. Albert, 1992. Phylogenetics of the cotton genus Gossypium: character-state weighted parsimony analysis of chloroplast-DNA restriction site data and its systematic and biogeographic implications. Syst. Bot. 17 115–143. [Google Scholar]
- Wendel, J. F., and R. C. Cronn, 2003. Polyploidy and the evolutionary history of cotton. Adv. Agron. 78 139–186. [Google Scholar]
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2004. Evolutionary changes in cis and trans gene regulation. Nature 430 85–88. [DOI] [PubMed] [Google Scholar]
- Yang, S., F. Cheung, J. J. Lee, M. Ha, N. E. Wei et al., 2006. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 47 761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, Y., and K. L. Adams, 2007. Extensive allelic variation in gene expression in Populus F1 hybrids. Genetics 177 1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]