Abstract
Given that plants are sessile organisms, traits involved in adapting to local environments and/or in monitoring the surrounding environment, such as having photoreceptors, are significant targets of natural selection in plant evolution. To assess the intraspecific adaptive evolution of photoreceptors, we investigated sequence variations in four phytochrome genes (PHYA–C and PHYE) of Cardamine nipponica (Brassicaceae), an endemic Japanese alpine plant. The genealogies of haplotypes and genetic differentiations showed inconsistent patterns of evolution across phytochromes, suggesting that evolutionary forces were distinct in phytochromes of C. nipponica. An overall low level of nucleotide diversity in phytochrome genes suggests that the evolution of phytochromes is constrained by purifying selection within C. nipponica, which is consistent with previous findings on phytochromes. However, PHYE alone exhibited a non-neutral pattern of polymorphisms (Tajima's D = 1.91, P < 0.05) and an accumulation of nonsynonymous substitutions between central and northern Japan. In particular, the PHY domain, which plays an important role in stabilizing the active form (Pfr) of phytochromes, harbored a specific nonsynonymous fixation between regions. Thus, our finding indicates that local adaptation is involved in the evolution of PHYE in C. nipponica and is the first to suggest the involvement of PHYE in local adaptation.
GIVEN that plants are sessile organisms, adapting to the surrounding environment and monitoring environmental changes such as temperature, aridity, and day length are important not only for survival, but also for reproductive success. Thus, natural selection has likely played a significant role in selecting for such traits in the evolution of plants. In particular, various developmental responses are influenced by the light environment (known as photomorphogenesis); for example, germination, de-etiolation, shade avoidance, and flowering are regulated by light signals (Whitelam and Devlin 1997; Whitelam et al. 1998; Smith 2000; Mathews 2006). Thus, plants have obtained sophisticated systems, including photoreceptors, to monitor light signals such as intensity, direction, quality, and duration. In particular, phytochromes, which sense red and far-red light, are among the most studied photoreceptors and play a major role in developmental pathways as well as in evolutionary history.
Phytochromes have two photoreversible conformations: an inactive red-light-absorbing form (Pr) and an active far-red-light-absorbing form (Pfr). Red light converts Pr to Pfr, while far-red light converts Pfr back to Pr. At least three phytochromes are widely known in angiosperms (PHYA–C; Mathews et al. 1995), and five have been identified in Arabidopsis thaliana (PHYA–E; Sharrock and Quail 1989; Clack et al. 1994). According to phylogenetic analyses, these gene families are clustered into two major groups, PHYA/C and PHYB/D/E (Alba et al. 2000), and the duplication of these two major clusters occurred prior to angiosperm radiation (Mathews et al. 1995). Further duplication resulted in PHYA and PHYC and PHYB/D and PHYE, although some groups, such as monocots and poplars, lack PHYE (Mathews and Sharrock 1996), and PHYD was duplicated from PHYB specifically in the Brassicaceae (Mathews and McBreen 2008). The evolutionary consequences following gene duplications have recently been reported. Positive selection was involved in the functional divergence following duplications in PHYA and PHYC and PHYB/D and PHYE, whereas purifying selection suppressed the divergence in each group (Yang and Nielsen 2002). Moreover, adaptive evolution was involved in the evolution of PHYA in early angiosperms (Mathews et al. 2003).
Because light signals differ among habitats, such as forest understory, canopy, and open meadow, as well as among populations at high and low latitudes and altitudes, the functions of phytochromes should be both a target for natural selection and adapted to the local light environment. This hypothesis was recently supported by studies of A. thaliana, which showed that amino acid substitutions in PHYA, PHYB, and PHYC may be responsible for intraspecific phenotypic differences among accessions (Maloof et al. 2001; Balasubramanian et al. 2006; Filiault et al. 2008). In addition to the conclusions of the Arabidopsis studies, the importance of PHYB2 for local adaptation was also suggested in studies of Populus tremula (Ingvarsson et al. 2006, 2008). Thus, determining polymorphisms in phytochrome genes and their geographic distribution could demonstrate the importance of phytochromes for local adaptation.
A previous phylogeographic study on Cardamine nipponica Franch. et Savat. (Brassicaceae), an endemic Japanese alpine plant, found a higher level of fixation of nonsynonymous substitutions among populations in a partial sequence of PHYE (Ikeda et al. 2008b), indicating the involvement of PHYE in local adaptation. In the Japanese archipelago, alpine flora are distributed on high mountaintops from central to northern Japan. Nearly half of the Japanese alpine flora species are also found in Arctic regions [such as Dryas octopetala (Rosaceae), Loiseleuria procumbens (Ericaceae), and Diapensia lapponica (Diapensiaceae)], indicating that Arctic plants significantly contributed to the Japanese alpine flora. According to recent phylogeographic investigations, most alpine plants show strong genetic differentiation between populations from central and northern Japan, suggesting a history of vicariance in the Japanese archipelago (Fujii et al. 1997, 1999; Senni et al. 2005; Fujii and Senni 2006; Ikeda et al. 2006, 2008a,b; Ikeda and Setoguchi 2007, 2009). Most importantly, the genetic structure of 10 nuclear genes in C. nipponica revealed that haplotypes of most loci were closely related in each region and diverged from those in the other region. Moreover, the isolation with migration model (Nielsen and Wakeley 2001; Hey and Nielsen 2004, 2007) demonstrated no migration between the two regions after the regions were split, suggesting that the divergence in functional genes occurred following the vicariance between regions (H. Ikeda, N. Fujii and H. Setoguchi, unpublished results). In a partial sequence of PHYE (∼700 of the 3700 bp), three nonsynonymous substitutions were fixed between central and northern Japan, whereas nine other loci showed few nonsynonymous substitutions between the two regions. Thus, local adaptation in PHYE may have occurred following the vicariance between central and northern Japan, while genetic drift following the vicariance happened to fix the nonsynonymous substitutions. Further investigations that evaluate evolutionary patterns along the functional domains of phytochromes, including determining the entire sequences of PHYE, may demonstrate the involvement of PHYE in local adaptation.
In this study, to test the hypothesis that PHYE has been involved in local adaptation between central and northern Japan, we determined the entire sequences of PHYE from the entire distribution range of C. nipponica and examined polymorphisms and their geographic structures. Furthermore, to assess whether the signature of local adaptation was exclusively detected in PHYE or whether other phytochromes were also involved, we determined the entire sequences of PHYA–C and examined their polymorphisms.
MATERIALS AND METHODS
Primer design for amplification of the entire coding region of PHYA–C and PHYE:
To obtain the total sequence of phytochrome genes from C. nipponica, genome walking was conducted following the method of Lian et al. (2006) using previously reported primers (Kuittinen et al. 2002; Ikeda et al. 2008b) instead of the compound SSR primers used in the original method (Lian et al. 2006). Homology of the sequenced fragments obtained from each genome walking was examined by a BLAST search, and all fragments were aligned with the corresponding PHY sequences of A. thaliana. Following genome walking and obtained sequences, we designed PCR primers that amplify all coding regions of each phytochrome and additional primers for sequencing the entire coding regions (supporting information, Table S1).
DNA extraction, PCR, and sequencing:
Previously extracted DNA samples of C. nipponica (Ikeda et al. 2008b) were used, from which 11 samples (one sample per population; Table 1) for PHYA–C and 19 samples (one or two samples per population) for PHYE were chosen as PCR templates and used in the following analysis. These samples covered the entire geographic range of this species and represented polymorphisms of the species because previous phylogeographic studies detected little genetic diversity within populations (Ikeda et al. 2008b). The summary of analyzed samples is shown in Table 1. Cardamine resedifolia was used as the outgroup; DNA from one individual was extracted using DNeasy (Qiagen, Hilden, Germany) and dissolved in 100 μl TE buffer.
TABLE 1.
Summary of populations and the number of samples analyzed in each gene
| Region | Population no. | Locality | Coordinates | Altitude (m) | PHYA | PHYB | PHYC | PHYE |
|---|---|---|---|---|---|---|---|---|
| North | 1 | Taisetsusan | 43°38′ N/142°55′ E | 2290 | 1 | 1 | 1 | 2 |
| 2 | Ashibetsudake | 43°14′ N/142°17′ E | 1726 | 1 | 1 | 1 | 2 | |
| 3 | Shokanbetsudake | 43°43′ N/141°31′ E | 1491 | 1 | 1 | 1 | 2 | |
| 4 | Gassan | 38°33′ N/140°01′ E | 1984 | 1 | 1 | 1 | 2 | |
| 5 | Iidesan | 37°51′ N/139°41′ E | 2105 | 1 | 1 | 1 | 2 | |
| South | 6 | Shiroumadake | 36°45′ N/137°45′ E | 2932 | 1 | 1 | 1 | 1 |
| 7 | Norikuradake | 36°06′ N/137°33′ E | 3026 | 1 | 1 | 1 | 1 | |
| 8 | Kisokomagatake | 35°47′ N/137°48′ E | 2956 | 1 | 1 | 1 | 2 | |
| 9 | Senjyoudake | 35°43′ N/138°11′ E | 3033 | 1 | 1 | 1 | 2 | |
| 10 | Kitadake | 35°40′ N/138°14′ E | 3193 | 1 | 1 | 1 | 2 | |
| 11 | Hakusan | 36°09′ N/136°46′ E | 2702 | 1 | 1 | 1 | 1 | |
| Outgroup | C. resedifolia | 1 | 1 | 1 | 1 |
PCR amplification was conducted in a total reaction volume of 10.0 μl, containing 5.9 μl of autoclaved ion-exchanged water, 0.4 mm dNTP mixture, 1× LA PCR Buffer II (Takara LA Taq; Takara, Kyoto, Japan), 0.5 unit LA Taq HS (Takara), 0.2 μm of each primer (Table S1), and 1.0 μl of DNA. Amplification was performed with an initial denaturation for 3 min at 94° followed by 45 cycles of denaturation for 45 sec at 94°, annealing for 45 sec at 56°, and extension for 6 min at 72°. PCR products were visualized on 1.0% TAE–agarose gels stained with ethidium bromide and were gel purified with glass powder using GeneCleanII (Bio 101, Vista, CA). Products were sequenced directly using the standard methods of the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied BioSystems, Foster City, CA) using all listed primers (Table S1). To avoid erroneous polymorphisms, all sequences with singletons were sequenced more than twice from independent PCR products.
Sequence alignment:
All sequences were aligned using Auto Assembler software (Applied BioSystems) together with the mRNA sequence of A. thaliana because insertions and deletions in introns made alignment difficult. Coding regions, including start and stop codons, were assigned on the basis of the sequence of A. thaliana. Because we found no dual peaks from any analyzed sequences, haplotypes of all sequences were determined directly. Amino acid sequences of all haplotypes, including C. resedifolia, were aligned together with those of A. thaliana (602188A, Q5G899, 2104420A), Solanum lycopersicum (Q41331, Q9ZS62, Q41335), Sorghum bicolor (Q6S542, Q6S525, P93528), and Triticum aestivum (Q5K5K6, A9JR06, Q8VWN1) from the database for PHYA–C and that of Ipomea nil (P55004), Vitis vinifera (A7QQJ6), A. thaliana (2017370B), and S. lycopersicum (Q9M6P6) and PHYB of A. thaliana from the database for PHYE using Clustal X (Thompson et al. 1997). To evaluate the functional importance of polymorphic sites, sites without variations across outgroup taxa were assigned as conserved sites.
Analysis of the phytochrome genes:
Phylogenetic relationships among haplotypes were constructed on the basis of DNA sequences using PAUP4.0b with a Branch and Bound search (Swofford 2003) following the maximum-likelihood model of evolution (PHYA, HKY + G; PHYB, F81; PHYC, F81; PHYE, HKY) determined by jModeltest (Guindon and Gascuel 2003; Posada 2008). The significance of each branch was tested with 1000 bootstrap resamplings following the maximum-likelihood method with a heuristic search using PAUP 4.0b10 (Swofford 2003). The minimum number of recombination events across analyzed sequences was estimated (RM) on the basis of the four-gamete test (Hudson and Kaplan 1985) using DnaSP (Rozas et al. 2003).
Nucleotide diversity for total (πTotal), nonsynonymous (πa), and synonymous sites (πs) within C. nipponica and each region and genetic divergence from C. resedifolia for total (KTotal), nonsynonymous (Ka), and synonymous sites (Ks) were estimated according to Jukes and Cantor (1969). To test whether the patterns of polymorphisms were compatible with neutral equilibrium, we applied Tajima's D test (Tajima 1989). The significance of D was evaluated with 10,000 coalescent simulations. The genetic differentiation between central and northern Japan was estimated by FST using the method of Hudson et al. (1992a). Significant genetic differentiation between these two regions was tested with KST* (Hudson et al. 1992b) and Snn (Hudson 2000) on the basis of 10,000 permutations, because these tests have the highest power to detect population differentiation (Hudson 2000). To evaluate the level of these summary statistics of PHYA–C and PHYE, an additional eight loci used in previous studies (H. Ikeda, N. Fujii and H. Setoguchi, unpublished results) were analyzed: COP1, a gene regulator downstream of phytochrome and cryptochromes; DET1, a gene regulator downstream of phytochromes and cryptochromes; GA1, which is involved in the biosynthesis of gibberellic acid; TFL1, which controls inflorescence meristem identity; CAL, which is essential for the proper identity of the floral meristem; F3H, which encodes an enzyme of the phenylpropanoid pathway; DFR, which encodes an enzyme of the phenylpropanoid pathway; and CHS, which is involved in secondary metabolism. Samples from the analyzed populations were also analyzed at these additional loci. In addition, the Hudson–Kreitman–Aguadé test (HKA; Hudson et al. 1987) was conducted to assess neutral evolution on the basis of the ratio of polymorphisms within C. nipponica to the divergence from the outgroup using the 12 loci. Moreover, the mean value of Tajima's D across the 12 loci was estimated, and the significance was evaluated using 10,000 coalescence simulations. These analyses were performed using the DnaSP (Rozas et al. 2003) and the HKA program (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm#HKA). Additionally, evidence of non-neutral evolution of the phytochrome genes was investigated using the McDonald–Kreitman (MK) test (McDonald and Kreitman 1991) using DnaSP (Rozas et al. 2003).
Data analysis for PHYE:
Because PHYE was assigned to an outlier locus due to highly divergent clusters of haplotypes (see results), we further evaluated the evolutionary history of this phytochrome gene. To determine regions with a signature for non-neutral evolution along the sequence, sliding-window analyses were performed for nucleotide diversity (π), genetic divergence (K), and Tajima's D using DnaSP (Rozas et al. 2003). A fixed window size of 350 bp with a step size of 5 bp was used because smaller window sizes yielded windows with no polymorphisms. The 97.5% confidence interval of Tajima's D was evaluated using 10,000 coalescent simulations. Regions of the functional domains in phytochromes were also shown in the window: the N-terminal extension (N), the Per/Arn/Sim-like (PAS) domain, the cGMP phosphodiesterase/adenyl cyclase/FhlA domain, the phytochrome (PHY) domain, the PAS-related domain, and the histidine-kinase-related domain (Montgomery and Lagarias 2002).
Whether the pattern of nonsynonymous and synonymous substitutions between interregional and interspecies branches followed neutral evolution was tested using the MK test. Furthermore, non-neutral evolution within the genealogy of haplotypes was assessed by the ratio of nonsynonymous to synonymous substitutions (dN/dS) using CODEML in PAML4.0 (Yang 2007). Because branches between central and northern Japan (interregional branches) were highly diverged (see results), positive selection on the interregional branches was tested using the branch-site model (Zhang et al. 2005). This model is powerful in detecting positive selection that acts on a few sites in sequences, the signal for which may be swamped by purifying selection (Zhang et al. 2005). The model estimates three ratios of nonsynonymous to synonymous substitutions (ω = dN/dS) and allows four site classes, in which different ω are allowed in the same site between the foreground (interregional) and background (intraregional and interspecific) branches. Thus, sites in the background have two ω (0 < ω0 < 1 and ω1 = 1), and an additional ω (ω2 > 1) is allowed for sites in the foreground. The fit of the data to this model (model A of Zhang et al. 2005) was tested against the fit of a model that varies in having ω2 = 1 (model A1 of Zhang et al. 2005). Model A was also tested against the nearly neutral model, which assumes that all sites have one of two ω (ω0, ω1), regardless of background or foreground, but this test does not distinguish between relaxed selective constraints and positive selection. Specific sites inferred to be under positive selection were identified using the Bayes empirical Bayes method (Yang et al. 2005).
The likelihood-ratio test was applied to test all models against null models. A phylogenetic tree was assumed [(north), (south), C. resedifolia] for the analysis because this relationship was significantly supported in the present phylogenetic analysis of haplotypes (bootstraps >95%) and because our data were intraspecific and detailed relationships would vary due to stochastic effects. The convergence of likelihood and ω was evaluated by running the analysis three times, with different initial values of ω (0, 1, 10).
RESULTS
Sequences of PHYA–C and PHYE genes in C. nipponica:
The locations of exons and introns were consistent with those in A. thaliana. Excluding the stop codon, the sequenced lengths of the coding regions of phytochrome genes of C. nipponica were 3716, 3974, 3578, and 3676 bp in PHYA, -B, -C, and -E, respectively. Identity of sequences in coding regions between C. nipponica and A. thaliana is ∼94, 88, 92, and 92% in PHYA, -B, -C, and -E, respectively. All sequences were deposited in the DNA Data Bank of Japan (AB438983–AB438994, AB456956–AB456982). As expected from the primarily selfing character in the previous study (Ikeda et al. 2008b), there was no recombination among analyzed sequences. Therefore, the following analyses of the DNA sequence, especially the neutral test, could not be influenced by recombination.
Genealogies among haplotypes:
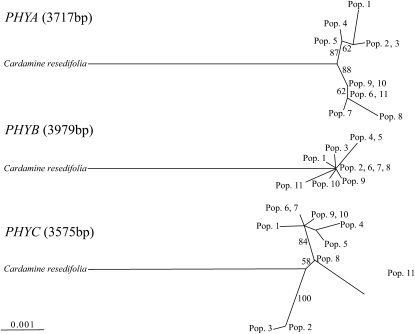
Genealogies among haplotypes based on DNA sequences are shown in Figure 1 and Figure 2. In PHYA and PHYE, two highly supported clusters of haplotypes (PHYA, >85%; PHYE, 100%), which correspond to the geographic regions of central and northern Japan, were detected (Figures 1 and 2). There were no such geographic structures in the genealogies of PHYB and PHYC. Haplotypes of PHYB gave a star-shaped tree, while haplotypes of PHYC formed a cluster (100%), which consisted of haplotypes detected from two neighboring populations (populations 2 and 3) whereas another highly supported cluster (84%) contained haplotypes from both central and northern Japan (Figure 1).
Figure 1.—
Maximum-likelihood tree of the entire PHYA–C coding region of C. nipponica, including the outgroup (C. resedifolia). Numbers next to branches indicate significance evaluated by 1000 bootstrap resamplings. The names of haplotypes are represented by populations in which these haplotypes were found.
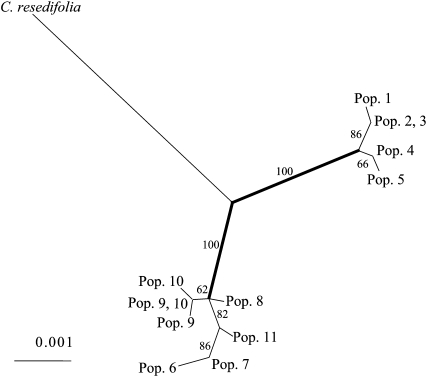
Figure 2.—
Maximum-likelihood tree of the entire PHYE coding region (3676 bp) of C. nipponica, including the outgroup (C. resedifolia). Numbers next to branches indicate the significance evaluated by 1000 bootstrap resamplings. The names of haplotypes are represented by populations in which these haplotypes were found. The thick line indicates the interregional branch.
Nucleotide diversity, genetic divergence, and population differentiation:
Nucleotide diversities varied among phytochrome genes, which were comparatively low across other loci of C. nipponica (Table 2). The level of diversity in phytochromes and other loci was lower than that detected in the functional genes of Arabidopsis species (A. thaliana: πTotal = 0.0019–0.0055; A. halleri: πTotal = 0.0018–0.0327; A. lyrata: πTotal = 0.0006–0.0324; Ramos-Onsins et al. 2004), although it was compatible with PHYB and PHYD in A. thaliana (πTotal = 0.0015 and 0.0024 for PHYB and PHYD, respectively; Mathews and McBreen 2008). Moreover, this low level of nucleotide diversity in phytochrome genes in C. nipponica was also consistent with previous findings in phytochrome genes in other species (Sorghum species: πTotal = 0.00099–0.00129, White et al. 2004; Pinus sylvestris: πTotal = 0.0004–0.0011, García-Gil et al. 2003).
TABLE 2.
Summary statistics within entire populations of C. nipponica and the northern and southern regions
| Locus | bp | Region | n | πTotal | πa | πs | Tajima's D |
|---|---|---|---|---|---|---|---|
| PHYA | 3716 | Total | 22 | 0.00126 | 0.00076 | 0.00327 | 0.491 |
| North | 10 | 0.00067 | 0.00048 | 0.00115 | 0.718 | ||
| South | 12 | 0.00046 | 0.00012 | 0.00180 | 0.092 | ||
| PHYB | 3974 | Total | 22 | 0.00054 | 0.00025 | 0.00134 | −0.741 |
| North | 10 | 0.00058 | 0.00013 | 0.00193 | 1.233 | ||
| South | 12 | 0.00038 | 0.00033 | 0.00037 | −0.313 | ||
| PHYC | 3578 | Total | 22 | 0.00173 | 0.00089 | 0.00517 | −0.086 |
| North | 10 | 0.00209 | 0.00097 | 0.00665 | 1.482* | ||
| South | 12 | 0.00110 | 0.00073 | 0.00274 | 0.780 | ||
| PHYE | 3673 | Total | 38 | 0.00312 | 0.00289 | 0.00314 | 1.909* |
| North | 20 | 0.00060 | 0.00046 | 0.00066 | 1.233 | ||
| South | 18 | 0.00103 | 0.00073 | 0.00219 | −0.386 | ||
| COP1 | 529 | Total | 22 | 0.00269 | 0.00000 | 0.00000 | 0.826 |
| North | 10 | 0.00303 | 0.00000 | 0.00000 | 0.505 | ||
| South | 12 | 0.00115 | 0.00000 | 0.00000 | −0.248 | ||
| DET | 577 | Total | 22 | 0.00228 | 0.00265 | 0.00000 | −0.120 |
| North | 10 | 0.00154 | 0.00358 | 0.00000 | 0.830 | ||
| South | 12 | 0.00189 | 0.00122 | 0.00000 | 0.322 | ||
| GA1 | 1225 | Total | 22 | 0.00317 | 0.00233 | 0.00317 | 0.625 |
| North | 10 | 0.00189 | 0.00157 | 0.00233 | 1.233 | ||
| South | 12 | 0.00154 | 0.00000 | 0.00317 | 0.497 | ||
| TFL1 | 1009 | Total | 22 | 0.00220 | 0.00168 | 0.00000 | 0.330 |
| North | 10 | 0.00229 | 0.00091 | 0.00000 | 1.233 | ||
| South | 12 | 0.00108 | 0.00113 | 0.00000 | 0.322 | ||
| CAL | 526 | Total | 22 | 0.00264 | 0.00000 | 0.00521 | −1.211 |
| North | 10 | 0.00339 | 0.00000 | 0.01074 | 0.023 | ||
| South | 12 | 0.00173 | 0.00000 | 0.00000 | −0.278 | ||
| F3H | 534 | Total | 22 | 0.00032 | 0.00050 | 0.00000 | −0.641 |
| North | 10 | 0.00067 | 0.00103 | 0.00000 | 0.015 | ||
| South | 12 | 0.00000 | 0.00000 | 0.00000 | NA | ||
| DFR | 522 | Total | 22 | 0.00126 | 0.00000 | 0.00000 | −0.518 |
| North | 10 | 0.00000 | 0.00000 | 0.00000 | NA | ||
| South | 12 | 0.00209 | 0.00295 | 0.00000 | 0.322 | ||
| CHS | 585 | Total | 22 | 0.00113 | 0.00060 | 0.00000 | −0.518 |
| North | 10 | 0.00213 | 0.00123 | 0.00000 | 0.624 | ||
| South | 12 | 0.00000 | 0.00000 | 0.00000 | NA |
The length of each locus (bp), the number of sequences (n), nucleotide diversity (π), and Tajima's D are shown. The nucleotide diversity was estimated for the entire (πTotal), nonsynonymous (πa), and synonymous sites (πs) separately. Asterisks (*) indicate significant values of Tajima's D (P < 0.05).
In particular, PHYB harbored few variations not only in C. nipponica (πTotal = 0.00054) but also in each region (πTotal = 0.00058 and 0.00038 for northern and central Japan, respectively). The level of diversity in PHYA was similar to that of PHYB in each region (πTotal = 0.00067 and 0.00046 for northern and central Japan, respectively), while it was higher in the entire C. nipponica population (πTotal = 0.00126). The diversity within regions was high in PHYC compared to the other phytochromes (Table 2), especially in northern Japan (πTotal = 0.00209). PHYE harbored the highest genetic diversity among phytochrome genes within C. nipponica (πTotal = 0.00312), while the diversity within each region was compatible with other phytochrome genes, especially in northern Japan (πTotal = 0.0060). In particular, the high level of genetic diversity in PHYE was due to the higher ratio of nonsynonymous to synonymous substitutions (πa/πs = 0.23, 0.19, 0.17, and 0.92 for PHYA, PHYB, PHYC, and PHYE, respectively).
The level of genetic divergence of PHYA–C and PHYE from C. resedifolia was similar across genes (KTotal = 0.0061–0.0075), particularly in synonymous sites for PHYA, -C, and -E (Ks = 0.012–0.013), and intermediate compared to other loci (Table 3). This indicates that the evolution of phytochrome genes occurred at similar rates in C. nipponica. Additionally, the ratio of nonsynonymous to synonymous divergences was compatible across phytochrome genes (Ka/Ks = 0.32, 0.57, 0.35, and 0.38 for PHYA, PHYB, PHYC, and PHYE, respectively). Thus, evolutionary rates of nonsynonymous and synonymous substitutions were not distinct among phytochrome genes. This suggests that the high level of nonsynonymous substitutions in PHYE was caused by a specific force that accumulated nonsynonymous substitutions.
TABLE 3.
Genetic differentiation (FST) between northern and central Japan and genetic divergence from C. resedifolia (K)
| Locus | FST | KST* | Snn | KTotal | Ka | Ks |
|---|---|---|---|---|---|---|
| PHYA | 0.7066 | ** | ** | 0.00664 | 0.00389 | 0.01226 |
| PHYB | 0.2138 | ** | ** | 0.00611 | 0.00456 | 0.00804 |
| PHYC | 0.1735 | ** | ** | 0.00641 | 0.00466 | 0.01316 |
| PHYE | 0.8422 | ** | ** | 0.00753 | 0.00512 | 0.01293 |
| COP1 | 0.3868 | ** | ** | 0.00396 | 0.00000 | 0.01592 |
| DET1 | 0.3813 | ** | ** | 0.00490 | 0.00549 | 0.00000 |
| GA1 | 0.6237 | ** | ** | 0.01242 | 0.00596 | 0.00835 |
| TFL1 | 0.3925 | ** | ** | 0.01437 | 0.00372 | 0.00794 |
| CAL | 0.1044 | * | * | 0.01290 | 0.00000 | 0.00273 |
| F3H | 0.1111 | P = 0.196 | P = 0.196 | 0.00393 | 0.00026 | 0.01911 |
| DFR | 0.1818 | ** | ** | 0.00841 | 0.00102 | 0.27540 |
| CHS | 0.2222 | * | * | 0.00923 | 0.00031 | 0.01236 |
The divergence from C. resedifolia was estimated for the entire (KTotal), nonsynonymous (Ka), and synonymous sites (Ks) separately. The significance of genetic differentiation was evaluated by KST and Snn (*P < 0.05, **P < 0.01).
The level of population differentiation varied across all phytochrome genes (Table 2): high in PHYA (FST = 0.71) and PHYE (FST = 0.84) and low in PHYB (FST = 0.21) and PHYC (FST = 0.17) compared to the other eight loci (Table 3). The overall high level of differentiation is consistent with the vicariance history between populations in central and northern Japan.
Neutrality test:
Significant deviation from neutral equilibrium was detected in PHYE for the entire region (D = 1.91; Table 2) and in PHYC for the northern region (D = 1.44; Table 2). The mean values of Tajima's D across the 12 loci did not deviate from neutral equilibrium for the entire region (D = 0.004, P = 0.41) or for the southern region (D = 0.111, P = 0.29), while a significant positive value was detected in the northern region (D = 0.830, P < 0.05).Thus, although the patterns of polymorphisms across genes in entire regions and in the southern region followed the expectation of neutral equilibrium, those in the northern region deviated from neutral equilibrium across genes. This multilocus departure from neutrality in the northern region could have been caused by genomewide effects, such as demographic changes and/or population subdivisions (Schmid et al. 2005). This indicates that the significant departure from neutral equilibrium in PHYC in the northern region was not likely caused by locus-specific effects, such as natural selection, but rather by effects across the whole genome.
In contrast, regardless of the neutrality of polymorphisms across genes for the entire region, PHYE alone showed a significant positive deviation from neutral equilibrium. This indicates that locus-specific effects, such as natural selection, may have shaped the pattern of polymorphisms in PHYE for entire populations and that PHYE was an outlier locus. Because the positive deviation in Tajima's D was caused by an excess of polymorphisms with intermediate frequency (Tajima 1989), diverged alleles (haplotypes) would persist in entire populations. As shown in the genealogy (Figure 2), the existence of two highly diverged clusters that correspond to the geographic distribution of haplotypes was consistent with the significant positive value of Tajima's D.
The HKA test showed no evidence of deviation from neutral evolution (χ2 = 4.75, P = 0.94). Moreover, no evidence of non-neutral evolution among phytochrome genes was detected by the MK test (Table 4), while PHYC and PHYE were close to a significant deviation (P = 0.09 and 0.08 for PHYC and PHYE, respectively).
TABLE 4.
Summary of the McDonald–Kreitman test
| Locus | Type of substitutions | Synonymous | Nonsynonymous | G-test |
|---|---|---|---|---|
| PHYA | Polymorphisms | 8 | 6 | P = 0.695 |
| Fixed differences | 8 | 8 | ||
| PHYB | Polymorphisms | 4 | 4 | P = 0.423 |
| Fixed differences | 6 | 12 | ||
| PHYC | Polymorphisms | 15 | 8 | P = 0.086 |
| Fixed differences | 6 | 10 | ||
| PHYE | Polymorphisms | 7 | 20 | P = 0.077 |
| Fixed differences | 8 | 7 |
The number of polymorphisms and fixed differences indicate the number of substitutions within C. nipponica and those fixed between C. nipponica and C. resedifolia.
Amino acid replacements:
Six, 4, 8, and 20 amino acid replacements were detected in PHYA, -B, -C, and -E, respectively. In PHYA, 3 of 6 replacements were located at conserved sites (Table 5). One of these 3 replacements was fixed between regions (E38D), while the remaining 2 were found exclusively in single populations (P191S, L424M). Three of 4 replacements in PHYB were at conserved sites (S415T, L482F, E815D), and all were found in single populations (Table 5). In PHYC, 3 of 8 replacements occurred at conserved sites (R7S, L104S, L492F) and 2 were in population 11 (R7S, L492F), while another was fixed in neighboring populations (populations 2 and 3; Table 5). In PHYE, 6 of 20 replacements were located at conserved sites (I153N, V303I, A483V, F508Y, V550I, N757H; Table 4). Five of these 6 were fixed between regions (except for I153N). Additionally, 11 of 20 replacements were fixed between regions (V303I, A483V, F508Y, V550I, Y689D, F695C, N757H, L822V, Y951D, D1058E, T1100K; Table 6). Overall, 17 of 38 replacements were found in single populations, of which 8 replacements were located at conserved sites. Although the functional deficiencies due to these replacements are not known, considering the phylogeographic history of this species (Ikeda et al. 2008b) and the small populations in the current distribution, reduced population size during the postglacial warming and subsequent genetic drift may have caused the fixation of these singleton mutations on each mountain. In particular, as expected from its distinct genetic structure and isolated history (Ikeda et al. 2008b), population 11 (Hakusan) harbored 5 singleton amino acid replacements.
TABLE 5.
Comparisons of amino acid substitutions across taxa in PHYA–C
| Site
|
Site
|
Site
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PHYA | 38 | 191 | 424 | 473 | 864 | 976 | PHYB | 415 | 482 | 815 | 1086 | PHYC | 7 | 14 | 99 | 104 | 341 | 370 | 492 | 881 |
| Population 1 | E | P | M | S | E | T | Population 1 | T | L | E | A | Population 1 | R | R | Q | L | S | M | L | A |
| Populations 2 and 3 | E | P | L | S | E | T | Populations 2, 6, 7, 8 | S | L | E | A | Population 2 | R | R | E | S | R | V | L | A |
| Population 4 | E | S | L | S | V | T | Population 3 | S | L | E | A | Population 3 | R | R | E | S | R | V | L | T |
| Population 5 | E | P | L | S | V | T | Populations 4 and 5 | S | L | E | A | Population 4 | R | R | Q | L | S | M | L | A |
| Populations 6 and 11 | D | P | L | S | V | A | Population 9 | S | L | D | A | Population 5 | R | R | Q | L | S | M | L | A |
| Population 7 | D | P | L | S | V | A | Population 10 | S | F | E | A | Populations 6 and 7 | R | R | Q | L | S | M | L | A |
| Population 8 | D | P | L | T | V | A | Population 11 | S | L | E | G | Population 8 | R | R | Q | L | R | V | L | A |
| Populations 9 and 10 | D | P | L | S | V | A | C. resedifolia | S | L | E | A | Populations 9 and 10 | R | R | Q | L | S | M | L | A |
| C. resedifolia | E | P | L | S | V | A | A. thaliana | S | L | E | A | Population 11 | S | S | Q | L | R | V | F | A |
| A. thaliana | E | P | L | S | V | A | S. lycopersicum | S | L | E | S | C. resedifolia | R | R | Q | L | R | V | L | A |
| S. lycopersicum | E | P | L | S | A | A | S. bicolor | S | L | E | S | A. thaliana | R | R | Q | L | S | V | L | A |
| S. bicolor | E | P | L | F | V | S | T. aestivum | S | L | E | S | S. lycopersicum | R | R | P | L | Q | L | L | K |
| T. aestivum | E | P | L | L | L | A | S. bicolor | R | R | P | L | D | V | L | T | |||||
| T. aestivum | R | K | A | L | D | V | L | T | ||||||||||||
Sites that are conserved across taxa are in italic type. The names of haplotypes are represented by populations in which the haplotypes were found.
TABLE 6.
Comparison of amino acid substitutions across taxa in PHYE
| Site
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PHYE | 122 | 153 | 300 | 303 | 380 | 483 | 508 | 550 | 689 | 695 | 731 | 757 | 822 | 833 | 880 | 930 | 951 | 984 | 1058 | 1100 |
| Population 1 | N | I | P | I | M | V | F | V | Y | F | I | N | L | E | T | A | Y | L | D | T |
| Populations 2 and 3 | N | I | P | I | M | V | F | V | Y | F | I | N | L | E | T | T | Y | L | D | T |
| Population 4 | N | I | P | I | M | V | F | V | Y | F | I | N | L | V | T | T | Y | L | D | T |
| Population 5 | N | I | P | I | I | V | F | V | Y | F | I | N | L | V | T | T | Y | L | D | T |
| Population 6 | N | N | P | V | M | A | Y | I | D | C | I | H | V | E | S | T | D | L | E | K |
| Population 7 | N | I | P | V | M | A | Y | I | D | C | I | H | V | E | S | T | D | L | E | K |
| Population 8 | D | I | P | V | M | A | Y | I | D | C | I | H | V | E | T | T | D | L | E | K |
| Population 9 | N | I | S | V | M | A | Y | I | D | C | I | H | V | E | T | T | D | F | E | K |
| Populations 9 and 10 | N | I | S | V | M | A | Y | I | D | C | I | H | V | E | T | T | D | L | E | K |
| Population 10 | N | I | S | V | M | A | Y | I | D | C | I | H | V | E | T | T | D | L | E | K |
| Population 11 | N | I | P | V | M | A | Y | I | D | C | M | H | V | E | T | T | D | L | E | K |
| C. resedifolia | N | I | P | V | M | A | Y | V | D | C | I | H | V | E | T | T | N | L | D | T |
| A. thaliana | D | I | P | V | M | A | Y | V | D | C | I | N | V | E | T | T | N | L | D | T |
| I. nil | P | I | P | V | M | A | F | V | — | Y | V | N | F | D | A | M | N | I | T | S |
| V. vinifera | T | I | A | V | M | A | F | V | — | Y | I | N | F | E | A | I | N | L | D | Q |
| S. lycopersicum | K | I | P | V | T | A | F | V | — | Y | I | N | F | E | V | M | N | L | N | K |
| PHYB (A. thaliana) | K | I | P | V | M | A | F | V | — | F | I | N | F | Q | A | M | S | V | S | A |
Sites that were conserved across taxa are in italic type. The names of haplotypes are represented by populations in which the haplotypes were found.
Molecular evolution of PHYE:
The ratio of synonymous and nonsynonymous substitutions between interregional branches (synonymous/nonsynonymous substitutions: 2/11; see Figure 2 legend) was significantly distinct from the interspecies branch (synonymous/nonsynonymous substitutions: 8/7, G-test = 4.61, P < 0.05). This indicates that the specific high level of nonsynonymous substitutions in PHYE may be attributable to the elevated nonsynonymous substitutions between haplotypes in central and northern Japan.
The evolutionary forces for these elevated nonsynonymous substitutions were evaluated by PAML analysis. The branch-site model that allows positive selection on sites of interregional branches fit significantly better than the nearly neutral model that assumed no positive selection at any sites (2Δℓ = 2 × 4.37 = 8.74, d.f. = 2, P < 0.05; Table 7). However, the model was not significantly better than the null model that assumed that ω2 = 1 (2Δℓ = 2 × 4.37 = 2.54, d.f. = 1, P = 0.11; Table 7). This indicated that the relaxed constraint on selection, rather than on positive selection, was the more likely explanation of the higher rate of nonsynonymous substitutions to synonymous substitutions on interregional branches. No site was significantly influenced by positive selection in interregional branches (>95%).
TABLE 7.
Results of the branch-site model analysis for PHYE
| Model | Estimated parameters | λ | P-value | ||
|---|---|---|---|---|---|
| Nearly neutral | ω0 = 0.000 | p0 = 0.70 | −4822.38 | ||
| ω1 = 1.000 | p1 = 0.30 | ||||
| Branch-site | ω0−b = 0.000 | ω0−f = 0.000 | p0 = 0.76 | −4818.01 | P1 < 0.05 |
| ω1−b = 1.000 | ω1−f = 1.000 | p1 = 0.19 | P2 = 0.11 | ||
| ω0−b = 0.000 | ω2 = 32.17 | p2A = 0.04 | |||
| ω1−b = 1.000 | ω2 = 32.17 | p2B = 0.01 | |||
| Null with ω2 = 1 | ω0−b = 0.000 | ω0−f = 0.000 | p0 = 0.70 | −4819.28 | |
| ω1−b = 1.000 | ω1−f = 1.000 | p1 = 0.30 | |||
| ω0−b = 0.000 | ω2 = 1.000 | p2A = 0.00 | |||
| ω1−b = 1.000 | ω2 = 1.000 | p2B = 0.00 | |||
ω0−b/ω1−b and ω0−f/ω1−f indicate two ratios of nonsynonymous to synonymous substitutions (ω0, ω1) in the background (the interspecific and intraregional branches) and in the foreground (the interregional branches), respectively. ω2 indicates the ratio of nonsynonymous to synonymous substitutions on positively selected sites in the foreground. p0, p1, p2A, and p2B indicate the frequency of sites that have a pair of ω's in the background and foreground. The P-values above and below indicate the significance of the branch-site model tested against the nearly neutral model (test 1) and the null model with ω2 = 1 (test 2), respectively.
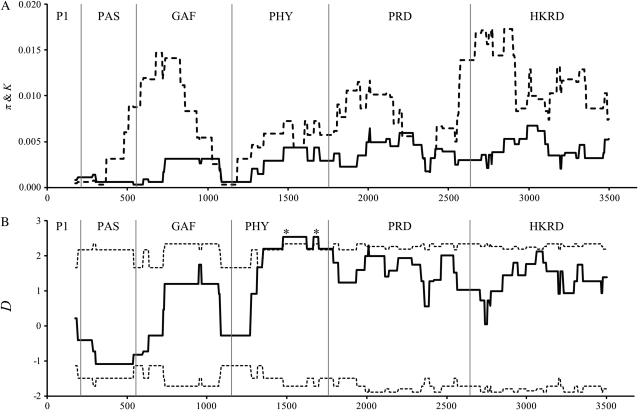
The sliding-window analysis showed the distribution of polymorphisms along functional domains (Figure 3). The patterns of polymorphisms deviated significantly from neutral equilibrium exclusively in the PHY domain (Figure 3B). As expected from the low divergence in the PHY domain (Figure 3A), one synonymous substitution was fixed between species, and three nonsynonymous substitutions were detected in C. nipponica. This pattern of synonymous and nonsynonymous polymorphisms and divergences in the PHY domain deviated significantly from the neutral expectation (χ2 = 4.00, P < 0.05), while the pattern of polymorphisms and divergence between the PHY and the remaining domains did not deviate from neutral evolution (HKA test; χ2 = 0.197, P = 0.66). Furthermore, all nonsynonymous substitutions were fixed between central and northern Japan (A483V, F508Y, V550I; Table 6).
Figure 3.—
Sliding-window plots of (A) nucleotide diversity within C. nipponica (π) and genetic divergence between C. nipponica and C. resedifolia (K) and (B) Tajima's D in PHYE. A 350-bp-wide window was moved along the sequence in 5-bp increments, and statistics were calculated for each window segment. The locations of functional domains are indicated within the plot. (A) Solid and dashed lines represent nucleotide diversity (π) and genetic divergence (K), respectively. (B) Solid and dashed lines represent the estimated values (D) and 97.5% confidence interval, respectively, obtained through 10,000 coalescent simulations. Asterisks (*) indicate the significant deviations of D.
DISCUSSION
Although the pattern of evolution varied among phytochromes of C. nipponica, the evolution of phytochromes was largely constrained by purifying selection. However, PHYE alone exhibited the signature of non-neutral evolution. Our finding of non-neutral evolution in PHYE indicates that local adaptation may be involved in the evolution of PHYE in C. nipponica. Considering the prominent role of PHYE at cool temperatures in A. thaliana (Halliday and Whitelam 2003; Heschel et al. 2007), and because C. nipponica grows in alpine habitats, PHYE may play an important role in C. nipponica. While the adaptive evolution of phytochromes has been previously suggested (White et al. 2004; Balasubramanian et al. 2006; Ingvarsson et al. 2006, 2008; Filiault et al. 2008), our study is the first to suggest the involvement of PHYE in local adaptation.
Inconsistent patterns of evolution across phytochromes of C. nipponica:
The present investigation of the molecular evolution of phytochrome genes detected inconsistent patterns of evolution across genes in C. nipponica, as shown in the level of nucleotide diversity (Table 2) and in the geographic structure of the haplotypes (Figures 1 and 2, Table 2). In contrast, the genetic divergence from the closely related species C. resedifolia was similar across genes, indicating that the inconsistent evolutionary pattern across genes may be the result of differing evolutionary forces among phytochromes. This indicates that functional constraints varied across phytochromes and that roles for local adaptation across these photoreceptors may also vary.
An overall low level of nucleotide diversity in phytochrome genes suggests that the evolution of phytochromes is constrained by purifying selection within C. nipponica, which is consistent with previous findings on the molecular evolution of phytochromes (Mathews and Sharrock 1996; Yang and Nielsen 2002; García-Gil et al. 2003; White et al. 2004; Filaullt et al. 2008; Mathews and McBreen 2008). In particular, functional constraints and purifying selection were strong in PHYB, which plays the largest role in red light perception and is the most important phytochrome in open habitats (Whitelam and Devlin 1997; Mathews 2005). Additionally, the rather high level of nucleotide diversity in PHYC is consistent with previous findings of a faster rate of evolution in PHYC in some tomato and sorghum (Alba et al. 2000; White et al. 2004).
Despite the strong functional constraints on and the subsequent purifying selection in phytochromes, previous reports have suggested the involvement of phytochromes in local adaptation (Balasubramanian et al. 2006; Ingvarsson et al. 2006, 2008; Filiault et al. 2008). In A. thaliana, amino acid replacements in PHYB were involved in differences in light sensitivity across accessions, suggesting the importance of PHYB in adaptation (Filiault et al. 2008). Moreover, haplotype groups of PHYC that were distinguished by eight amino acids were significantly involved in a latitudinal cline in flowering time (Balasubramanian et al. 2006). In P. tremula, PHYB2 was mapped to a linkage group containing QTL for bud set and bud flush, important adaptive traits in forest trees (Frewen et al. 2000; Chen et al. 2002). The distribution of four SNPs on PHYB2 was significantly correlated with latitude (Ingvarsson et al. 2006), from which two nonsynonymous SNPs were associated with bud-set variation (Ingvarsson et al. 2008).
Plausible local adaptation in PHYE:
In our investigation, PHYE alone showed a signature of non-neutral evolution that was caused by a high level of nonsynonymous substitutions (Table 2), especially haplotypes between central and northern Japan. Although maximum-likelihood tests showed that the accumulated nonsynonymous substitutions were attributable to the relaxation of selective constrains (Table 7), interpretation of this model base analysis comes with a caveat. Because the model assumes that sequences were sampled from divergent species, applying it to intraspecific data, as well as data from a single population, may not correctly evaluate the natural selection correctly (Kryazhimskiy and Plotkin 2008). In our case, nonsynonymous substitutions accumulated significantly on the interregional branch compared to the interspecies branch (Figure 2). Moreover, the distribution of amino acid replacements along the functional domain indicates the adaptive importance of nonsynonymous substitutions. In particular, three of the five replacements at conserved sites were located in the PHY domain (A483V, F508Y, V550I; Table 6), where a significant departure from neutrality was detected (Figure 3B). The low level of nucleotide diversity and divergence in this domain indicates a slow rate of mutations (Figure 3A), consistent with the lack of other substitutions in this domain. Thus, regardless of the slow mutation rate in the PHY domain, amino acid replacements were fixed between central and northern Japan (Table 6).
Physiological studies have shown that the PHY domain is important for stabilizing the active form of phytochrome (Pfr) and that amino acid replacements in this domain caused the phenotypic differences in A. thaliana (Oka et al. 2004, 2008). Thus, fixed differences in the PHY domain may cause functional differences in PHYE as well as subsequent phenotypic differences between central and northern Japan. Previous studies on natural variation in phytochrome genes have suggested that amino acid replacements in phytochromes were associated with phenotypic differences and local adaptation (Maloof et al. 2001; Balasubramanian et al. 2006; Ingvarsson et al. 2006, 2008; Filiault et al. 2008). Similarly, the accumulation of amino acid replacements in the conserved sites in the functionally important domain (PHY) may indicate the involvement of PHYE in local adaptation between central and northern Japan.
Although few phenotypic differences were observed in monogenic phyE mutants in normal growth conditions in A. thaliana (Devlin et al. 1998), PHYE plays a prominent role in germination and flowering at cooler temperatures (Halliday and Whitelam 2003; Heschel et al. 2007). Because C. nipponica is an alpine plant growing at high altitudes (2000–3000 m), it experiences cool temperatures throughout its life cycle. Thus, physiological mechanisms active at cool temperatures are important for C. nipponica's survival, suggesting the importance of PHYE function. As a result, the evolution of PHYE should be strongly constrained. Nevertheless, amino acid substitutions at conserved sites in an important domain (PHY) persisted between regions, suggesting that they were associated with functional divergence, as well as with adaptation to the local environment. Because the stability of Pfr is involved in the perception of light signals (Oka et al. 2004, 2008), divergence in photosensory function may be responsible for the local adaptation between central and northern Japan.
Although alpine flora do not extend across a wide geographic range in Japan (35–45° N), day length is different between the southernmost and northernmost populations (∼0.5–1 hr in the summer; National Astronomical Observatory 2002). The altitudes of alpine populations and of the mountains are also different across ranges; i.e., mountains in central Japan are nearly 3000 m high, whereas those in northern Japan are ≤2000 m. Therefore, the lower altitude of populations at higher latitudes results in lower light intensity in northern Japan compared to the populations in central Japan, which are located at higher altitudes and lower latitudes. Consequently, the light environment would be variable between populations in central and northern Japan. Further investigation of the functions of PHYE in C. nipponica and its effects on phenotype may demonstrate the adaptive importance of this gene in the local environment.
Most alpine plants in the Japanese archipelago show strong differentiation between populations from central and northern Japan (Fujii et al. 1997, 1999; Senni et al. 2005; Fujii and Senni 2006; Ikeda et al. 2006, 2008a,b,c; Ikeda and Setoguchi 2007). However, on the basis of geographic separation alone, explaining genetic differentiation is difficult, as some alpine plants exhibit homogenous genetic structures throughout the Japanese archipelago (Fujii et al. 1996; Senni et al. 2005; Ikeda and Setoguchi 2006). Thus, genetic differentiation between central and northern Japan may persist in some species because of functional differences involved in local adaptation that prohibited potential gene exchange between regions. In fact, the latitudinal cline of flowering in A. thaliana, mostly attributable to two genes (FLC and FRI), is also influenced by the amino acid substitution in PHYC (Balasubramanian et al. 2006). If the differences in photosensory functions of PHYE in C. nipponica cause changes in flowering time in the same environment, gene exchanges following range expansion could be prevented. If that were so, the signature of local adaptation in PHYE could be a candidate for explaining the persistence of genetic differentiations in C. nipponica, which may further explain genetic differentiation in other alpine plants.
Acknowledgments
We thank H. Tachida (Kyushu University) for analyzing sequence data, A. Nagatai (Kyoto University) for advice on phytochrome functional structure, Y. Mitsui for advice on conducting the experiments, and J. Lihova and K. Marhold for leaf materials of C. resedifolia. This study was supported by a Grant-in-Aid for Scientific Research (H.S.) and a Grant-in-Aid for Japan Society for the Promotion of Science Fellows (H.I.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.102152/DC1.
References
- Alba, R., P. M. Kelmenson, M. M. Cordonnier-Pratt and L. H. Pratt, 2000. The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol. Biol. Evol. 17 362–373. [DOI] [PubMed] [Google Scholar]
- Balasubramanian, S., S. Sureshkumar, M. Agrawal, T. P. Michael, C. Wessinger et al., 2006. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat. Genet. 38 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. H. H., G. T. Howe and H. D. Bradshaw, 2002. Molecular genetic analysis of dormancy-related traits in poplars. Weed Sci. 50 232–240. [Google Scholar]
- Clack, T., S. Mathews and R. A. Sharrock, 1994. The phytochrome apoprotein family in Arabidopsis is encoded by 5 genes: the sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25 413–427. [DOI] [PubMed] [Google Scholar]
- Devlin, P. F., S. R. Patel and G. C. Whitelam, 1998. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10 1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiault, D. L., C. A. Wessinger, J. R. Dinneny, J. Lutes, J. O. Borevitz et al., 2008. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc. Natl. Acad. Sci. USA 105 3157–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frewen, B. E., T. H. H. Chen, G. T. Howe, J. Davis, A. Rohde et al., 2000. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 154 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, N., and K. Senni, 2006. Phylogeography of Japanese alpine plants: biogeographic importance of alpine region of central Honshu in Japan. Taxon 55 43–52. [Google Scholar]
- Fujii, N, K. Ueda and T. Shimizu, 1996. Intraspecific sequence variation of chloroplast DNA in Japanese alpine plants. J. Phytogeogr. Taxon 44 72–81. [Google Scholar]
- Fujii, N., K. Ueda, Y. Watano and T. Shimizu, 1997. Intraspecific sequence variation of chloroplast DNA in Pedicularis chamissonis Steven (Scrophulariaceae) and geographic structuring of the Japanese “Alpine” plants. J. Plant Res. 110 195–207. [Google Scholar]
- Fujii, N., K. Ueda, Y. Watano and T. Shimizu, 1999. Further analysis of intraspecific sequence variation of chloroplast DNA in Primula cuneifolia Ledeb. (Primulaceae): implication for biogeography of the Japanese alpine flora. J. Plant Res. 112 87–95. [Google Scholar]
- García-Gil, M. R., M. Mikkonen and O. Savolainen, 2003. Nucleotide diversity at two phytochrome loci along a latitudinal cline in Pinus sylvestris. Mol. Ecol. 12 1195–1206. [DOI] [PubMed] [Google Scholar]
- Guindon, S., and O. Gascuel, 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52 696–704. [DOI] [PubMed] [Google Scholar]
- Halliday, K. J., and G. C. Whitelam, 2003. Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol. 131 1913–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heschel, M. S., J. Selby, C. Butler, G. C. Whitelam, R. A. Sharrock et al., 2007. A new role for phytochromes in temperature-dependent germination. New Phytol. 174 735–741. [DOI] [PubMed] [Google Scholar]
- Hey, J., and R. Nielsen, 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila psudoobscura and D. persimilis. Genetics 167 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., and R. Nielsen, 2007. Integration within the Felsenstein equation for improvaed Markov chain Monte Carlo methods in population genetics. Proc. Natl. Acad. Sci. USA 104 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2000. A new statistic for detecting genetic differentiation. Genetics 155 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Slatkin and W. P. Maddison, 1992. a Estimation of levels of gene flow from DNA sequence data. Genetics 132 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. b A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9 138–151. [DOI] [PubMed] [Google Scholar]
- Ikeda, H., and H. Setoguchi, 2006. Phylogeography of Arcterica nana (Maxim.) Makino (Ericaceae) suggests another range expansion history of Japanese alpine plants. J. Plant Res. 119 489–495. [DOI] [PubMed] [Google Scholar]
- Ikeda, H., and H. Setoguchi, 2007. Phylogeography and refugia of the Japanese endemic alpine plant Phyllodoce nipponica Makino (Ericaceae). J. Biogeogr. 34 169–176. [Google Scholar]
- Ikeda, H., and H. Setoguchi, 2009. The homogenous genetic structure and inferred unique history of range shifts during the Pleistocene climatic oscillations of Arcterica nana (Maxim.) Makino (Ericaceae). J. Plant Res. 122 141–151. [DOI] [PubMed] [Google Scholar]
- Ikeda, H., K. Senni, N. Fujii and H. Setoguchi, 2006. Refugia of Potentilla matsumurae (Rosaceae) located at high mountains in the Japanese archipelago. Mol. Ecol. 15 3731–3740. [DOI] [PubMed] [Google Scholar]
- Ikeda, H., K. Senni, N. Fujii and H. Setoguchi, 2008. a Survival and genetic divergence of an arctic-alpine plant, Diapensia lapponica subsp. obovata (Fr. Schm.) Hultén (Diapensiaceae), in the high mountains of central Japan during climatic oscillations. Plant Syst. Evol. 272 197–210. [Google Scholar]
- Ikeda, H., K. Senni, N. Fujii and H. Setoguchi, 2008. b Consistent geographic structure among multiple nuclear sequences and cpDNA polymorphisms of Cardamine nipponica Franch. et Savat. (Brassicaceae). Mol. Ecol. 17 3178–3188. [DOI] [PubMed] [Google Scholar]
- Ikeda, H., K. Senni, N. Fujii and H. Setoguchi, 2008. c Post-glacial range fragmentation is responsible for the current distribution of Potentilla matsumurae Th. Wolf (Rosaceae) in the Japanese archipelago. J. Biogeogr. 35 791–800. [Google Scholar]
- Ikeda, H., K. Senni, N. Fujii and H. Setoguchi, 2009. High mountains of the Japanese archipelago as refugia for arctic-alpine plants: phylogeography of Loiseleuria procumbens (L.) Desvaux (Ericaceae). Biol. J. Linn. Soc. (in press).
- Ingvarsson, P. K., M. V. Garcia, D. Hall, V. Luquez and S. Jansson, 2006. Clinal variation in phyB2, a candidate gene for day-length-induced growth cessation and bud set, across a latitudinal gradient in European aspen (Populus tremula). Genetics 172 1845–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson, P. K., M. V. Garcia, V. Luquez, D. Hall and S. Jansson, 2008. Nucleotide polymorphisms and phenotypic associations within and around the phytochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes, T. H., and C. R. Cantor, 1969. Evolution of protein molecules, pp. 32–132 in Mammalian Protein Metabolis, edited by H. N. Munro. Academic Press, New York.
- Kryazhimskiy, S., and J. B. Plotkin, 2008. The population genetics of dN/dS. PLoS Genet. 4 e1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuittinen, H., M. Aguadé, D. Charlesworth, A. D. E. Haan, B. Lauga et al., 2002. Primers for 22 candidate genes for ecological adaptations in Brassicaceae. Mol. Ecol. Notes 2 258–262. [Google Scholar]
- Lian, C., M. A. Wadud, Q. Geng, K. Shimatani and T. Hogetsu, 2006. An improved technique for isolating codominant compound microsatellite markers. J. Plant Res. 119 415–417. [DOI] [PubMed] [Google Scholar]
- Maloof, J. N., J. O. Borevitz, T. Dabi, J. Lutes, R. B. Nehring et al., 2001. Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29 441–446. [DOI] [PubMed] [Google Scholar]
- Mathews, S, 2005. Phytochrome evolution in green and nongreen plants. J. Hered. 96 197–204. [DOI] [PubMed] [Google Scholar]
- Mathews, S., 2006. Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15 3483–3503. [DOI] [PubMed] [Google Scholar]
- Mathews, S., and K. McBreen, 2008. Phylogenetic relationships of B-related phytochromes in the Brassicaceae: redundancy and the persistence of phytochrome D. Mol. Phylogenet. Evol. 49 411–423. [DOI] [PubMed] [Google Scholar]
- Mathews, S., and R. A. Sharrock, 1996. The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol. Biol. Evol. 13 1141–1150. [DOI] [PubMed] [Google Scholar]
- Mathews, S., M. Lavin and R. A. Sharrock, 1995. Evolution of the phytochrome gene family and its utility for phylogenetic analysis of angiosperms. Ann. Mo. Bot. Gard. 82 296–321. [Google Scholar]
- Mathews, S., J. G. Burleigh and M. J. Donoghue, 2003. Adaptive evolution in the photosensory domain of phytochrome A in early angiosperms. Mol. Biol. Evol. 20 1087–1097. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351 652–654. [DOI] [PubMed] [Google Scholar]
- Montgomery, B. L., and J. C. Lagarias, 2002. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci. 7 357–366. [DOI] [PubMed] [Google Scholar]
- National Astronomical Observatory, 2002. Chronological Scientific Tables. Maruzen, Tokyo.
- Nielsen, R., and J. Wakeley, 2001. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, Y., T. Matsuchita, N. Mochizuki, T. Suzuki, S. Tokutomi et al., 2004. Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell 16 2104–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, Y., T. Matsushita, N. Mochizuki, P. H. Quail and A. Nagatani, 2008. Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet. 4 e1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D., 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25 1253–1256. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins, S. E., B. E. Stranger, T. Mitchell-Olds and M. Aguadé, 2004. Multilocus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics 166 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP: DNA polymorphisms analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schmid, K. J., S. Ramos-Onsins, H. Ringys-Beckstein, B. Weisshaar and T. Mitchell-Olds, 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphisms. Genetics 169 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senni, K., N. Fujii, H. Takahashi, T. Sugawara and M. Wakabayashi, 2005. Intraspecific chloroplast DNA variation of the alpine plants in Japan. Acta Phytotax. Geobot. 56 265–275. [Google Scholar]
- Sharrock, R. A., and P. H. Quail, 1989. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3 1745–1757. [DOI] [PubMed] [Google Scholar]
- Smith, H., 2000. Phytochromes and light signal perception by plants: an emerging synthesis. Nature 407 585–591. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2003. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b. Sinauer Associates, Sunderland, MA.
- Tajima, F., 1989. The effect of change in population size on DNA polymorphism. Genetics 123 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J., T. Gibson, F. Plewniak, F. Jeanmougin and D. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, G. M., M. T. Hamblin and S. Kresovich, 2004. Molecular evolution of the phytochrome gene family in sorghum: changing rates of synonymous and replacement evolution. Mol. Biol. Evol. 21 716–723. [DOI] [PubMed] [Google Scholar]
- Whitelam, G. C., and P. F. Devlin, 1997. Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 20 752–758. [Google Scholar]
- Whitelam, G. C., S. Patel and P. F. Devlin, 1998. Phytochromes and photomorphogenesis in Arabidopsis. Philos. Trans. R. Soc. Lond. B 353 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 2007. PAML4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2002. Codon-substituion model for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19 908–917. [DOI] [PubMed] [Google Scholar]
- Yang, Z., W. S. W. Wong and R. Nielsen, 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22 1107–1118. [DOI] [PubMed] [Google Scholar]
- Zhang, J., R. Nielsen, and Z. Yang, 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22 2472–2479. [DOI] [PubMed] [Google Scholar]