Abstract
Gene conversion between duplicated genes has been implicated in homogenization of gene families and reassortment of variation among paralogs. If conversion is common, this process could lead to errors in gene tree inference and subsequent overestimation of rates of gene duplication. After performing simulations to assess our power to detect gene conversion events, we determined rates of conversion among young, lineage-specific gene duplicates in four mammal species: human, rhesus macaque, mouse, and rat. Gene conversion rates (number of conversion events/number of gene pairs) among young duplicates range from 8.3% in macaque to 18.96% in rat, including a 5% false-positive rate. For all lineages, only 1–3% of the total amount of sequence examined was converted. There is no increase in GC content in conversion tracts compared to flanking regions of the same genes nor in conversion tracts compared to the same region in nonconverted gene-family members, suggesting that ectopic gene conversion does not significantly alter nucleotide composition in these duplicates. While the majority of gene duplicate pairs reside on different chromosomes in mammalian genomes, the majority of gene conversion events occur between duplicates on the same chromosome, even after controlling for divergence between duplicates. Among intrachromosomal duplicates, however, there is no correlation between the probability of conversion and physical distance between duplicates after controlling for divergence. Finally, we use a novel method to show that at most 5–10% of all gene trees involving young duplicates are likely to be incorrect due to gene conversion. We conclude that gene conversion has had only a small effect on mammalian genomes and gene duplicate evolution in general.
THE evolutionary processes affecting duplicated genes have been of great interest since Ohno (1970) suggested that duplicates play a major role in the evolution of new traits. Genome sequencing has revealed that gene duplication is widespread in eukaryotic genomes (Zhang 2003), and functional studies of many gene duplicates have supported Ohno's claims about its importance in evolution (reviewed in Hahn 2009). Elucidating how gene duplicates evolve over time is therefore fundamental to our understanding of organismal evolution and adaptation.
Several studies have recently begun assessing the role gene conversion plays in the evolution of duplicated genes. Gene conversion, the nonreciprocal transfer of genetic information between homologous sequences, is a type of concerted evolution thought to be responsible for the homogenization of small segments of DNA, generally smaller than several hundred base pairs (Chen et al. 2007). This is in contrast to unequal crossing over, which is usually implicated in homogenizing larger tracts of DNA. Gene conversion is often categorized on the basis of the location of donor and recipient sequences and can generally be classified as either allelic (conversion between alleles on sister chromatids or homologous chromosomes) or nonallelic (conversion between paralogous sequences either on the same chromosome or between chromosomes). In this article, we discuss only the effects of conversion events that occur between duplicated loci (nonallelic or “ectopic” gene conversion).
If widespread, gene conversion between paralogs could greatly impact the evolution of gene families by homogenizing variation among duplicates, thus slowing evolutionary divergence. This pattern has been demonstrated, for example, in the rDNA gene family (Arnheim et al. 1980) and visual pigment genes in Old World monkeys (e.g., Winderickx et al. 1993; Zhou and Li 1996). Conversely, it has been suggested that gene conversion may generate diversity among paralogs through reassortment of genetic variation in the major histocompatibility complex gene family (e.g., Weiss et al. 1983; Ohta 1997; Martinsohn et al. 1999). In addition, gene conversion between allelic sequences has been found to be biased such that G or C alleles preferentially convert A or T alleles (Galtier et al. 2001), resulting in more substitutions of G and C over time. This bias is a consequence of GC-biased repair of mismatches in heteroduplex intermediates during recombination, though there have so far been few studies showing that this mechanism affects ectopic gene conversion (Galtier 2003; Kudla et al. 2004; Benovoy et al. 2005).
Recent studies have begun to assess genomewide rates of conversion between duplicates, attempting to address whether gene family evolution is influenced largely by conversion or by other processes (e.g., Nei and Rooney 2005). Most of these studies indicate that gene conversion may not be so extensive as to have significant effects on gene family evolution. Using a statistical method for inferring conversion events based on the distribution of differences between duplicates (i.e., the software package GENECONV; Sawyer 1989), Drouin (2002) found a genomewide rate of gene conversion (number of conversion events/number of gene pairs) of 7.8% among gene families with more than two members in the yeast Saccharomyces cerevisiae. The same method found a conversion rate of 0.88% in humans (Benovoy and Drouin 2009). The rate of gene conversion detected in the Caenorhabditis elegans genome using a similar method was only 2% (Semple and Wolfe 1999). Using more limited “quartet” methods—which require two related paralogs in each of two species—Wang et al. (2007b) found that ∼8% of Oryza sativa japonica paralogs on chromosomes 11 and 12 have been affected by gene conversion since the split with O. sativa indica. A similar study in humans (Jackson et al. 2005) also using a quartet method estimated a conversion rate of 5% among a subset of gene families. Finally, using a composite method that includes both quartet-based and GENECONV analyses, Ezawa et al. (2006) detected evidence for conversion in 18% of mouse and rat gene families (quartets) and 13% of mouse gene pairs.
The only study to find extremely high rates of gene conversion compared 68 pairs of duplicates in S. cerevisiae, using yet another method intended to detect conversion indirectly (Gao and Innan 2004). This study found that 81% of paralogs in yeast have been recently converted. If gene conversion rates are this high, methods that estimate the rate of gene duplication based on the number of highly similar pairs of paralogs in a genome (e.g., Lynch and Conery 2000) will badly overestimate this rate (Lynch and Conery 2000; Gao and Innan 2004). This is because even ancient paralogs will appear to be recently duplicated if conversion has homogenized their sequences.
Experimental evidence has demonstrated that even slight increases in divergence between homologous sequences can greatly reduce the frequency of conversion (Lukacsovich and Waldman 1999). In this study, therefore, we focus on patterns of gene conversion among young, lineage-specific duplicates. While studies of gene conversion in human (Jackson et al. 2005; Benovoy and Drouin 2009) or mouse and rat (Ezawa et al. 2006) have been performed previously, these studies used either more limited methods or data sets that included much older, more divergent paralogs. As young, lineage-specific paralogs are the most likely to undergo conversion, focusing our study on these duplicates will not only provide an upper bound on rates and effects of conversion genomewide but will also give us more power to detect patterns of conversion. Here, we estimate independent rates of ectopic gene conversion among young duplicates in four mammalian lineages (human, macaque, mouse, and rat) using the method implemented in GENECONV (Sawyer 1989). This method does not require multiple coparalogs in multiple species and can therefore be used to study gene conversion genomewide. To ensure the accuracy of our results, we also use simulations to determine the power of GENECONV to detect gene conversion events within our data and to determine the false-positive rate. Finally, we use a novel method to show that at most 5–10% of all gene trees involving young duplicates are likely to be incorrect due to gene conversion among paralogs and, therefore, that estimates of gene duplication are not greatly affected by conversion.
METHODS
Simulation of gene conversion:
While the power and false-positive rate of GENECONV has been tested in other studies (Posada and Crandall 2001; Posada 2002), the simulated and empirical data sets used were significantly different from those used in this study (e.g., no alignments of only two sequences were included). We therefore determined GENECONV's power and false-positive rate among simulated sequences that more accurately resemble our data. Simulated sequences were generated in PAML using the program Evolver (Yang 2007). Each data set consisted of 1000 duplicates of two sequences representing a coding region of 1500 nucleotides. Duplicates were built under a pattern of two site classes: dN/dS = 0 and dN/dS = 1, 0.5 frequency each (dN is the number of nonsynonymous substitutions per nonsynonymous site; dS is the number of synonymous substitutions per synonymous site). Divergence (dS) was fixed at 0.01, 0.02, 0.05, 0.075, 0.1, and 0.18 in different data sets. These divergences were chosen to be representative of those found in our data set. Note that while likelihood estimates of dS correct for multiple substitutions, the correction is negligible at such low divergences, making these values of dS approximately equal to the true proportion of synonymous substitutions. When a third sequence was added to the alignment, its divergence from each of the duplicates was twice the divergence between the duplicates. Converted tracts of 45, 90, 150, 252, 402, and 501 bp were then transferred from donor to recipient sequences at random. The conversion tract lengths for the simulations were chosen on the basis of the tract lengths observed in our data and in other studies (e.g., Semple and Wolfe 1999; Drouin 2002). Codon frequency was uniform (1/61) and transition/transversion rate ratio was fixed at κ = 2. No rate variation among sites was used, though the effect of such variation—if it affects both synonymous and nonsynonymous mutation rates—will be to inflate the false-positive rate.
Detecting conversion using GENECONV:
GENECONV v.1.81 (http://www.math.wustl.edu/∼sawyer/geneconv) (Sawyer 1989) was used to identify gene conversion events. Significance is determined based on 10,000 permuted data sets. GENECONV determines both global and pairwise P-values, the former corrected for the number of sequences in the alignment. Because we sought to compare gene families of various sizes, we used pairwise P-values to determine significance comparably across families. Calculating conversion rates with pairwise P-values (number of pairs with significant pairwise P-values/number of total pairs analyzed) indicates the percentage of all gene pairs with evidence for conversion. GENECONV was run using all default settings except for the addition of the option to display pairwise P-values (--ListPair) and the option to include monomorphic sites in the calculation when there were only two sequences in an alignment (--Include-monosites). This last option removes controls for constant sites but is necessary for analyzing an alignment with only two sequences. Significant “Pairwise Inner” fragments were considered gene conversion events. No mismatches were allowed in conversion tracts. Only duplicate pairs with at least three differences between the two sequences were considered for analysis. Analysis of average conversion tract lengths and the distribution of tract lengths included only conversion events that do not cross intron/exon boundaries or either end of the gene coding sequence, as our study does not determine to what extent the conversion tracts extend into introns or flanking sequences. All conversion events, however, were used for calculation of the total proportion of sequence converted. Subsequent analyses (position of tract in gene; GC content of converted vs. nonconverted pairs and conversion tracts vs. flanking regions; divergence of flanking regions of converted pairs vs. nonconverted pairs; correlation between probability of conversion and meiotic recombination rate) were performed using in-house perl scripts.
Alignment and analysis of mammalian gene duplicates:
We used Ensembl v41 gene models for human, macaque, mouse, and rat. Construction of the gene trees for each gene family and inference of duplications from gene trees are described in Hahn et al. (2007). Briefly, 9920 gene trees were constructed from protein alignments (including homologs from an outgroup, the dog genome), followed by gene-tree/species-tree reconciliation conducted using NOTUNG (Chen et al. 2000). Duplication events specific to each lineage (i.e., in mouse after the split with rat, in rat after the split with mouse, in human after the split with macaque, and in macaque after the split with human) were identified for each tree. Following identification of duplication events, cDNA sequences of lineage-specific paralogs were aligned by first aligning the protein sequences with ClustalW and then threading the nucleotide sequences through the protein alignments. Families containing transposable elements mistakenly annotated as genes were filtered out.
Since duplication events can incorrectly appear to be lineage specific when a copy is lost in an outgroup, we further filtered the duplicates on the basis of branch lengths for our analysis of conversion. We required the distance (dS) between any two paralogs to be less than twice the distance since the speciation event separating sister lineages (i.e., human–macaque and mouse–rat). This requirement simply identified and removed those duplicates that only appeared to be lineage specific artifactually and that are, in reality, more ancient duplicates. The average dS values for each of the four lineages were taken from the genomic average of 9448 one-to-one orthologs (Wang et al. 2007a): human, dS = 0.032; macaque, dS = 0.038; mouse, dS = 0.095; rat, dS = 0.095. For example, this requirement means that for two paralogs to be considered lineage specific along the human branch, their divergence must be less than (2) × (0.032) = 0.064. Nucleotides present in only one gene in an alignment and the corresponding gaps in all other genes were removed before analysis with GENECONV. Gaps aligned with sequence present in at least two genes, however, were maintained.
Gene tree vs. CAFE analysis:
To compare the number of lineage-specific duplications inferred by gene tree analyses and copy number analyses, we considered the 9920 gene families used above. For each of these families we counted the number of lineage-specific duplicates inferred from the gene tree along the branch leading to each of human, macaque, mouse, and rat using NOTUNG (Chen et al. 2000). We compared these counts for each family to the number of lineage-specific duplicates inferred from the number of copies in each lineage using CAFE (Hahn et al. 2005; De Bie et al. 2006). The number of families along each lineage with a greater number of duplicates inferred by the gene tree method was divided by the total number of families with two or more genes in that lineage, resulting in the proportion of trees possibly affected by gene conversion (see results).
RESULTS
Assessing false-positive and false-negative rates:
Among simulated sequences representative of our data set, we determined that GENECONV has higher statistical power to detect recent gene conversion when the divergence between the duplicates is higher and when the conversion tract is longer (supporting information, Table S1). At the highest tested divergence, 0.18 substitutions per site, GENECONV detected only 21.6% of conversions when the tract was 45 bp but detected all conversions when the tract was at least 90 bp (in the 1500-bp sequence). At the lowest divergence, 0.01 substitutions per site, however, GENECONV only detected 37.1% of conversion events at even the largest tract length, 501 bp. These simulations indicate that GENECONV is able to detect almost all conversion events that are > ∼200 bp when duplicates are at least as divergent as ∼0.05 substitutions per site. Addition of a third sequence to the alignment (with no additional conversion event simulated) had no effect on the power of GENECONV to detect conversion between the original two sequences.
We also performed simulations to determine GENECONV's false-positive rate under the default conditions (three or more sequences) and when including “monomorphic” sites (two sequences). It has been suggested previously that the false-positive rate may be particularly high when only two sequences are present in an alignment (Drouin 2002; Mondragon-Palomino and Gaut 2005). In our simulations of alignments with only two sequences, the false-positive rates (number of conversion events detected/number of gene pairs) for the divergences of 0.016, 0.05, and 0.1 were 5.7%, 4.9%, and 4.4%, respectively. The average conversion tract length detected was negatively correlated with the divergence of the duplicates. The overall proportion of total sequence implicated in a (false) conversion event was therefore highest (0.45%) for the lowest divergence (0.016). When a third sequence was added to the alignment and GENECONV was run under default conditions, the false-positive rate was still <5%: at a divergence of 0.05, the fraction of false positives per pairwise comparison was 2.7% with three sequences, compared to 4.9% with two sequences.
These simulations indicate that GENECONV has reasonable power to detect true conversion events in our data, though comparison of very young duplicates is undoubtedly underpowered. In addition, we find no evidence that the false-positive rate is aberrantly high when only two sequences are present in an alignment. The rate of false positives of GENECONV appears to be what is expected when a significance threshold of P < 0.05 is used.
Conversion rates and patterns in mammalian genomes:
To obtain independent estimates of gene conversion in each of the four species, we compared only lineage-specific paralogs within each lineage (methods). Higher divergence between paralogs leads to less frequent gene conversion as well as shorter conversion tracts (Lukacsovich and Waldman 1999); we are therefore confident that an analysis focused on less divergent paralogs captures the majority of gene conversion events occurring in these genomes. It also provides an upper bound on the rate and effects of gene conversion genomewide. Our final data consisted of 261 alignments of lineage-specific duplicates (549 pairwise comparisons) in humans, 206 alignments (363 pairs) in macaque, 629 alignments (1913 pairs) in mouse, and 603 alignments (1171 pairs) in rat.
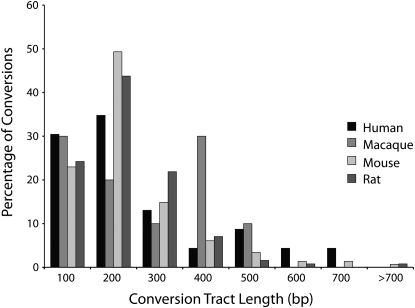
Among all lineage-specific gene pairs analyzed, we found the rate of gene conversion (number of conversion events/number of gene pairs) to be 12.57% in human, 8.26% in macaque, 14.58% in mouse, and 18.96% in rat at P < 0.05 (see Table S2 for a list of predicted conversion events between gene pairs). The actual rates, however, are likely even lower as these values include a false-positive rate of 5% at this P-value. The distribution of conversion tract lengths illustrates that most conversion events extend < ∼500 bp (Figure 1); it also reflects the poor power of GENECONV to detect conversions < ∼100 bp in length. The average length of the conversion tracts is 210 bp in human, 229 bp in macaque, 190 bp in mouse, and 172 bp in rat. Because the method used to detect conversion looks for long stretches of identity that must be bounded on either side by a difference between the paralogs, the conversion tract lengths detected by GENECONV are maximum estimates of the size of the tract. The positions of conversion tracts within genes were uniformly distributed, with the start of most tracts in the first 25% of the gene sequence. The overall proportion of total sequence that has been converted is 2.16% in human, 1.76% in macaque, 2.57% in mouse, and 2.15% in rat. This indicates that gene conversion among duplicates is likely to affect a mere 1–3% of total sequence within recently duplicated mammalian genes (and even smaller amounts among older duplicates).
Figure 1.—
Lengths of conversion events detected in the human, macaque, mouse, and rat genomes. Bin values on the x-axis represent the maximum tract length included in each bin. Only conversion events that do not cross intron/exon boundaries are included. The underrepresentation of conversion events <100 bp likely reflects the low power of GENECONV to detect short conversions.
Biased gene conversion between allelic sequences has been shown to lead to an increase in the GC content of conversion tracts (Galtier and Duret 2007). There have been few studies, however, to investigate the effects of nonallelic gene conversion on GC content (Galtier 2003; Kudla et al. 2004; Benovoy et al. 2005). Among alignments in our analysis with only two sequences, the average GC content within conversion tracts was not significantly greater than the average GC content of nonconverted flanking sequence and was actually slightly lower in some lineages: 52.0% vs. 53.8% in human, 50.6% vs. 49.6% in macaque, 46.8% vs. 47.3% in mouse, and 47.5% vs. 47.3% in rat (paired t-test, P > 0.05 for all). This comparison could potentially miss an increase in GC content in converted tracts, however, as it compares different regions of genes (conversion tracts vs. flanking sequences). We therefore also compared the GC content of a conversion tract with the same gene segment from nonconverted paralogs when there were more than two sequences in an alignment. Again, there was no significant trend toward higher GC content in converted sequences vs. nonconverted sequences: 54.0% vs. 52.5% in human, 49.4% vs. 49.2% in macaque, 45.9% vs. 45.7% in mouse, and 47.4% vs. 47.4% in rat (paired t-test, P > 0.05 for all).
While gene conversion is known to occur more frequently between more similar duplicates (Lukacsovich and Waldman 1999), the distribution of the divergences of nonconverted gene pairs compared to those of converted pairs (excluding the conversion tract) does not clearly demonstrate such a pattern (Figure S1). One reason for the apparent lack of the expected pattern is GENECONV's poor power to detect conversion events when divergence between duplicates is very low. It is also of course true that conversion between highly similar genes will often have no homogenizing effect, as there may be no nucleotide differences in the conversion tracts to begin with.
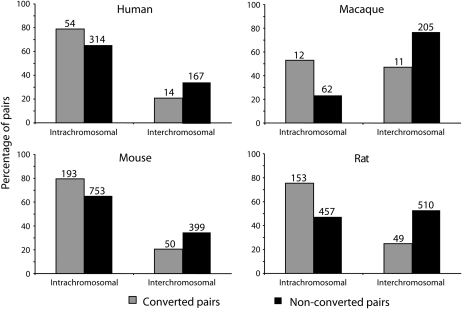
Many recent gene duplication events result in paralogs that reside on different chromosomes (Figure 2), though there is evidence for an expansion in intrachromosomal duplications along the human lineage (She et al. 2006). The majority of duplicated genes that have undergone gene conversion are located on the same chromosome in all four species (Figure 2). The excess of intrachromosomal conversion relative to the number of intrachromosomal duplicates is statistically significant in every genome (Fisher's exact test, all P < 0.05). In addition, intrachromosomal conversion occurs at a disproportionately higher frequency between duplicates that are close together (<50 kb apart), and there is a significantly negative correlation (P < 0.05) between rates of conversion and intrachromosomal distance in human, mouse, and rat (Figure S2). However, neighboring paralogs are more likely to be recently duplicated and thus less divergent (Katju and Lynch 2003), and it is possible that interchromosomal duplicates may on average be more divergent, confounding the factors of chromosomal location, physical distance, and divergence. Linear regressions demonstrate that while chromosomal location (intrachromosomal vs. interchromosomal) is still a significant predictor of conversion after correcting for divergence (P < 0.01 for all genomes), physical distance between intrachromosomal duplicates is not a significant predictor of conversion once divergence is accounted for (P > 0.1 for all genomes).
Figure 2.—
Overrepresentation of intrachromosomal gene pairs among pairs showing evidence of gene conversion. The percentage of converted pairs that are present on the same chromosome (intrachromosomal) and the percentage present on different chromosomes (interchromosomal) are shaded; percentages of nonconverted pairs are solid. The number of pairs in each category is given above the bars. In all four genomes, conversion appears to occur preferentially between intrachromosomal duplicates.
For intrachromosomal duplicates, we also hypothesized that gene conversion might be influenced by the orientation of duplicates relative to each other. We therefore classified each pair of intrachromosomal duplicates as head to tail, head to head, or tail to tail. If duplicates are arranged randomly, we expect 50% in a head-to-tail orientation and 25% in each of head-to-head and tail-to-tail orientations. Among all mammalian duplicates we found a significant excess of head-to-tail arrangements for intrachromosomal paralogs within 50 kb of each other (Fisher's exact test, all P < 0.05; Figure S3), though there was only an excess for all intrachromosomal paralogs in rat and mouse. Contrary to our expectations, however, there was no excess of gene conversion associated with any specific orientation of paralogs in any of the four genomes (Figure S4). These patterns of correlation between conversion and chromosomal location, distance between paralogs, and gene orientation largely agree with those found previously for conversion events between older paralogs in mouse (Ezawa et al. 2006) and human (Benovoy and Drouin 2009), though these studies did not consider the confounding effects of divergence and physical distance.
While meiotic recombination is responsible for both allelic gene conversion and crossovers, the relationship between meiotic recombination rate and ectopic gene conversion is unclear. We therefore looked for a relationship between human recombination rates based on the deCODE map (Kong et al. 2002) and the frequency of gene conversion among human paralogs.
The proportion of converted vs. nonconverted duplicated pairs shows no correlation with recombination rates for pairs <1 or <5 Mb apart (r = −0.007 and r = 0.039). Similar results are obtained using all duplicated pairs and averaging the recombination rates of the two genes (r = 0.024). This is contrary to the results of Benovoy and Drouin (2009), who found a significant positive correlation between meiotic recombination rate and frequency of gene conversion in humans. This difference in results could be due to a difference in methods or recombination rates used.
Effect of conversion on gene trees and estimates of duplication rates:
Lynch and Conery (2000) proposed a method to estimate rates of gene duplication by counting the number of very young duplicates (i.e., dS < 0.01) in a genome and dividing by the total number of genes. This method therefore assumes that low divergence between duplicates reflects recent duplication events and is not due to gene conversion among paralogs (Lynch and Conery 2000). A study of gene conversion in yeast has cast doubt on the results of this method by showing extremely high rates of conversion in this species, implying that actual rates of gene duplication are much lower than previously thought (Gao and Innan 2004). However, the yeast study only indirectly inferred gene conversion and was limited to 68 pairs of duplicates; its results were also in conflict with previous studies of the rate of gene conversion in yeast that used GENECONV (Drouin 2002).
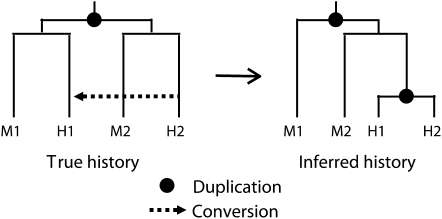
We have recently introduced a method for estimating rates of gene duplication and loss that only relies on changes in the number of paralogous genes among species and not on sequence identity (Hahn et al. 2005). This method will not overestimate rates of gene duplication due to gene conversion, as the number of duplicates in a genome does not change because of conversion (Hahn et al. 2007). For example, if human and macaque each had two duplicate copies of a gene and other mammals had only one copy, this method (as implemented in the program CAFE) (De Bie et al. 2006) would infer a single duplication in the human–macaque ancestor, regardless of the similarity between the human paralogs. We can therefore use this method to confirm that gene conversion among young duplicates in mammalian genomes is not leading to widespread error in gene trees and duplication estimates. To do this we compared the number of lineage-specific duplications inferred from gene trees—constructed from the protein sequences of the genes—to the number inferred by CAFE in all gene families with a size of at least two (methods). If gene conversion has recently homogenized pairs of duplicates, gene tree-based methods will overestimate the number of duplication events. This is because conversion will cause the intraspecific duplicates to be more similar to each other, leading to an estimation of two recent duplication events, one in each lineage, rather than one duplication event that preceded speciation (Figure 3).
Figure 3.—
Duplication followed by gene conversion can lead to an overestimation of the number of duplications in a gene family. The true history of a gene family is shown on the left, with a single duplication event in the human–macaque ancestor followed by speciation giving rise to two macaque paralogs (M1 and M2) and two human paralogs (H1 and H2). If the human paralogs subsequently undergo gene conversion such that H2 converts H1, phylogenetic analysis of the gene family will yield the inferred tree illustrated on the right, where H1 and H2 are highly similar sequences. This tree would imply two duplication events as shown.
In all four lineages, the percentage of gene families where the number of duplications inferred by gene trees was greater than the number inferred by CAFE (i.e., families where gene conversion may be affecting the tree) was very low: 227/3378 (6.7%) in human, 276/3560 (7.8%) in macaque, 301/3505 (8.6%) in mouse, and 328/3388 (9.7%) in rat. We should not assume, however, that all of the cases where the gene tree has inferred more duplications are due to gene conversion (i.e., the CAFE estimate is correct while the gene tree estimate is incorrect), as some are undoubtedly due to true parallel duplications or multiple duplications coupled with gene loss (i.e., the gene tree estimate is correct while the CAFE estimate is incorrect). To provide a rough estimate of the rate of parallel duplication vs. gene conversion, we examined the seven cases where a gene family had exactly two gene copies in both human and macaque, independent duplications had been implied by the gene tree, and where all four genes have been assigned to a chromosomal location. Of the seven cases, only three show both duplicates maintained on homologous chromosomes between species. The remaining four families have one ortholog on homologous chromosomes between human and macaque (likely the single gene present in the most recent common ancestor) while the additional copies are on nonhomologous chromosomes between species. While gene conversion followed (or preceded) by translocation cannot be ruled out in these four cases, we believe it is more likely that they represent parallel duplications in the two lineages. It is therefore likely that the percentage of families where gene conversion might lead to an overestimation of duplications is even <5–10%, perhaps less than half this value.
DISCUSSION
This study shows that the overall impact of conversion among young gene duplicates in mammalian genomes is likely to be minimal. This conclusion is consistent with that of Nei and Rooney (2005), who suggested that the contribution of gene conversion to gene family evolution is minor in the long term. We found rates of conversion between recently duplicated genes in human, macaque, mouse, and rat to be low: <5–15% of duplicate pairs showed evidence of conversion (when the 5% false-positive rate is considered). We also found no increase in GC content of converted sequences, indicating that biased gene conversion is not a significant driver of nucleotide content evolution in gene duplicates in these genomes. On the whole, only 3–6% of the total sequence analyzed was involved in a conversion event, meaning only 1–3% of sequence was actually converted (a recipient of gene conversion). These numbers are comparable to the 2–13% conversion frequencies observed previously for the yeast, nematode, mouse, and rice lineages (Semple and Wolfe 1999; Drouin 2002; Ezawa et al. 2006; Wang et al. 2007b), indicating that gene conversion is likely to be far from ubiquitous in most genomes. In particular, our estimate for the percentage of gene pairs undergoing conversion in mouse, 14.56%, is highly consistent with the percentage estimated by Ezawa et al. (2006), 13%. This is striking when we consider the different methodologies and data sets used—our study was limited to lineage-specific duplicates while the Ezawa et al. data excluded lineage-specific duplicates and focused on duplicates that arose in the mouse–rat ancestor.
Our estimate for conversion rate among young duplicates in human (12.57%), on the other hand, is much larger than the 0.88% frequency recently estimated by Benovoy and Drouin (2009). This is to be expected, however, as Benovoy and Drouin included duplicate pairs with at least 60% protein identity over at least 50% of the sequence. Inclusion of more divergent duplicates should lower the observed conversion rate, as the young duplicates in our study likely undergo the highest rates of conversion of any duplicates in the genome. In addition, Benovoy and Drouin utilized GENECONV's global P-values in their calculation of conversion rate, which makes direct comparison with our values difficult but which is also likely to decrease the observed conversion rate.
While we believe our study provides an important estimate of the upper bound of the frequency and effects of conversion among duplicates in these four mammalian genomes, there are some limitations to our analysis. Our method is underpowered for detecting conversion events between duplicates < ∼5% divergent, though such conversion events are likely to have the smallest impact on the genome as they will lead to few substitutions in the converted copies. However, this lack of power at very low divergences is potentially responsible for the slightly lower conversion rates in human and macaque compared to mouse and rat, as there are more lineage-specific duplicates with higher divergence in the rodent lineages (methods). In addition, our estimates of tract length (and therefore total sequence involved in conversions) are likely to be somewhat overestimated, as conversion tracts identified by GENECONV must necessarily be bounded by differences between duplicates; in actuality, however, the conversion tract may have been shorter. Because we did not allow mismatches within gene conversion tracts detected by GENECONV, our analysis may miss older events where one or more mutations have occurred after conversion. This would cause our numbers to be underestimates of the actual conversion rates in these genomes. However, because our study is focused on conversion events between recent duplicates, we believe this is not likely to be a significant source of error. Finally, GENECONV does not take into account purifying selection that may be acting differentially on different gene segments. If selection were maintaining identical sequences between duplicates in one part of the gene but relaxed selection were allowing mutations in another region, this could lead to the appearance of gene conversion. However, we believe this type of false positive is unlikely in our data, as our analysis included not only nonsynonymous sites but synonymous sites as well. Because the large majority of synonymous mutations are believed to be silent, purifying selection should generally not affect mutations at synonymous sites. Situations where an identical stretch of coding sequence between duplicates has been maintained by purifying selection at both nonsynonymous and synonymous sites must therefore be very rare, if they occur at all, in these data.
Perhaps most importantly, our comparison of the number of duplications inferred by gene trees compared to the number inferred by copy number demonstrates that gene conversion does not lead to widespread gene tree inconsistencies and large overestimates of the gene duplication rate. Even if we have missed conversion events between young duplicates using GENECONV, or conversion has occurred across the full length of two paralogs, the comparison of gene trees and copy number indicates that the overall effects of gene conversion must be minimal. Simply the fact that copy numbers do change at such high rates—even in yeast (Hahn et al. 2005)—supports the original contention of Lynch and Conery (2000) that rates of gene duplication are high.
While our results emphasize the minor impact of gene conversion genomewide, other studies have highlighted the important role gene conversion can play in duplicate gene evolution in certain gene families (e.g., Hoffmann et al. 2008). Those studies, in the context of our results, imply that variation in the frequency and selective advantage of conversion among gene families may be high. Despite these rare cases, however, when all gene families with young duplicate genes are considered, gene conversion clearly does not play a major role across the genome.
Acknowledgments
We thank Mira Han for sharing her data. This work was supported by a grant from the National Science Foundation (DBI-0543586) to M.W.H.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101428/DC1.
References
- Arnheim, N., M. Krystal, R. Schmickel, G. Wilson, O. Ryder et al., 1980. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc. Natl. Acad. Sci. USA 77 7323–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovoy, D., and G. Drouin, 2009. Ectopic gene conversions in the human genome. Genomics 93 27–32. [DOI] [PubMed] [Google Scholar]
- Benovoy, D., R. T. Morris, A. Morin and G. Drouin, 2005. Ectopic gene conversions increase the G+C content of duplicated yeast and Arabidopsis genes. Mol. Biol. Evol. 22 1865–1868. [DOI] [PubMed] [Google Scholar]
- Chen, J.-M., D. N. Cooper, N. Chuzhanova, C. Ferec and G. P. Patrinos, 2007. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8 762–775. [DOI] [PubMed] [Google Scholar]
- Chen, K., D. Durand and M. Farach-Colton, 2000. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 7 429–447. [DOI] [PubMed] [Google Scholar]
- De Bie, T., N. Cristianini, J. P. Demuth and M. W. Hahn, 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22 1269–1271. [DOI] [PubMed] [Google Scholar]
- Drouin, G., 2002. Characterization of the gene conversions between the multigene family members of the yeast genome. J. Mol. Evol. 55 14–23. [DOI] [PubMed] [Google Scholar]
- Ezawa, K., S. Oota and N. Saitou, 2006. Genome-wide search of gene conversions in duplicated genes of mouse and rat. Mol. Biol. Evol. 23 927–940. [DOI] [PubMed] [Google Scholar]
- Galtier, N., 2003. Gene conversion drives GC content evolution in mammalian histones. Trends Genet. 19 65–68. [DOI] [PubMed] [Google Scholar]
- Galtier, N., and L. Duret, 2007. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 23 273–277. [DOI] [PubMed] [Google Scholar]
- Galtier, N., G. Piganeau, D. Mouchiroud and L. Duret, 2001. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics 159 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L., and H. Innan, 2004. Very low gene duplication rate in the yeast genome. Science 306 1367–1370. [DOI] [PubMed] [Google Scholar]
- Hahn, M. W., 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. (in press). [DOI] [PubMed]
- Hahn, M. W., T. De Bie, J. E. Stajich, C. Nguyen and N. Cristianini, 2005. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 15 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. W., J. P. Demuth and S. G. Han, 2007. Accelerated rate of gene gain and loss in primates. Genetics 177 1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, F. G., J. C. Opazo and J. F. Storz, 2008. Rapid rates of lineage-specific gene duplication and deletion in the alpha-globin gene family. Mol. Biol. Evol. 25 591–602. [DOI] [PubMed] [Google Scholar]
- Jackson, M. S., K. Oliver, J. Loveland, S. Humphray, I. Dunham et al., 2005. Evidence for widespread reticulate evolution within human duplicons. Am. J. Hum. Genet. 77 824–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju, V., and M. Lynch, 2003. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165 1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31 241–247. [DOI] [PubMed] [Google Scholar]
- Kudla, G., A. Helwak and L. Lipinski, 2004. Gene conversion and GC-content evolution in mammalian Hsp70. Mol. Biol. Evol. 21 1438–1444. [DOI] [PubMed] [Google Scholar]
- Lukacsovich, T., and A. S. Waldman, 1999. Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics 151 1559–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Martinsohn, J. T., A. B. Sousa, L. A. Guethlein and J. C. Howard, 1999. The gene conversion hypothesis of MHC evolution: a review. Immunogenetics 50 168–200. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., and B. S. Gaut, 2005. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol. Biol. Evol. 22 2444–2456. [DOI] [PubMed] [Google Scholar]
- Nei, M., and A. P. Rooney, 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, Berlin.
- Ohta, T., 1997. Role of gene conversion in generating polymorphisms at major histocompatibility complex loci. Hereditas 127 97–103. [DOI] [PubMed] [Google Scholar]
- Posada, D., 2002. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol. Biol. Evol. 19 708–717. [DOI] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98 13757–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, S. A., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6 526–538. [DOI] [PubMed] [Google Scholar]
- Semple, C., and K. H. Wolfe, 1999. Gene duplication and gene conversion in the Caenorhabditis elegans genome. J. Mol. Evol. 48 555–564. [DOI] [PubMed] [Google Scholar]
- She, X., G. Liu, M. Ventura, S. Zhao, D. Misceo et al., 2006. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res. 16 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. Y., H. C. Chien, N. Osada, K. Hashimoto, S. Sugano et al., 2007. a Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 5 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., H. Tang, J. E. Bowers, F. A. Feltus and A. H. Paterson, 2007. b Extensive converted evolution of rice paralogs and the road to regaining independence. Genetics 177 1753–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E. H., A. Mellor, L. Golden, K. Fahrner, E. Simpson et al., 1983. The structure of a mutant H-2 gene suggests that the generation of polymorphism in H-2 genes may occur by gene conversion-like events. Nature 301 671–674. [DOI] [PubMed] [Google Scholar]
- Winderickx, J., L. Battisti, Y. Hibiya, A. G. Motulsky and S. S. Deeb, 1993. Haplotype diversity in the human red and green opsin genes: evidence for frequent sequence exchange in exon 3. Hum. Mol. Genet. 2 1413–1421. [DOI] [PubMed] [Google Scholar]
- Yang, Z. H., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang, J., 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18 292–298. [Google Scholar]
- Zhou, Y.-H., and W.-H. Li, 1996. Gene conversion and natural selection in the evolution of X-linked color vision genes in higher primates. Mol. Biol. Evol. 13 780–783. [DOI] [PubMed] [Google Scholar]