Abstract
One of the major difficulties for many laboratories setting up proteomics programs has been obtaining and maintaining the computational infrastructure required for the analysis of the large flow of proteomics data. We describe a system that combines distributed cloud computing and open source software to allow laboratories to set up scalable virtual proteomics analysis clusters without the investment in computational hardware or software licensing fees. Additionally, the pricing structure of distributed computing providers, such as Amazon Web Services, allows laboratories or even individuals to have large-scale computational resources at their disposal at a very low cost per run. We provide detailed step by step instructions on how to implement the virtual proteomics analysis clusters as well as a list of current available preconfigured Amazon machine images containing the OMSSA and X!Tandem search algorithms and sequence databases on the Medical College of Wisconsin Proteomics Center website (http://proteomics.mcw.edu/vipdac).
Keywords: mass spectrometry, data analysis, search algorithms, software, cloud computing
Introduction
High throughput proteomics research has been kept out of the reach of many laboratories because of the high costs of instrumentation and setting up and maintaining the computational infrastructure required for the analysis of the resulting data1. Modern mass spectrometers are capable of generating data many times faster than a typical single desktop computer is able to analyze it. To overcome this problem, institutions or larger laboratory groups have invested in computation clusters composed of large numbers of processors. The creation of these types of computational resources requires not only a significant investment in hardware, but also software license fees, additional space to house the cluster, and trained personnel to administer it. In most laboratories it is difficult to manage the workflow in such a way that the cluster is fast enough to reduce “analytical backlog” during periods of peak activity without having significant underutilization during slower periods. We have brought together two recent developments, open source proteomics search programs and distributed on demand or `cloud' computing, to allow for the construction of a highly flexible, scalable, and very low cost solution to proteomics data analysis, the Virtual Proteomics Data Analysis Cluster (ViPDAC).
Until recently, the standard programs used for proteomics data analysis were almost exclusively commercial, proprietary, closed source and expensive. Some programs were tightly linked to a specific instrument manufacturer and could not be used easily with data from other instruments. License fees for these commercial applications typically have rivaled or exceeded the cost of the computer hardware required to run them. Several groups have introduced open source alternatives to commercial proteomics database search software. One group from the National Institutes of Health (NIH), has developed and distributed an open source alternative to commercial proteomics search programs entitled the Open Mass Spectrometry Search Algorithm (OMSSA)2. The source code for OMSSA has been placed in the public domain by the authors. Since its introduction in 2004, OMSSA has continued to be developed and supported by Geer laboratory at the NIH, and an OMSSA results browser with a graphical user interface (GUI) has been recently introduced3. A second open source proteomics database search program is X!Tandem. X!Tandem was developed by the Bevis laboratory at the University of Manitoba and has been released as open source software under the “Artistic license”4. Both OMSSA and X!Tandem have been widely accepted by the proteomics community for the analysis of high throughput proteomics data. Additionally, proteomics journals have begun to specifically request that the algorithms used for data analysis be available to the general public, not only so that others can replicate the analysis, but also so that the algorithm and its implementation can be independently verified5-7.
Although the use of open source proteomics database search programs can significantly reduce the cost of proteomics data analysis by eliminating licensing fees, a substantial investment in computer hardware and the personnel to administer and maintain it is still required. Recently, an alternative to purchasing and maintaining computation clusters has become widely available5. Amazon and other vendors have developed systems in which computer hardware is `virtualized', allowing both physical computers and data storage to be replaced with virtual equivalents available through a high-speed network connection6. Because of this, many organizations have been able to satisfy their computing needs without incurring the costs of purchasing and maintaining computer equipment. In addition, since the virtual computers exist only while they are in use, it is possible to very flexibly expand or shut down compute clusters and pay only for the computational time used.
The combination of open source software and virtual computer resources allows for the construction of proteomics data analysis systems that are both low cost to set up and low cost to operate. We have estimated that the cost to perform a database search for a typical mass spectrometer run using this virtual system is on the order of $1 US. We have developed of a set of tools that allow typical scientific end users to easily set up and customize their own virtual proteomics analysis system using both the OMSSA and X!Tandem database search engines running on the Amazon Web Services (AWS) systems.
Methods
The standard Ubuntu public Amazon Machine Image (AMI) (8.04 LTS (Long Term Support) Hardy), available from Amazon, was customized by the installation of Ruby (1.8.6) and Rails (2.1.1). In addition, the OMSSA (2.1.4) and X!Tandem (08-02-01-3) database search programs and their dependencies were also installed on the AMI. A selection of common protein databases was also included to allow the user to be able to analyze data for a wide range of biological experiments. The resulting ViPDAC AMI is available from the Amazon community public AMI list7.
The data used to demonstrate the use of the ViPDAC was generated locally from the Sigma USP1 48 human protein standard mix (Sigma-Aldrich) or as part of the publicly available data from the Association of Biomolecular Resource Facilities (ABRF) study of the same set of proteins (Sigma) as previously described 8 and analyzed with a Thermo Fisher LTQ mass spectrometer. Spectra were extracted from raw data files using the Thermo Fisher provided extract_ms.exe program and the resulting .dta files combined into .mgf files by a local script based on a program provided by Matrix Science9. To instantiate the head node, the Firefox browser10 with the ElasticFox plugin was used11. OMSSA parameters used for the demonstration searches were -zc 1 -zh 3 -zt 2 -tom 0 -te 2.5 -v 3 -e 0 -zcc 1 -tez 1 -mv 1,3 -to 0.8 -tem 0 -i 1,4 -d 25.H_sapiens.fasta. Complete details, including all required computer code are available at the Medical College of Wisconsin Proteomics Center website (http://proteomics.mcw.edu/vipdac).
Results and Discussion
The major advantage of cloud computing is that it allows for the storage and analysis of data without the need to own and maintain physical computers and storage devices. In this work we describe an implementation of virtual proteomics data analysis cluster based on the cloud computing resources provided by Amazon6. Although Amazon is currently the major supplier of cloud computing services, this approach could easily be generalized to other cloud-computing providers.
The cloud data storage component provided by Amazon is called the Amazon Simple Storage System or S36. S3 allows users to transfer data into or out of `buckets' of data storage located on Amazon's servers. The users are charged a very small fee for uploading and downloading data, as well as a small fee for monthly storage. The typical cost for transferring and storage of the results of a single mass spectrometer run is on the order of several cents, so even large number of data sets can be transferred and stored for minimal cost. Once data has been uploaded to a user's S3 storage area, data transfers to and from the virtual computer nodes created by the user are free. All of the data transfer and storage is secure and controlled by the use of user specific access keys.
The computational resources provided by the Amazon cloud computing system are called the Amazon Elastic Compute Cloud (EC2)12. The EC2 system is very flexible in that the user can call into existence one or multiple instances of a choice of five different size virtual computers. The sizes run from the `small' instance, similar in computational power and storage to a typical 32 bit single core desktop computer, to the `extra large' instance, similar to a large 64 bit 8 core server computer. Each instance is billed based on the number of hours or parts thereof that it exists. The typical mass spectrometer MudPIT13 run requires several core-hours of computational time, which currently costs on the order of $0.40 to $2.50 US. Messages are passed between the head node and worker nodes to inform the worker nodes of packages of data that need to be processed and the head node of results that are complete.
The first step in the process of setting up a virtual proteomics data analysis cluster is to build an image of an individual virtual computer. These computer images are known as Amazon Machine Images, or AMIs, and they contain the operating system, applications and databases used for the analysis. To do this, a local computer can be set up and customized and then the contents of the computer converted to a compressed .zip file and the file transferred to the S3 storage system as an AMI. More typically, a predefined AMI containing the operating system and other common components preinstalled can be called into existence, customized by user, and saved as a new AMI to the S3 system. The user customization required for proteomics data analysis includes the installation of analysis software and the required databases. After configuration, when these saved AMIs are called back into existence, they are fully loaded with the analysis software and databases installed and are ready to be used to analyze data. Additionally, AMIs stored in S3 can be made to be either private or publicly available. Making an AMI publicly available, or publishing the AMI, allows the customized AMI to be called into existence and used by other users. As part of this project, we have published an AMI configured for proteomics data analysis that is pre-built with open source OMSSA and X!Tandem database search programs and commonly used protein databases. A current list of public AMIs configured by our group for proteomics data analysis along with detailed instructions on how to implement the data stream on the on the S3 can be found at: http://proteomics.mcw.edu/vipdac.
Integration of AWS components to create a virtual cluster
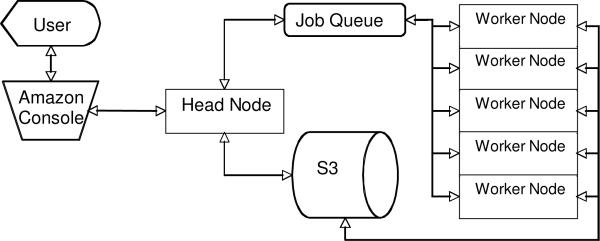
The combination of software running on a `head node' computer, the S3 storage and preconfigured AMI worker nodes running in the EC2 form the components of a Virtual Proteomics Data Analysis Cluster (ViPDAC). The overall design and workflow of the ViPDAC is illustrated in Figure 1. There are four main components to the ViPDAC system: a single head node, a number of worker nodes and the S3 storage system. Although the head node could be either a physical computer owned by the user or a virtual computer running in the EC2 environment, for the current implementation of ViPDAC we have configured it as a virtual computer. When the user launches the first instance of the ViPDAC AMI using the ElasticFox plug-in or Amazon AWS Console (Figure 2), the first node to be launched configures itself to be the head node. The head node launches a web interface and additional `worker' nodes can be launched by the user through the web interface. The user uploads the spectral data and sets the parameters for the analysis also using the head node web interface (Figure 2). When the data upload is complete, the head node formats the data by dividing it into packets and bundles each data packet with a file specifying the parameter to be used in the search. The head node then sends messages informing the worker nodes of the location in S3 of the data packets they are to analyze and collects the messages returned by the worker nodes as to the status of the individual packets.
Figure 1.
Flowchart of ViPDAC Workflow
Figure 2.
UML Case Study Diagram for ViPDAC
To build the predefined AMIs for the ViPDAC, we have used the Ubuntu public AMI available from Amazon as a base. On to this pre-existing AMI, we have installed both OMSSA and X!Tandem programs and their dependencies. In addition, we have also included the current versions of a selection of the most commonly used proteomics databases from IPI, UniProt and SGD18-20. Additional custom databases can be installed by the user through the head node website. These databases are kept in the users S3 area and secure and not publicly accessible. A collection of programs that divide the data into chunks and package the data with an appropriate parameter file into zip files is also included on the AMI as well as software to aggregate and filter the results14.
Zip files containing the data are uploaded to S3 by the web interface running on the head node and messages are placed into a queue informing the worker nodes that packets of data are available to be processed. The worker nodes carry out the database searches using the parameters included in the packet and the databases preinstalled in their AMI. Each of the worker nodes then transfers the results it has produced in a file to the S3 storage system and places a message in the queue informing the head node that the analysis of the packet is complete. Worker nodes continue to process packets of data until the queue is exhausted and the analysis complete. After it has received messages indicating that all the packets have been processed and therefore the analysis is complete, the head node then instructs one of the worker nodes to download the results from the S3 storage system and assemble the results from the multiple packets into a single result file. The resulting files are equivalent to those produced by the standard database analysis program and can be visualized using the standard tools provided by the authors of the search tools. An .ez2 proteomics data results file compatible with the Visualize program, a component of the MCW Proteomics Data Analysis Suite, is also generated. The results can be downloaded from the head node website or through other tools that access the users S3 area.
Using ViPDAC for proteomics analysis
The workflow of the ViPDAC is illustrated in Figure 1 and the UML use case diagram in Figure 2. In the first step in the proteomics data analysis workflow, the user formats the MS2 data to be searched as simple .mgf (Mascot Generic Format) files (Figure 2, step 2). This can be done either from .dta files with a simple Perl script such as merge.pl that is available from the Matrix Science website9, dta_merge_OMSSA.pl that is distributed with OMSSA, the Mascot Distiller application from Matrix Science, or other open source solutions such as the DTASuperCharge program distributed as part of the MSQuant package developed by the Mann laboratory22.
The second step in the data analysis workflow is to instantiate the head node virtual computer. It should be noted that prior to this step the user must have registered for an AWS account which has access to the EC2 and S3 services and received the public and private key that goes along with this account (URL). Using either the Elastic Fox plug-in for the Firefox web browser11 or the Amazon AWS Console the user can the initiate ViPDAC AMI (Figure 2, step 3). The user enters their Amazon account credentials and selects the ViPDAC AMI from their saved AMIs. This launches the head node and starts the web server on the head node. The user can choose from the databases that are preloaded onto the image, as well as uploading custom databases (Figure 2, step 4). Once uploaded, these databases are stored in the user's personal S3 area and will become available for future runs. Similarly, the user can upload datafiles that are also transferred to the users S3 area. The user can then create parameter files to be used with either OMSSA or X!Tandem or use a previously saved parameter set (Figure 2, step 5). The parameters available include modifications to be considered in the search and other search parameters. The user supplies a name for the file, allowing this set of parameters to be used for multiple searches. After the parameter file has been set up, the user can then use the `Jobs' link from the main menu to open the job creation page. Here the user specifies the search engine, data file and parameters file to be used. When the user submits the job, the head node then instructs a worker node to package the data into chunks of 200 spectra with the parameter files. The head node then places the data packages in the queue in S3 and sends messages directing the worker nodes to process these packets. Since each packet contains both the data and the parameters files, the individual worker nodes can search the data in the packet independently (Figure 2, step 6). When the worker node completes the search task for its packet, it creates and saves a file containing the results to S3. Since this activity is usually transparent to the user, a job-monitoring screen is used to follow the progress of the search. Data packets that are shown in red are pending, packets that are shown in yellow are active, and packets that are shown in green are complete, allowing the user to monitor the job in real time. When all packets are complete and all data packets have been searched, the head node then sends a message for a worker node to integrate the results of all of the individual searches. The system has been designed such that if the results file for an individual data packet is not returned by one of the worker nodes, the data packet is placed back in the queue and will be analyzed be a different node. This allows the system to be both fault tolerant and self healing so that even if individual nodes fail or are taken offline, the analysis of the data will still be completed. One of the worker nodes takes up the task of combining the data from the individual results file and also runs a script that filters and annotates the results to produce an .ez2 file. At the completion of the run, a web page displaying the statistics for the run as well as a download link for the results is displayed (Figure 2, step 6). The results files remain on the user's S3 account and can also be downloaded from there even after the head node has been terminated.
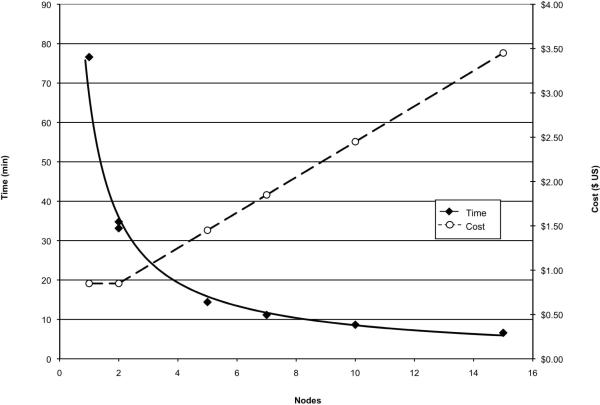
To compare the speed of an OMSSA search using ViPDAC to an identical search done on a typical desktop computer, we searched a LC-MS/MS analysis of the USP1 human protein reference sample (Sigma) performed on a ThermoFisher LTQ mass spectrometer. The single run comprising 22,385 spectra was searched against the current UniProt Human database (release 14.3) containing 39,514 proteins. Searching on both the desktop and ViPDAC cluster implementation of OMSSA both identified the same set of 98 proteins, including the 48 expected proteins plus known contaminants and `bonus' proteins15. The time to run the OMSSA search on a physical computer (Intel Core 2 Duo, 2.167 GHz, 3 GB RAM and 64-bit architecture) was 70 minutes. For comparison, we performed OMSSA searches with the same spectral data, sequence database and parameters on a variety of ViPDAC configurations (Figure 3). In all of the configurations used, all of the nodes were `High-CPU Medium Instances' equivalent to 1.7 GB of memory, 5 EC2 Compute Units (2 virtual cores with 2.5 EC2 Compute Units each), 350 GB of instance storage, and a 32-bit architecture. In the case of the 1 node `cluster', the single node performed the functions of both the head node and the worker node. The single node cluster took 76 minutes to complete the analysis, slightly slower than did the physical computer. This is expected since there is additional time to split and combine the data that is incurred by the virtual computer that is not incurred by the physical computer. Adding a second worker node to the cluster yields a significant reduction in analysis time to 34 minutes. Adding additional nodes reduces the total analysis time, but the benefits decrease with each additional node until the total time asymptotically approaches the fixed times for the data transfer and manipulation at the beginning and end of the run (Figure 3). For comparison, an equivalent run using X!Tandem took 14.2 minutes on a desktop computer and 11.65 minutes using ViPDAC with a single node.
Figure 3.
Time and Cost of Different ViDAC Configurations
Wall clock time to complete the analysis is indicated in filled diamonds and solid lines. Cost of analysis is indicated by open circles and dashed lines. The number of nodes is the total number of nodes functioning as both the head node and as worker nodes. Data from two independent runs for a two node clusters is shown. Costs are in US dollars as of 11/1/2008.
The cost of running the ViPDAC search with a single worker node is shown in Table 1. The primary cost of the search is the hourly charge for each of the nodes ($0.40) and the cost to transfer the data into and out of the cloud ($0.27). The charges are billed in terms of the number of fixed units, i.e., terabytes of data transferred, used over a period of a month so that performing multiple searches in a given month will be more cost efficient than the single search presented here which only uses a small fraction of some units. For this example, we have assumed that the analysis was the only activity for the billing period of one month. Since only a small fraction of the billable units for data transfer and storage were used in a single run, performing additional runs within the billing period will reduce the cost per run to closer to $0.50 US per run. To put the cost to analyze a single run by ViPDAC in perspective with respect to the cost of the experiment, it is less than 0.5% of the typical fee charged by most core proteomics laboratories to perform a single LC-MS/MS run or roughly equivalent to a handful of disposable microfuge tubes or pipette tips. Since the major contributor to the cost of a ViPDAC run is the number of node-hours used, for runs that are completed within the 1-hour minimum billing unit, the cost is linear with the number of nodes. As shown in Figure 3, completing the run in less time sometimes offsets the cost of using more nodes. In this example, the cost of the analysis using a single node or a dual node cluster is the same since the dual node cluster completes the analysis within one hour and the single node cluster uses more than one hour and therefore is billed for two node-hours.
Table 1.
Analysis Cost Breakdown Using a Single Worker Node
| Charge | Amount Used | Unit Size | Units | Cost/Unit | Cost |
|---|---|---|---|---|---|
| EC2 | |||||
| EC2 - Data Transfer In | 156 MB | 1 GB | 1 | $0.10 | $0.10 |
| EC2 - Data Transfer Out | 3.3 MB | 1 GB | 1 | $0.17 | $0.17 |
| High CPU Instance (Medium) | 2 instance-hr | 1 instance-hr | 2 | $0.20 | $0.40 |
| S3 | |||||
| Request - Tier 1 | 227 | 1,000 | 1 | $0.01 | $0.01 |
| Request - Tier 2 | 394 | 10,000 | 1 | $0.01 | $0.01 |
| C3 Data Transfer In | 191 MB | No charge | |||
| C3 Data Transfer Out | 798 MB | No charge | |||
| Storage | 36.6 MB | 1 GB | 1 | $0.15 | $0.15 |
| Total | $0.84 |
Amount for requests is the number of message and as indicated for data and computation.
Unit is the metric that Amazon uses to assess charges. Charges are assessed for any partial unit usage.
For users with simple analysis needs and a limited number of runs to analyze, a typical workstation in the laboratory may be sufficient if short turnaround times are not required. As the number of runs and the complexity of the analysis increases, the benefits of using a computational cluster become apparent. As an example of a `case-study' in which increased computational effort provides greater biological insight, we examined a sample of urine from a rat that had been exposed to 10 Gy of ionizing radiation directed to the kidney. Since from previous work examining the effects of total body irradiation showed that there were changes in proteases present in the urine16, we performed an unconstrained or `no enzyme' search so that cleavages other than those catalyzed by trypsin could also be observed. Running a search of a 1,000 spectra portion of the 81,000 spectra in the file using OMSSA on a typical desktop computer took 2.1 hours. In contrast, running the identical search using ViPDAC with a single node cluster took 1.6 hours and the entire dataset took 6.8 hours with a 20-node ViPDAC cluster. For the entire dataset, we estimate it would take approximately 170 hours to run the analysis on a desktop computer compared to the 6.8 hours it took using ViPDAC. Considering that this single sample is only one of the approximately 20 biological and technical replicates in the study, the total computation time for the study using the desktop computer would be estimated to be 140 days as compared to 5.7 days using a 20 node ViPDAC. Adding more protein modifications to the search would make the search slower and the difference between a desktop computer and ViPDAC even greater.
There are several potential considerations in implementing proteomics data analysis using virtual clusters. One concern is availability of the Amazon EC2 resource. Historically, Amazon has maintained a very high level of service continuity. When there was a failure of AWS in February 2008, it lasted only a few hours17 and this did cause a significant impact to many businesses that depend on the AWS service as a central part of their business model. Since ViPDAC is built to be `self healing', this unlikely event would only cause the automatic resubmission of the data chunks that were being processed at the time of the failure when ViPDAC is restarted so that no data and very little analysis time would be lost. Amazon has just moved EC2 out of `beta testing' and now provides an optional service level agreement, guaranteeing 99.95% availability, other vendors of distributed computing resources also offer service level agreements guaranteeing minimum levels of access to their systems18. As a further backup the laboratory could also keep a computer configured to run analyses locally in the unlikely event of a extended AWS service outage. Another concern with cloud computing is data security and safety. Data placed in S3 is protected by secure access keys and can be kept private unless it is made publicly available by the user12. Since the data stored in S3 is replicated at multiple Amazon sites, loss of data is not expected and many businesses choose to deposit data in S3 for this reason18 but backing up important data to a local server is still prudent. Also, unlike submitting data for analysis to public websites such as the Matrix Science19 or GPM sites20, with ViPDAC the user's data is never out of the control of the user and is not added to public repositories and strict data confidentiality is maintained. Furthermore, there are no limitations on the size or number of searches that can be performed using ViPDAC.
The ViPDAC method for proteomics data analysis has several key advantages over acquiring and maintaining a local computer cluster to carry out proteomics data analysis. First, there is no initial cost or investment in computational infrastructure and no ongoing cost associated with its maintenance and upgrades. This also avoids the need to find appropriate facilities to house the cluster that meets its needs for power, cooling and network connectivity as well as technical personnel to administer it. Second, it avoids both initial cost of licensing software and the time and effort required to install and maintain it. Use of the Amazon Web Services for proteomics data analysis also allows the exact cost of the analysis to be determined and assigned to an individual project. In comparison, it is often very difficult to calculate costs for the analysis of a particular run based on local infrastructure and personnel costs. Third, the ViPDAC AMI provides a standardized platform that can be used by multiple laboratories making it easer to integrate and compare search results. Fourth, it provides data security in that all analyses are done using the user's own secure account credentials and results are stored in the user's own S3 search space, though it is unclear if this would be appropriate for the storage of protected patient information without additional encryption. Lastly, since it is also possible for a user to save a copy of the ViPDAC AMI in their own S3 storage space, the ViPDAC AMI can be further customized to meet specialized needs or to add additional programs. These customized AMIs could then be either kept private or shared with other users through the Amazon community AMI list.
Cloud computing is clearly an emerging area of great interest in the computing industry and beyond and ViPDAC illustrates its applicability to the many computationally intensive applications found in bioinformatics. The importance of freely available, open-source algorithms that can be deployed on these virtual clusters also cannot be overstated. These approaches will put significant computational resources within the reach of any proteomics researcher. Our hope is that this will accelerate their research and potentially enable new analyses previously thought too computationally intensive to consider. We welcome any comments and suggestions via our website (http://proteomucs.mcw.edu/vipdac) or the public AMI discussion forums available on Amazon.
ACKNOWLEDGMENT
This work was supported by the NHLBI Proteomics Center contract NIH-N01 HV-28182 to ASG. We would also like to thank Drs. Shama Mirza and Mukut Shama for graciously allowing us to use their datasets to demonstrate ViPDAC.
REFERENCES
- 1.Deutsch EW, Lam H, Aebersold R. Data analysis and bioinformatics tools for tandem mass spectrometry in proteomics. Physiol Genomics. 2008;33(1):18–25. doi: 10.1152/physiolgenomics.00298.2007. [DOI] [PubMed] [Google Scholar]
- 2.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3(5):958–64. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 3.Geer LY. http://pubchem.ncbi.nlm.nih.gov/omssa/browser.htm
- 4.The G. P. M. Organization TANDEM project. http://www.thegpm.org/TANDEM/instructions.html
- 5.Aaron W. Computing in the clouds. netWorker. 2007;11(4):16–25. [Google Scholar]
- 6.Amazon Amazon Elastic Compute Cloud (Amazon EC2) http://aws.amazon.com/ec2/
- 7.Amazon, Amazon Web Services Developer Community Amazon Machine Images (AMIs) 2008
- 8.Pellitteri-Hahn MC, Warren MC, Didier DN, Winkler EL, Mirza SP, Greene AS, Olivier M. Improved mass spectrometric proteomic profiling of the secretome of rat vascular endothelial cells. J Proteome Res. 2006;5(10):2861–4. doi: 10.1021/pr060287k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matrix Science Finnigan Xcalibur. http://www.matrixscience.com/help/instruments_xcalibur.html
- 10.Mozilla Firefox Release Notes. http://en-us.www.mozilla.com/en-US/firefox/3.0.3/releasenotes/
- 11.Amazon Elasticfox Firefox Extension for Amazon EC2. http://developer.amazonwebservices.com/connect/entry.jspa?externalID=609
- 12.Amazon Amazon Simple Storage Service (Amazon S3) http://aws.amazon.com/s3/
- 13.Wolters DA, Washburn MP, Yates JR. An automated multidimensional protein identification technology for shotgun proteomics. Analytical Chemistry. 2001;73(23):5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 14.Medical College of Wisconsin Virtual Proteomics Data Analysis Cluster. http://github.com/mcwbbc/vipdac/tree/master
- 15.ABRF ABRF2007 BIC Final Correct Proteins_List. www.abrf.org/ResearchGroups/ProteomicsInformaticsResearchGroup/Studies/ABRF2007_BIC_Final_Correct_Proteins_List.xls
- 16.Sharma M, Halligan BD, Wakim BT, Savin VJ, Cohen EP, Moulder JE. The urine proteome as a biomarker of radiation injury. Proteomics - Clinical Applications. 2008;2(78):1065–1086. doi: 10.1002/prca.200780153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins J. Amazon S3/EC2/AWS outage this morning.. http://radar.oreilly.com/2008/02/amazon-s3-ec2-aws-outage-this.html
- 18.Hess K. Cloud Computing: Resistance is Pointless. http://www.linux-mag.com/id/7162
- 19.Matrix Science Inc Mascot. http://www.matrixscience.com/search_form_select.html
- 20.The GPM http://www.thegpm.org/