Abstract
The hy1 mutants of Arabidopsis thaliana fail to make the phytochrome-chromophore phytochromobilin and therefore are deficient in a wide range of phytochrome-mediated responses. Because this defect can be rescued by feeding seedlings biliverdin IXα, it is likely that the mutations affect an enzyme that converts heme to this phytochromobilin intermediate. By a combination of positional cloning and candidate-gene isolation, we have identified the HY1 gene and found it to be related to cyanobacterial, algal, and animal heme oxygenases. Three independent alleles of hy1 contain DNA lesions within the HY1 coding region, and a genomic sequence spanning the HY1 locus complements the hy1–1 mutation. HY1 is a member of a gene family and is expressed in a variety of A. thaliana tissues. Based on its homology, we propose that HY1 encodes a higher-plant heme oxygenase, designated AtHO1, responsible for catalyzing the reaction that opens the tetrapyrrole ring of heme to generate biliverdin IXα.
A complex network of receptors and signal-transduction pathways mediates plant responses to changing light environments. The best-characterized component of this network is the phytochrome (phy) family of photoreceptors that regulates plant morphogenesis and growth from seed germination and deetiolation to flowering and senescence (1–3). phys are homodimeric chromoproteins; each subunit is composed of the linear tetrapyrrole chromophore (3E)-phytochromobilin (PΦB) covalently bound to a ≈120-kDa polypeptide (4). PΦB is linked via a thiol-ether bond to a specific Cys in the apoprotein, occurring autocatalytically by using a lyase activity residing within the Phy polypeptide (5). This bilin–protein complex can exist as one of two spectrally distinct forms, a red light-absorbing form (Pr) and a far-red light-absorbing form (Pfr), that are interconvertible by the absorption of red light (R) and far-red light (FR), respectively (2). In higher plants, the Phy apoproteins are encoded by a small gene family, with the holoprotein derived from each isoform having both distinct and overlapping functions in light perception (1, 3, 6).
Ultimately, holo-phy assembly requires the convergence of two pathways, one that generates the various Phy polypeptides and another that produces PΦB. As a consequence, it has been possible to genetically uncouple these pathways from each other. Whereas mutations within the genes encoding the various apoproteins affect responses controlled by the corresponding phy isoforms, mutations in genes required for PΦB synthesis globally attenuate all phy responses. For example, Arabidopsis thaliana mutants in four of five Phy-apoprotein loci, phyA, phyB, phyD, and phyE, have been described and shown to affect the photomorphogenic responses specifically controlled by that isoform (7–10). In contrast, mutations in A. thaliana, tomato, tobacco, and pea that attenuate PΦB synthesis behave as phy-deficient plants (11–15).
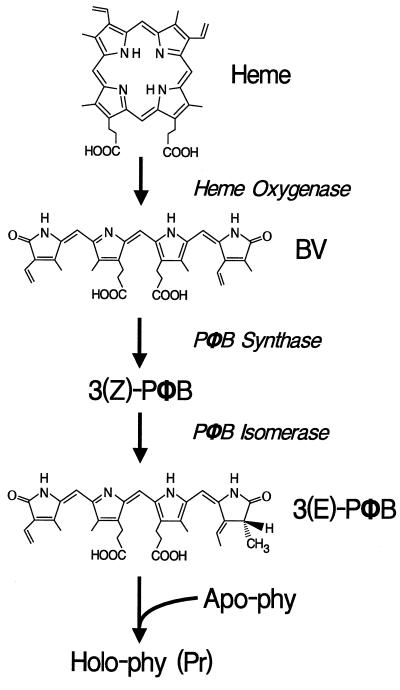
Based on metabolic studies in algae and the biochemical analysis of pea and tomato PΦB-deficient mutants, it has been proposed that PΦB is produced in the plastid from 5-aminolevulinic acid in a pathway that branches off from that used to synthesize chlorophyll (Fig. 1 and refs. 4 and 16). The first unique reaction in the PΦB pathway is the oxidative conversion of heme to biliverdin IXα (BV), presumably by a heme oxygenase (HO). The PΦB-deficient mutants of pea (pcd1) and tomato (yg-2) appear to be blocked at this step (15, 17). Next, BV is reduced by a PΦB-synthase to create 3(Z)-PΦB, and, finally, 3(Z)-PΦB is isomerized to form 3(E)-PΦB, which is then attached to the apoprotein (18). The pea pcd2 and tomato au mutants appear deficient in the conversion of BV to 3(Z)-PΦB (17, 19). Mutants that affect the conversion of 3(Z)-PΦB to 3(E)-PΦB have not been identified to date (20), but it is possible that apo-Phy itself potentiates this isomerization before coupling the chromophore.
Figure 1.
Proposed pathway for phytochrome-chromophore biosynthesis in higher plants (adapted from refs. 15 and 19).
In A. thaliana, the long-hypocotyl mutants hy1 and hy2 are unable to synthesize PΦB (11, 20). They are deficient in a wide range of phy-mediated responses (for examples, see refs. 21 and 22), including deetiolation under FR (a phyA-specific response) and R (primarily a phyB response) (11, 12). Although spectrally active chromoproteins cannot be detected, etiolated seedlings of hy1 and hy2 contain near-normal immunodetectable levels of the PhyA polypeptide, suggesting that the Phy apoproteins are synthesized but not assembled with PΦB (23). Parks and Quail (24) showed that feeding these mutants BV fully rescues hy1 and partially rescues hy2. Because heme is an essential cofactor, we assume that both hy1 and hy2 are compromised downstream of heme biosynthesis. It has been proposed that hy1 is affected in the oxygenase step and hy2 is affected in the synthase step as heme is converted to 3(Z)-PΦB (Fig. 1 and ref. 20).
Despite our biochemical understanding of the PΦB-biosynthetic pathway in higher plants, none of the enzymes (or their corresponding genes) that catalyze the synthesis of PΦB from heme have been identified to date. Because defects in these proteins/genes affect a broad range of phy responses, their characterization should be instrumental in defining the regulation of holo-phy synthesis and for generating crop plants generally deficient in phy signaling. Such a comprehensive repression of phy action may be advantageous in various agronomic applications by making plants less responsive to light (25, 26). To help define how PΦB is synthesized, we have begun to characterize the A. thaliana HY1 locus. Here, we report the identification of the HY1 gene and demonstrate that it encodes a protein related to HOs that are required in bacteria, algae, and animals for the conversion of heme to BV. Further analysis of this protein should be instrumental in understanding HOs from plants and defining steps that potentially regulate PΦB synthesis.
MATERIALS AND METHODS
Plant Material and Growth Conditions.
A. thaliana ecotypes Columbia (Col) and Landsberg erecta (Ler), hy1-1 in the Ler background, and hy1–100 [also know as hy6 (cs236)] and hy1–2 [also known as hy (cs3163)] in the Col background were obtained from Rick Amasino and Brian Parks (University of Wisconsin-Madison), and the Arabidopsis Biological Resource Center, respectively. For hypocotyl-growth assays, seeds were surface-sterilized and sown on half-strength Murashige and Skoog medium without sucrose or vitamins (MS; GIBCO/BRL), 2.5 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.7), and 8 g/liter agar. Plates were stored in the dark for 2 days at 4°C, placed in darkness at 25°C for 1 day, and then irradiated for 5 days with 22 μmol·m−2⋅sec−1 continuous white light at 25°C (27). Hypocotyl lengths were measured from computer-scanned images of the seedlings.
Isolation of the HY1 Candidates AtHO1 and AtHO2.
Bacteria artificial chromosome (BAC) clones surrounding the ER locus (28) (AtDB, Stanford University; http://genome-www.stanford.edu/Arabidopsis/) were analyzed by the blast computer algorithm (29) for translated sequences related to that of Synechocystis sp. 6803 HO1 (30). AtHO1 and AtHO2 were PCR-amplified from genomic DNA prepared as described (31) from Ler, hy1–1, hy1–100, and hy1–2. The primers for AtHO1 (annotated as F18A8.4), ATTATGAGTATTATTATTTTTAAATCTCTC and TAAGATGCCATACAAGTTGAGGAATATAAG, annealed immediately after the proximal annotated gene (F18A8.3) and just before the distal gene (F18A8.5), respectively. Likewise for AtHO2 (annotated as T9J22.22), the primers TGCCTCTTCAAATACCCTGGCTGTTCCAGC and CTAAAGAGATCAATTATTGTATCTTGTATATTG annealed just after T9J22.21 and just before T9J22.23, respectively. The resulting ≈3.3-kb AtHO1 and ≈4.7-kb AtHO2 PCR products were gel-purified and directly sequenced. An A. thaliana AtHO1 cDNA (80G1T7) was identified in the Arabidopsis Genome Initiative expressed sequence tag (EST) collection and sequenced. The AtHO2 cDNA was PCR-amplified from a cDNA library prepared from flower mRNA (32) with oligonucleotides AGTGAAGGCAGCGTCTATCTTGGTCGTCGG and CTGGTGCCGGAAACTGTTAACTTTAAAACC, which anneal just outside of the predicted coding region. This PCR product was gel-purified and directly sequenced. Sequence analyses were performed by using the Genetics Computer Group software package (GCG).
hy1–1 Complementation.
Ler genomic DNA encompassing AtHO1 (see above) was PCR-amplified by using Pfu polymerase (Stratagene) and cloned into the EcoRV site of pPZP211 (33), resulting in pPZP211-AtHO1. Ler wild-type and hy1–1 plants were transformed by the floral-dip method (34) with the Agrobacterium tumefaciens strain ABI harboring either pPZP211, pPZP211-AtHO1, or PZP211-smGFP (35). T1 seeds were plated on GM-agar plates [4 g/liter MS salts/2.5 mM 2-(N-morpholino)ethanesulfonic acid/10 g/liter sucrose/6 g/liter (wt/vol) agar, pH 5.7] supplemented with 40 mg/liter kanamycin to select for transformants. After a 2-day treatment at 4°C, seedlings were grown for 14 days in 0.16 μmol·m−2⋅s−1 red light.
RNA Isolation and RNA Gel-Blot Analyses.
Total RNA was isolated by the RNA-Isolator protocol (Genosys, The Woodlands, TX) from either axenically grown 6-day-old seedlings or roots, rosette leaves, stems, and flowers of soil-grown plants. Total RNA (15 μg) was fractionated on 1.5% agarose-formaldehyde gels and transferred and fixed onto nylon membranes (Zeta-Probe; Bio-Rad). Equal loading was checked by ethidium-bromide staining. The membranes were probed sequentially with coding DNA from AtHO1 and β-tubulin (36) labeled with 32P by random priming. The blots were hybridized by using standard techniques (37) and washed at 65°C with 2× SSC (20× SSC = 3 M sodium chloride/0.3 M sodium citrate) and 0.1% SDS, and the signals were visualized by phosphorimage analysis using optiquant software (Packard). Signal intensities obtained with the AtHO1 and β-tubulin probes were expressed as a ratio.
RESULTS
Phenotypic Analysis of hy1 Mutants.
The hy1 mutants were isolated by screening mutagenized seedlings for those with a long hypocotyl when grown under white light (11). The hy1–1 allele was isolated from a fast-neutron mutagenized population of Ler; it has been used widely in biochemical and physiological studies as a mutant “lacking” the activity of all phys (12, 21, 24, 38). Other alleles of hy1 were isolated from ethyl methanesulfonate-mutagenized populations by using similar, long-hypocotyl screens [e.g., hy6 (12) and hy (39)] or using screens designed to identify phy-signaling components [e.g., ted4 (40)].
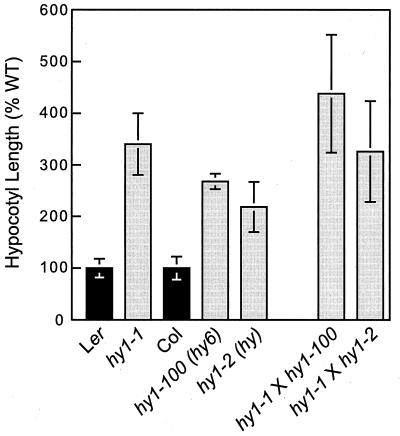
Although originally designated as a distinct locus, hy6 since has been reported to be an allele of hy1 and is now called hy1–100 (ref. 20 and M. Koornneef, personal communication). Similarly, the hy mutant isolated by Rédei (39) also has been reported to be an allele of hy1 (M. Koornneef, personal communication). To confirm these results, we performed allelism tests; hy1–1 was used as the pollen donor to generate crosses with hy6 (hy1–100) and hy. As can be seen in Fig. 2, the F1 progeny from these crosses still exhibited the hy1 defect; these compound heterozygotes had long hypocotyls similar to their homozygous-mutant parents. Based on this observation, we concluded that hy1–100 and hy are indeed alleles of hy1, and we propose that hy be renamed hy1–2. While conducting the allelism tests, we observed that the average hypocotyl length differed between the independently isolated mutants of HY1. The hy1–1 mutant was the most severely affected, followed by hy1–100 and hy1–2 (Fig. 2). Because hy1 mutations are recessive, they are likely loss-of-function alleles, with the most dramatically affected hy1–1 mutant possibly representing the null phenotype.
Figure 2.
Hypocotyl-growth responses of wild-type and hy1 mutants of A. thaliana. Wild-type Ler and Col, the homozygous hy1 mutants [hy1–1, hy1–100, and hy1–2 (formally called hy)], and the compound-heterozygous progeny from crosses of hy1–1 with hy1–2 and hy1–100 were irradiated for 5 days with ≈22 μmol m−2⋅s−1 of continuous white light after germination. Each bar represents the average hypocotyl length (±SD) of at least 10 seedlings. Values are represented as the percentage of the mean of the respective wild-type parent. For the compound heterozygotes containing two wild-type backgrounds, the average of the Ler and Col values was used.
Identification of Two Putative Heme Oxygenase Genes That Map Near the HY1 Locus.
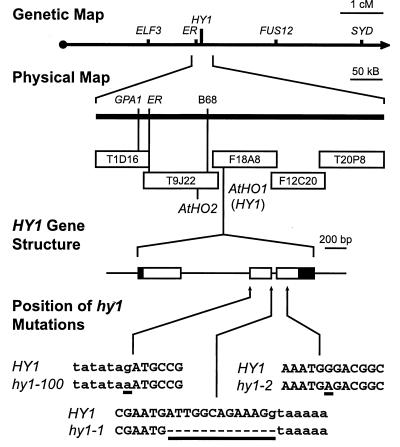
To help define the HY1 locus, its position in the A. thaliana genome was determined. Previous mapping efforts have established that HY1 is located on the right arm of chromosome 2 at 48 cM and is tightly linked to the er mutation (39) (Fig. 3). The ER gene product has been isolated (28), and its sequence has been placed onto the physical map in close association with the recombinant-inbred marker B68 (The Institute for Genomic Research at http://www.tigr.org) (Fig. 3). A large section of chromosome 2 surrounding ER has been sequenced, resulting in a contig that currently spans several hundred kilobases on both the centromeric and telomeric sides of ER (“ER contig”; The Institute for Genomic Research at http://www.tigr.org). Given its large size, we predicted that the HY1 locus would be present within this sequenced region.
Figure 3.
Positional identification of the HY1 locus. The genetic map of chromosome 2 as viewed from the A. thaliana DNA database defined the genetic position of HY1 as linked to ER. Assembly of the physical map of the ER contig by overlapping genomic bacteria artificial chromosome clones identified the exact position of ER and two possible candidate genes for HY1 (AtHO1 and 2) based on its expected homology to Synechocystis HOs. The gene structure of AtHO1 was deduced by comparing the genomic sequence with that of a full-length cDNA. Open and solid boxes denote coding region and untranslated regions, respectively. Lines indicate introns and nontranscribed regions. DNA sequences surrounding the position of the three hy1 mutations are indicated and aligned with the wild-type (HY1) AtHO1 sequence. Underlined nucleotides denote sequence differences between wild type and the mutants. Upper- and lowercase letters represent exon and intron sequences, respectively.
Recently, Cornejo et al. (30) identified an HO protein from Synechocystis sp. 6803 that catalyzes the enzymatic reaction believed to be absent in hy1 backgrounds (20). Using this cyanobacterial HO1 gene as a blast query, we searched the ER contig for putative HO genes from A. thaliana. Two predicted ORFs with significant derived amino acid sequence homology to SynHO1 were identified approximately 40 kbp apart: one was annotated as F18A8.4 and the other was annotated as T9J22.23, and they subsequently are referred to as AtHO1 and AtHO2, respectively (Fig. 3). Several cDNAs related to AtHO1 were present in the EST database, confirming that it is a transcribed region. No ESTs were identified corresponding to the AtHO2 locus, but our subsequent identification of its transcript in a cDNA library confirmed that AtHO2 is expressed (data not shown). Searching the entire A. thaliana database with AtHO1 or AtHO2 failed to detect any additional sequences related to these two genes. Potential orthologs to AtHO1 were evident in the loblolly pine and rice EST collections, indicating that similar proteins likely are present in all higher plants.
Conceptual translation of the longest cDNA corresponding to AtHO1 (80G1T7) revealed a 1,072-bp ORF encoding a protein of 282 aa (≈33 kDa). By comparing the cDNA sequence to that derived from the genome, the organization of the AtHO1 locus was determined. The coding region starts with a perfect match to the consensus Kozak translational initiation sequence (41) (data not shown) and is interrupted by two introns of 655 and 75 bp (Fig. 3). The deduced amino acid sequence contains a predicted chloroplast-transit peptide of 54 aa (42) (Fig. 4). The potential chloroplast location of AtHO1 is consistent with biochemical evidence that higher-plant HO activity is plastid-localized (15, 17). For AtHO2, we determined that the coding region is interrupted by three introns of 77, 782, and 84 bp (data not shown). Interestingly, the first two exon–intron boundaries of AtHO1 and AtHO2 splice within a conserved amino acid sequence, suggesting that these two genes evolved from a common progenitor, possibly by gene duplication. The ORF for AtHO2 encodes a 299-aa protein (≈35 kDa) that also begins with a predicted chloroplast transit sequence (42) (Fig. 4). It is noteworthy that the 42-bp insertion (codons 107–120) that encodes the Glu/Asp repeat in AtHO2 (Fig. 4) is present in the cDNA, indicating that this sequence is translated and not an intron bracketed by nonconsensus splice junctions.
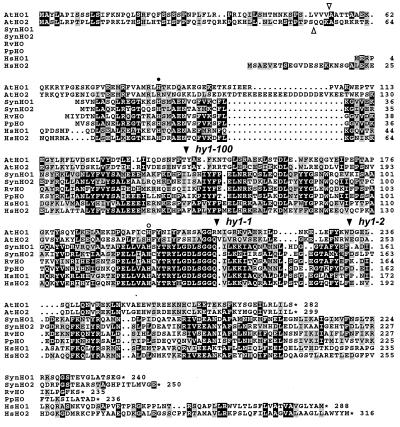
Figure 4.
Amino acid sequence comparison of AtHO1 and AtHO2 with HOs from cyanobacteria, alga, and humans. The alignment was created by using macboxshade 2.7 (Institute for Animal Health, Pirbright Surrey, U.K.). Identical and similar residues are in reverse type or shaded boxes, respectively. The open triangles identify the predicted cleavage sites for the transit peptide of AtHO1 and AtHO2, and the solid triangles denote the sites altered in the hy1–1, hy1–100, and hy1–2 mutants. The closed and open circles mark the His residues important in animal HOs for heme binding and protein structure, respectively. Numbers at the right indicate the respective amino acid residues. Asterisks identify the positions of stop codons. Sequences include Synechocystis (Syn) HO1 (D90091) and HO2 (D90912), Rhodella violacea (Rv) HO1 (AF000717), Porphyra purpurea (Pp) HO (P51271), and human (Hs) HO1 (P09601) and HO2 (P30519).
The predicted AtHO1 and AtHO2 proteins are highly related to each other throughout their entire peptide sequence (48/65% identity/similarity). The proposed transit peptide is conserved but a slightly different processing site is predicted (Fig. 4). Alignments showed that their closest relatives are HOs from cyanobacteria, algae, and animals. The best matches are with two Synechocystis HOs, followed by HOs from the alga Rhodella violacea and Porphyra purpurea, and two HOs from humans (Fig. 4). Although the overall similarity is only 20–30% between AtHO1 and 2 and the related HOs, a number of conserved amino acid clusters can be seen throughout their coding regions. One cluster contains a positionally conserved His (His-198 in AtHO1) shown to be important for structural stability in human HOs (43). A second cluster near the N terminus of human HOs (residues 76–96 in AtHO1) is required for the oxidative cleavage and employs a His to bind the heme substrate (44). This His is present within AtHO1 (His-86), but is notably absent from AtHO2 (Fig. 4). Theoretically, the Lys residue at this position in AtHO2 could substitute for His in this reaction given its similar charge. Animal HOs contain a C-terminal hydrophobic extension that is thought to anchor the enzyme to the microsomal membrane (44). Consistent with their behavior as soluble enzymes in the chloroplast stroma (15, 17), neither AtHO1 nor 2 contains a similar hydrophobic extension (Fig. 4).
Three hy1 Alleles Contain Mutations in AtHO1.
To determine whether one of these AtHO genes is mutated in the hy1 alleles, we PCR-amplified and sequenced the corresponding genomic DNA from Ler wild type and hy1–1. For the regions analyzed here, the AtHO1 gene is identical between the Ler and Col ecotypes except for a single T-to-C transition 321-bp upstream of the initiation codon and a single translationally silent C-to-T change within codon 204 (data not shown). The AtHO2 sequence was identical between Ler and the hy1–1 mutant (data not shown). In contrast, the hy1–1 allele generated by fast-neutron mutagenesis had a 13-bp deletion in AtHO1 (from nucleotides 1291–1303) (Figs. 3 and 4). This deletion created a frameshift and possibly an altered acceptor site at the splice junction of the second intron; if transcribed and translated, a truncated protein would be synthesized missing 71 aa at the C-terminal end (Figs. 3 and 4). Likewise, the AtHO1 gene was sequenced in the ethyl methanesulfonate-generated hy1–100 and hy1–2 backgrounds. hy1–2 contained a G-to-A transition at position 1426, creating a nonsense mutation that would truncate the polypeptide 50 aa from the C terminus (Figs. 3 and 4). hy1–100 contained a G-to-A transition at position 1079. This mutation altered the exon-acceptor site of the first intron, which could create a misspliced mRNA containing a frameshift beginning at codon 141 (Figs. 3 and 4).
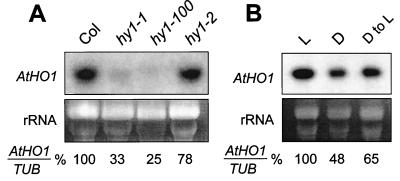
By RNA gel-blot analysis, we observed that two of the three hy1 mutations affected the steady-state levels of the AtHO1 mRNA. As can be seen in Fig. 5A, a ≈1.2-kb transcript was detected in total RNA from wild-type seedlings. This mRNA was at near-normal levels in hy1–2 but was reduced significantly in the hy1-l and hy1–100 lines (as compared with a β-tubulin standard). In none of the three hy1 mutants did we observe any abundant mRNAs with sizes distinct from the wild-type AtHO1 transcript, despite the possibility that the mutations affect mRNA splicing. Based on the observations that all three hy1 alleles contain mutations altering the sequence of the predicted protein and/or processing of the mRNA, we conclude that the HY1 locus encompasses the AtHO1 gene.
Figure 5.
Analysis of AtHO1 mRNA levels in wild-type A. thaliana and the hy1 mutants. (A) RNA gel-blot analysis of total RNA (15 μg) isolated from Col wild-type plants and the hy1 mutants, hy1–1, hy1–100, and hy1–2. (B) AtHO1 mRNA levels in 6-day-old light-grown (L) or dark-grown (D) seedlings, and dark-grown seedlings exposed to continuous white light for 1 day (D to L). (Upper) RNA gel blot probed with an AtHO1 coding-region fragment. (Lower) Ethidium bromide-stained gel before transfer of the RNA onto the membrane. RNA loading was normalized by reprobing the blot with a β-tubulin cDNA (TUB). The ratio of the signals obtained with the AtHO1 and TUB probes was expressed as a percentage of that obtained for wild-type (A) or light-grown (B) plants and is shown below each lane.
The Isolated AtHO1 Gene Complements the hy1–1 Mutation.
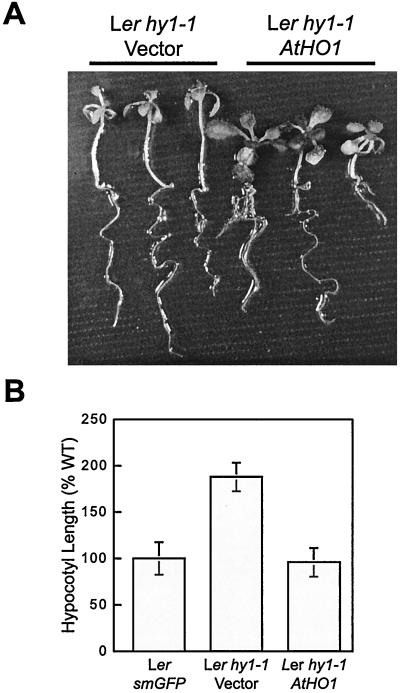
As a final step to establish that the HY1 locus encodes the AtHO1 protein, we tested whether a Ler genomic fragment containing the entire AtHO1 gene could complement hy1–1. This fragment, containing the entire AtHO1 coding region, 1.1 kb of 5′ flanking, and 0.6 kb of 3′ flanking sequence was PCR-amplified and transformed into the Ler ecotype homozygous for hy1–1. The seedlings then were grown on kanamycin-containing medium, and the phenotype of antibiotic-resistant plants was compared with that of hy1–1 plants transformed with an empty vector or with that of Ler plants transformed with a nonrelated gene [smGFP (35)]. As can be seen in Fig. 6, progeny seedlings from hy1–1 plants transformed with an empty vector were phenotypically similar to the hy1–1 parent displaying elongated hypocotyls, a reduced growth rate, and pale green leaves when grown in the light. In contrast, hy1–1 plants transformed with a vector harboring the AtHO1 gene exhibited wild-type photomorphogenesis, comparable to that of Ler plants expressing smGFP (Fig. 6 and data not shown). Using PCR primers that could detect the 13-bp sequence missing in the hy1–1 mutant as compared with the wild-type HY1 locus, we confirmed that the phenotypically rescued plants contain copies of both the wild-type and mutant versions of the AtHO1 gene (data not shown).
Figure 6.
Transgenic complementation of the hy1 mutant phenotype. (A) Ler plants homozygous for the hy1–1 mutation were transformed with an empty vector or one containing a genomic fragment encompassing the entire AtHO1 gene. T1 seeds were germinated on media containing kanamycin and grown for 14 days under continuous red light. Independently transformed antibiotic-resistant plants representative of hy1–1 harboring the empty vector or the AtHO1 gene are shown from the left to right, respectively. (B) The average hypocotyl length of hy1–1 seedlings transformed with the empty vector or one containing AtHO1 and the wild-type Ler parent transformed with a vector containing an unrelated protein (smGFP) (35). Each bar represents the average hypocotyl length (±SD) of at least 10 seedlings.
AtHO1 Is Expressed Throughout the Plant.
The AtHO1 mRNA was readily detected by RNA gel-blot analysis in all A. thaliana tissues examined, including roots, rosettes, stems, and flowers (data not shown). It was present in 6-day-old etiolated seedlings (Fig. 5B) but was reduced as compared with light-grown plants, suggesting that the AtHO1 gene is modestly light-regulated. When etiolated plants were exposed to continuous white light for 1 day, we observed a slight increase in the steady-state level of the AtHO1 mRNA (Fig. 5B).
DISCUSSION
The hy1 mutants have been analyzed in considerable detail at both the biochemical and phenotypic levels and shown to be reduced in the activities of all phys as a result of a defect in PΦB synthesis (11, 20, 24). Using available genomic and physical maps surrounding the HY1 locus and candidate-gene isolation based on the expected similarity of the HY1 protein to HOs (30), we identified the encoded protein (designated AtHO1) and show that its sequence is related to bacterial, algal, and animal HOs. Proof that the AtHO1 gene is mutated in the hy1 backgrounds was provided by results demonstrating that (i) three independent alleles of hy1 all bear nucleotide substitutions/deletions within the transcribed region of AtHO1, (ii) two of these three mutations affect the abundance of the AtHO1 mRNA, and (iii) transformation of hy1–1 plants with a wild-type genomic fragment of AtHO1 rescued the mutant phenotype. The predicted presence of a chloroplast transit sequence in AtHO1 is also consistent with the expected location of the plant enzyme that produces BV from heme (4). Our sequence data on the three hy1 alleles (hy1–1, hy1–100, and hy1–2) predict that these mutations either affect mRNA splicing or introduce nonsense codons. These defects could direct the synthesis of truncated AtHO1 proteins missing up to 145 aa from the C terminus of the 228-residue mature protein (minus the transit sequence).
Given its homology to HOs, we propose that AtHO1 is responsible for the oxidation reaction that opens the ring of heme to generate BV, the precursor of PΦB (Fig. 1). This activity would explain why hy1 plants exhibit near-complete rescue of photomorphogenesis when fed BV (24). A majority of the phenotypes in hy1 plants should be caused by a block in holo-phy assembly, thus creating phy-deficient plants. However, loss of AtHO1 activity also could alter heme homeostasis, which, in turn, could induce part of the inferred “phy-deficient” phenotypes by altering the levels of heme, heme intermediates, and/or heme products (such as chlorophyll), as has been suggested for the tomato PΦB-deficient mutants (45). RNA gel-blot analysis shows that the AtHO1 mRNA is present in most, if not all, tissues. Its levels are not dramatically light-regulated (Fig. 5), consistent with the near-constitutive synthesis of all A. thaliana phys, except phyA (1, 47, 48).
Certainly, the phenotype of hy1 indicates that AtHO1 plays a major role in PΦB synthesis in etiolated and developing seedlings. However, as hy1 and other mutant seedlings compromised in PΦB synthesis mature, most phy-deficient phenotypes become less pronounced (20). In tomato, for example, adult plants of the yg-2 mutants actually return to a near-normal phenotype and regain many of their phy-regulated photoresponses (20, 45). This recovery suggests that another source of BV becomes available that can supply adequate levels of PΦB for assembling active phys. One possibility is that chlorophyll breakdown generates PΦB-related compounds by a side reaction in tetrapyrrole metabolism. Another intriguing possibility is that a second HO protein assumes this responsibility later in plant development. One likely candidate in A. thaliana is AtHO2, which, based on its sequence similarity to AtHO1, is expected to perform the same enzymatic reaction(s) and is also likely to be chloroplast-localized. The presence of AtHO2 and possibly other A. thaliana paralogs may explain why hy1 mutants are not lethal and retain some, albeit diminished, phy responses (11, 12, 21, 22). Studies are now underway to examine the function of AtHO2 in this regard.
Although it is likely that AtHO1 is an HO, we have not yet shown that the protein catalyzes this reaction in vitro. In preliminary studies using a full-length recombinant AtHO1 protein (including the predicted transit sequence) expressed from E. coli (S.J.D., S. Beale, and R.D.V., unpublished data), both crude extracts and purified AtHO1 failed to generate meso-BV from meso-heme by a standard in vitro assay (30). This was not completely surprising given the difficulties that have been reported by others in their attempts to generate active human and Synechocystis HOs by recombinant methods (30, 44). Moreover, it is possible that plant HOs function as a complex with other factors in synthesizing PΦB and may be unstable or inactive in their absence (20).
Animal HOs play an important role in the degradation of heme to protect cells from oxidative damage and to replenish iron pools (48). This catabolic role is in contrast to the proposed biosynthetic function of cyanobacterial and algal HOs (30), which are thought to be important in generating the phycocyanobilin and phycoerythrobilin accessory pigments of the photosynthetic apparatus, and the chromophore for the lower-plant phys, likely phycocyanobilin (30). In higher plants and cyanobacteria, we cannot exclude the possibility that HOs have a dual function, one that generates linear tetrapyrroles and another that recycles heme and chlorophylls. This catabolic function could be especially important for recycling chlorophyll from damaged photoreaction centers and during chloroplast senescence.
The discovery that HY1 encodes a heme oxygenase has revealed the mechanism whereby this locus participates in phy-chromophore biosynthesis. Further characterization of AtHO1 and 2 should help understand how heme levels in general are controlled within the plant and help determine whether PΦB synthesis is regulated to coordinate its production with that of apo-Phy synthesis. With the knowledge that HOs are encoded by a gene family in A. thaliana, it now may be possible to generate a true phy-null plant by reverse genetic strategies. The availability of the AtHO genes also should be instrumental in defining the synthetic defects in the hy1-like mutants from pea (pcd1) (15), tomato (yg-2) (17), and tobacco (pew1) (14) by standard molecular approaches.
Acknowledgments
We thank Dr. Richard Amasino for supplying the Col and Ler ecotypes, Dr. Brian Parks for providing the hy1–1 seed, the Arabidopsis Biological Resource Center for providing the hy1–100 and hy1–2 (hy) seeds, the Arabidopsis thaliana DNA database and The Institute for Genomic Research for generating the genetic and physical maps described here, Drs. Juan Cornejo and Samuel Beale for their assistance with in vitro heme-oxygenase assays, and Dr. Maarten Koornneef for his helpful advise in locating multiple hy1 alleles. This work was supported by a National Institutes of Health predoctoral fellowship to S.J.D. (5 T32 GM07133) and a Department of Energy grant (DE-FG02–88ER13968) to R.D.V.
ABBREVIATIONS
- phy
phytochrome
- PΦB
3(E)-phytochromobilin
- BV
biliverdin IXα
- HO
heme oxygenase
- EST
expressed sequence tag
Note Added in Proof
Muramoto et al. (49) have independently reported the cloning of the HY1 locus from A. thaliana. Like our work, theirs shows that the locus encodes the heme oxygenase, designated here as AtHO1.
Footnotes
References
- 1.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 2.Kendrick R E, Kronenberg G H M. Photomorphogenesis in Plants. 2nd Ed. Dordrecht, The Netherlands: Kluwer; 1994. [Google Scholar]
- 3.Quail P H. Philos Trans R Soc London B Biol Sci. 1998;353:1399–1403. doi: 10.1098/rstb.1998.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terry M J, Wahleithner J A, Lagarias J C. Arch Biochem Biophys. 1993;306:1–15. doi: 10.1006/abbi.1993.1473. [DOI] [PubMed] [Google Scholar]
- 5.Lagarias J C, Lagarias D M. Proc Natl Acad Sci USA. 1989;86:5778–5780. doi: 10.1073/pnas.86.15.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitelam G C, Patel S, Devlin P F. Philos Trans R Soc London B Biol Sci. 1998;353:1445–1453. doi: 10.1098/rstb.1998.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parks B M, Quail P H. Plant Cell. 1993;5:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somers D E, Sharrock R A, Tepperman J M, Quail P H. Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aukerman M J, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino R M, Sharrock R A. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devlin P F, Patel S R, Whitelam G C. Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koornneef M, Rolff E, Spruit C J P. Z Pflanzenphysiologie. 1980;100:147–160. [Google Scholar]
- 12.Chory J, Peto C A, Ashbaugh M, Saganich R, Pratt L H, Ausubel F M. Plant Cell. 1989;1:867–880. doi: 10.1105/tpc.1.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López-Juez E, Nagatani A, Buurmeijer W F, Peters J L, Furuya M, Kendrick R E, Wesselius J C. J Photochem Photobiol. 1990;4:391–405. [Google Scholar]
- 14.Kraepiel Y, Jullien M, Cordonnier-Pratt M M, Pratt L. Mol Gen Genet. 1994;242:559–565. doi: 10.1007/BF00285279. [DOI] [PubMed] [Google Scholar]
- 15.Weller J L, Terry M J, Rameau C, Reid J B, Kendrick R E. Plant Cell. 1996;8:55–67. doi: 10.1105/tpc.8.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elich T D, Lagarias J C. Plant Physiol. 1987;84:304–310. doi: 10.1104/pp.84.2.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terry M J. J Biol Chem. 1996;271:21681–21686. doi: 10.1074/jbc.271.35.21681. [DOI] [PubMed] [Google Scholar]
- 18.Terry M J, McDowell M D, Lagarias J C. J Biol Chem. 1995;270:11111–11119. doi: 10.1074/jbc.270.19.11111. [DOI] [PubMed] [Google Scholar]
- 19.Weller J L, Terry M J, Reid J B, Kendrick R E. Plant J. 1997;11:1177–1186. [Google Scholar]
- 20.Terry M J. Plant Cell Environ. 1997;20:740–745. [Google Scholar]
- 21.Kim B C, Tennessen D J, Last R L. Plant J. 1998;15:667–674. doi: 10.1046/j.1365-313x.1998.00246.x. [DOI] [PubMed] [Google Scholar]
- 22.Liscum E, Briggs W R. Plant Physiol. 1996;112:291–296. doi: 10.1104/pp.112.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parks B M, Shanklin J, Koornneef M, Kendrick R E, Quail P H. Plant Mol Biol. 1989;12:425–437. doi: 10.1007/BF00017582. [DOI] [PubMed] [Google Scholar]
- 24.Parks B M, Quail P H. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagarias D M, Crepeau M W, Maines M D, Lagarias J C. Plant Cell. 1997;9:675–688. doi: 10.1105/tpc.9.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robson P R H, McCormac A C, Irvine A S, Smith H. Nat Biotech. 1996;14:995–998. doi: 10.1038/nbt0896-995. [DOI] [PubMed] [Google Scholar]
- 27.Jordan E T, Cherry J R, Walker J M, Vierstra R D. Plant J. 1996;9:243–257. doi: 10.1046/j.1365-313x.1996.09020243.x. [DOI] [PubMed] [Google Scholar]
- 28.Torii K U, Mitsukawa N, Oosumi T, Matsuura Y, Yokayama R, Whittier R F, Komeda Y. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornejo J, Willows R D, Beale S I. Plant J. 1998;15:99–107. doi: 10.1046/j.1365-313x.1998.00186.x. [DOI] [PubMed] [Google Scholar]
- 31.Cone K C, Frisch E B, Phillips T E. Maize Genetics Cooperation News Letter. 1989;63:68. [Google Scholar]
- 32.Weigel D, Alvarez J, Smyth D R, Yanofsky M F, Meyerowitz E M. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- 33.Hajdukiewicz P, Svab Z, Maliga P. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 34.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 35.Davis S J, Vierstra R D. Plant Mol Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- 36.Callis J, Carpenter T, Sun C-W, Vierstra R D. Genetics. 1995;139:921–939. doi: 10.1093/genetics/139.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 38.Okamuro J K, Szeto W, Lotys-Prass C, Jofuku K D. Plant Cell. 1997;9:37–47. doi: 10.1105/tpc.9.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rédei G P. Genetics. 1965;51:857–872. doi: 10.1093/genetics/51.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pepper A E, Chory J. Genetics. 1997;145:1125–1137. doi: 10.1093/genetics/145.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 42.Emanuelsson, O., Nielsen, H. & von Heijne, G. (1999) Protein Sci., in press. [DOI] [PMC free article] [PubMed]
- 43.Matera K M, Zhou H, Migita C T, Hobert S E, Ishikawa K, Katakura K, Maeshima H, Yoshida T, Ikeda-Saito M. Biochemistry. 1997;36:4909–4915. doi: 10.1021/bi962321m. [DOI] [PubMed] [Google Scholar]
- 44.Ishikawa K, Matera K M, Zhou H, Fujii H, Sato M, Yoshimura T, Ikeda-Saito M, Yoshida T. J Biol Chem. 1998;273:4317–4322. doi: 10.1074/jbc.273.8.4317. [DOI] [PubMed] [Google Scholar]
- 45.Terry M J, Kendrick R E. Plant Physiol. 1999;119:143–152. doi: 10.1104/pp.119.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goosey L, Palecanda L, Sharrock R A. Plant Physiol. 1997;115:959–969. doi: 10.1104/pp.115.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clack T, Mathews S, Sharrock R E. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- 48.Platt J L, Nath K A. Nat Med. 1998;4:1364–1365. doi: 10.1038/3947. [DOI] [PubMed] [Google Scholar]
- 49.Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman H M. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]