Abstract
The majority of existing computational tools rely on sequence homology and/or structural similarity to identify novel microRNA (miRNA) genes. Recently supervised algorithms are utilized to address this problem, taking into account sequence, structure and comparative genomics information. In most of these studies miRNA gene predictions are rarely supported by experimental evidence and prediction accuracy remains uncertain. In this work we present a new computational tool (SSCprofiler) utilizing a probabilistic method based on Profile Hidden Markov Models to predict novel miRNA precursors. Via the simultaneous integration of biological features such as sequence, structure and conservation, SSCprofiler achieves a performance accuracy of 88.95% sensitivity and 84.16% specificity on a large set of human miRNA genes. The trained classifier is used to identify novel miRNA gene candidates located within cancer-associated genomic regions and rank the resulting predictions using expression information from a full genome tiling array. Finally, four of the top scoring predictions are verified experimentally using northern blot analysis. Our work combines both analytical and experimental techniques to show that SSCprofiler is a highly accurate tool which can be used to identify novel miRNA gene candidates in the human genome. SSCprofiler is freely available as a web service at http://www.imbb.forth.gr/SSCprofiler.html.
INTRODUCTION
MicroRNAs (miRNAs) belong to a recently identified group of the large family of noncoding RNAs (1). The mature miRNA is usually 19–27 nt long and is derived from a larger precursor that folds into an imperfect stem-loop structure. The mode of action of the mature miRNA in mammalian systems is dependent on complementary base pairing primarily to the 3′-UTR region of the target mRNA, thereafter causing the inhibition of translation and/or the degradation of the mRNA.
According to recent estimates, while over 30% of vertebrate genomes is transcribed (2), only 1% consists of coding genes, suggesting that the rest must be various types of noncoding RNA genes. In addition, 701 human miRNA hairpin sequences are currently contained in the miRNA registry (miRBase, release 12.0), of which 92% have been experimentally verified, and it is anticipated that there may be thousands more. A recent estimate of the total number of miRNA genes in the human genome provided by the study of Miranda et al. (3) is in the range of ∼55 000, a number significantly larger than the experimentally verified human miRNAs currently in the registry. Searching through the entire genome of human and/or other species for novel miRNA genes is a complicated task for which fast, flexible and reliable identification methods are required. Currently available experimental approaches working towards this goal are complex and sub-optimal (4). Inefficiencies result from various sources, including difficulty in isolating certain miRNAs by cloning due to low expression, stability, tissue specificity and technical difficulties of the cloning procedure while selecting the right genomic region to investigate is often a very challenging task of its own. Computational prediction of miRNA genes from genomic sequences is an alternative technique which offers a much faster, cheaper and effective way of identifying putative miRNA genes. Moreover, by predicting the location of miRNA genes, these methods enable experimentalists to concentrate their efforts on genomic regions more likely to contain novel miRNA genes, thus facilitating the discovery process.
Accurate prediction of new miRNAs requires the consideration of certain characteristic properties of these molecules based on either experimental (5–7), or computational evidence (8–12) which can be used to build a classification scheme or predictive model. These general features include sequence composition, secondary structure and species conservation. MiRNA gene prediction can be achieved via the use of supervised algorithms that are trained on known miRNA biological features and then used to identify putative miRNAs, or un-supervised algorithms such as alignment or conservation. The prediction methodology can also vary significantly between different studies. It can be performed by: scanning for hairpins within sequences that are conserved between closely related organisms like Caenorhabditis elegans and C. briggsae (10,13), looking for regions of homology between known miRNAs and other sites within aligned genomes, as for example between human and mouse (14) or looking for conserved regions of synteny—conserved clustering of miRNAs in the genomes of closely related organisms (14). Profile-based detection (15) and secondary structure alignment (16) of miRNAs have also been suggested using sequences across multiple, highly divergent, organisms (i.e. mouse and fugu). Support vector machines that take into account multiple biological features such as free energy, paired bases, loop length and stem conservation have also been used to predict novel miRNAs (8,9,17). Many of these prediction methods undertake a pipeline approach, whereby cut-offs are assigned and sequences are eliminated as the pipeline proceeds (10,13). The drawback of these approaches is that they lose numerous true miRNAs along the line due to stringent cut-offs. Other approaches use homology to detect novel miRNAs based on their similarity to previously identified miRNAs (14–16). These methods obviously fail when scanning distantly related sequences and when novel miRNAs lack detectable homologs. Two studies (12,18) used Hidden Markov Models (HMMs) and Bayesian classifiers, respectively, to simultaneously consider sequence and structure information for the identification of miRNA precursors (pre-miRNAs). However, conservation information, a very important characteristic of the majority of miRNA precursors, was not integrated in those algorithms. Finally, in a more recent study (19), an HMM approach that simultaneously considered structure and conservation features of miRNA genes was shown to achieve very high performance on identifying miRNAs in the human genome.
In addition to computational tools, large scale, high throughput methods such as tiling arrays or deep sequencing have recently been used for the identification of novel miRNA genes (20–22). These methods are particularly useful as they can provide a very sophisticated and accurate expression map for small RNAs in the genome. Moreover, if such data is coupled to computational tools, it can facilitate rapid and precise detection of novel miRNAs, while at the same time giving greater credence to computational predictions.
MiRNAs have been suggested to play a key regulatory role in numerous processes, including cancer (23,24). For example, the expression levels of let-7 (25), miR-15a/miR-16-1 cluster (26) and neighboring miR-143/miR-145 (27), are found to be reduced in some malignancies, while other miRNAs such as the miR-17-92 cluster (28–30) and miR-155/BIC (31), are overexpressed in various cancers. Additionally it was recently shown that a high percentage of miRNA genes are located in cancer-associated genomic regions (CAGRs), thus implicating miRNAs in tumorigenic events (32). CAGRs take the form of (i) minimal regions of loss of heterozygosity (LOH), suggestive of the presence of tumor suppressor genes; (ii) minimal regions of amplification, suggestive of the presence of oncogenes; and (iii) common breakpoint regions in or near possible oncogenes or tumor suppressor genes. The identification of novel miRNA genes within these regions is very important as it may reveal putative gene players that exert a regulatory effect on different types of cancer, contribute to the better understanding of molecular pathways responsible for oncogenesis and provide potential targets for therapeutic intervention.
In this work, we present an efficient and freely available prediction tool (SSCprofiler) where Profile HMMs are trained to recognize key biological features of miRNAs such as sequence, structure and conservation in order to identify novel miRNA precursors. We first use our method to learn with high accuracy the characteristic features of 249 human miRNA precursors and then apply the trained model on CAGRs in search of novel miRNA genes. Predictions are ranked according to expression information from a recently published full genome tiling array (21) and the top four scoring candidates are verified experimentally using northern blot.
MATERIALS AND METHODS
Datasets
The sequences of human pre-miRNAs used to train and test the HMMs were downloaded from the miRNA registry (version 12.0) (http://microrna.sanger.ac.uk/sequences/). For the training/validation sequences BLASTclust (33) was initially performed to cluster all miRNA sequences into groups by precursor similarity and the most conserved member (according to multiz files) was used to represent the cluster. This procedure was done to eliminate redundant pre-miRNAs and avoid over-representation of similar miRNA precursors. Following a set of filtering criteria detailed below, a total of 249 sequences (originally listed in version 8.0) were used for training/validation while a total of 219 sequences (not in version 8.0) were used as a blind test set. The negative miRNA sequences were derived from 3′-UTR regions of the human genome (release—May 2004) since no true miRNA has yet been reported to reside within these regions. They were generated by using a sliding window of 104 nt, shifted 11 nt at a time, over the 3′-UTR regions. RNAfold was executed for every shift and the free energy of the secondary structure was noted. Only sequences whose energy did not exceed a threshold of –14.44 kcal/mol and had at least 14% of their nucleotides conserved, were selected. This generated over 35 000 negative sequences.
Biological features
SSCprofiler takes into account three different biological features: sequence, structure and conservation of miRNA precursors. In this study, conservation was retrieved from the multiz (34) full genome alignment files of the human May 2004 hg17 genome assembly and seven other vertebrate genomes: Mouse May 2004 (mm5), Rat June 2003 (rn3), Dog July 2004 (canFam1), Chicken February 2004 (galGal2), Fugu August 2002 (fr1), Zebrafish November 2003 (danRer1). Chimp data were not included due to high percentage similarity (∼95%) with humans. RNA secondary structure prediction was performed using the RNAfold function of the Vienna-RNA (35) package. A fixed window (104 nt) was used to align all sequences in order to generate a multiple sequence alignment (msa) required to train the HMM (see Training and Validation of the HMMs). This was achieved by enlarging sequences that fell shorter than this window using flanking genomic nucleotides and trimming sequences that exceeded the defined msa window. The window length was consequently used as the length of the training model and as the window size for querying genomic sequences.
Filtering
To minimize the search space and reduce computational load, the data were first filtered using various secondary structure features of miRNA precursors. Filtering results were displayed as histograms that show the relative distributions of the positive and negative data with respect to eight features:
Hairpin—the number of hairpins
Bulges—the number of bulges
Loops—the number of loops
Asymmetry—difference in loops + bulges on either side of the hairpin.
Bulges-loops—sum of loop and bulge count
Hairpin length—length of the hairpin
Folding min energy—min energy as defined by RNAfold
Conservation—according to multiz full genome alignment files
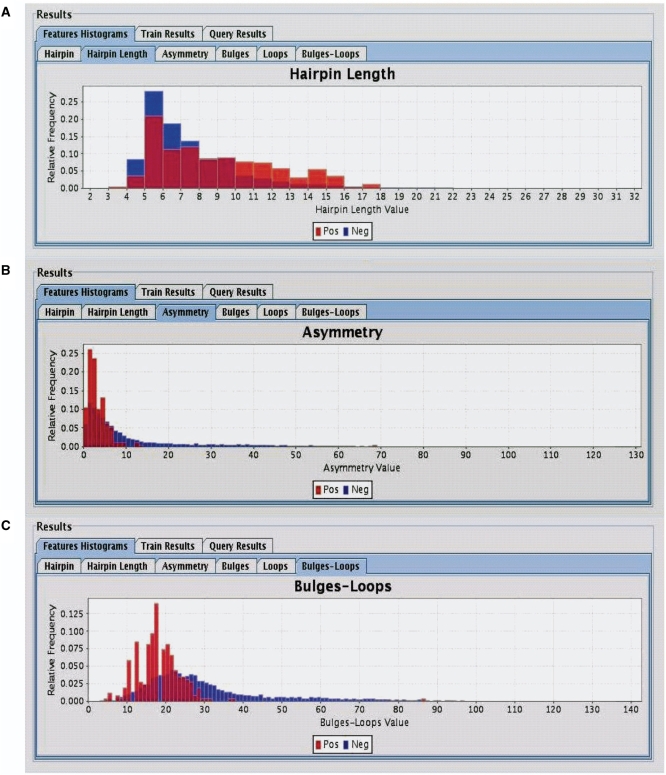
Illustration of data distributions for the various filtering parameters was done to facilitate the filtering process by enabling the adjustment of cut-off values according to the specific dataset. Cut-off values for each of these features are modifiable both prior and after the training procedure (see ‘Results’ section, Figure 4).
Figure 4.
Histograms of the distributions of human miRNA (Red–Positive) and negative sequences (Blue–Negitive), as displayed by SSCprofiler. Only three of the eight filtering parameters are shown here. (A) Hairpin length, (B) Asymmetry and (C) Bulges-loops count. Looking at the distributions of positive and negative data, it is possible for the user to select cut-offs that separate the two distributions which can be used for filtering the data.
Combining sequence, structure and conservation
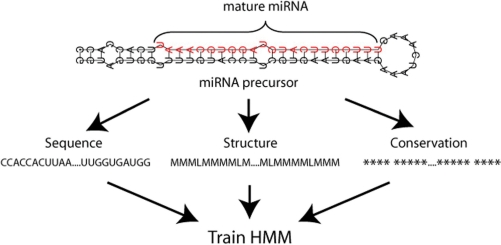
In order to simultaneously consider multiple biological features, a 16 character code was developed that integrates sequence, structure and conservation information for every nucleotide position in a given genomic sequence. Specifically, each position in the genomic sequence is replaced by 1 of 16 letters, depending on three factors: (i) Sequence (A, C, U, G), (ii) Structure, (M = match, L = loop) and (iii) Conservation (* = conserved, ‘‘= not-conserved) as detailed in Table 1.
Table 1.
The 16-letter code that was used to integrate sequence, structure and conservation information
| Conservation and structure | Sequence |
|||
|---|---|---|---|---|
| A | C | G | U | |
| *M | L | M | N | P |
| ‘‘M | C | D | E | F |
| *L | Q | R | S | T |
| ‘‘L | G | I | H | K |
Profile HMMs
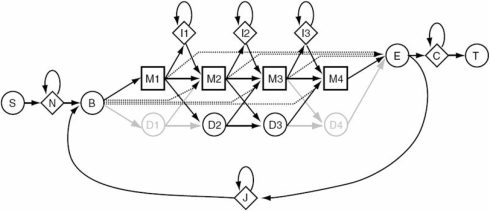
The HMMER (36) software package was used to build a HMM capable of predicting RNA or DNA Profiles. HMMs are generative probabilistic models which are frequently used to address serious theoretical problems. For correct statistical inference, it is necessary to be able to calculate a probability distribution P(S|M) for the probability of sequences S given a model M, and have this quantity sum to one over the ‘space’ of all sequences. Generative models work by recursive enumeration of possible sequences from a finite set of rules—rules that in an HMM are represented by states, state transitions and symbol emission probabilities. HMMER uses a Profile HMM architecture called Plan 7 which is illustrated in Figure 1. Profile HMMs are statistical models of multiple sequence alignments. They capture position-specific information about how conserved each column of the alignment is, and which residues are most likely.
Figure 1.
The HMMER Plan 7 architecture. Squares indicate match states (modeling consensus positions in the alignment). Diamonds indicate insert states (modeling insertions relative to consensus) and special random sequence emitting states. Circles indicate delete states (modeling deletions relative to consensus) and special begin/end states. Arrows indicate state transitions. Figure was adopted from Eddy SR, 1998.
Training and validation of the HMMs
Machine learning algorithms such as HMMs require carefully chosen training and validation data sets in order to achieve maximum performance. SSCprofiler allows for a user-defined partitioning of imported data into training and validation sets in order to perform a boosting validation. This is done by randomly dividing the positive data into K subsets, some of which are used for training and others for validation. The negative data is only used for validation purposes and is not included in the training sets. This partitioning is repeated 100 times and an average validation performance is reported. The training/validation results are displayed as sensitivity and specificity plots in order to obtain an indication of how well the trained HMMs perform on the specific dataset (Supplementary Figure S1). The x-axis of these plots displays the HMM score threshold and the y-axis is the average sensitivity and specificity for every score over the 100 validation runs. Training is performed on the biological feature(s) selected before hand. An overview of the training procedure is shown schematically in the flowchart of Figure 2. At the end of the training/validation procedure all true miRNAs are combined to train a final model which is subsequently used for scanning genomic regions. The HMM score at which average sensitivity and specificity values are ‘optimal’ can be selected by the user and it is used as a cut-off or threshold for classifying sequences as positives (true miRNAs) or negatives.
Figure 2.
The supervised procedure of training HMMs for miRNA precursor identification. Biological features of miRNA biogenesis and conservation across other organisms are used as input for training. Initially, secondary structure prediction is performed using the program RNAfold. Every nucleotide position is henceforth represented by an ‘M’ for match and an ‘L’ for loop. This information is aligned with conservation and sequence information for every nucleotide position. The 16-character code shown in Table 1 is then used to represent each position in this alignment with a single letter. The resulting strings of characters for true miRNAs are aligned with respect to their hairpins and used as a training set for the HMM. Once trained, the HMMs, can be utilized to analyze sequences of desired length and assign a likelihood score. The higher the score the greater the chances of a candidate sequence being a true miRNA precursor.
Assessing the expression level of predicted candidates using tiling array data
To provide additional support for computational predictions, SSCprofiler enables the detection of regions in the candidate(s) that are expressed in HeLa and/or HepG2 cells according to the recently published full genome tiling array that provides an expression map at 5-nt resolution in these two cell lines (21). SSCprofiler allows for the expression threshold to be adjusted ranging from 1 to 2000 in order to retain candidates which exceed a given expression cut-off. For the results reported here, a value of 200 was used as a cut-off.
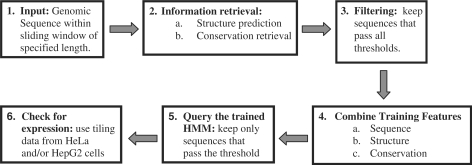
Scanning genomic regions for profiles
The process of scanning genomic regions for miRNA precursor profiles involves six steps, illustrated in Figure 3. Step 1: A sliding window of selected length is passed along the genomic sequence shifting 1 nt at a time. Step 2: For every window shift, sequence structure and conservation information is retrieved according to the selected training features; i.e. structure prediction is performed and conservation is obtained from the multiz files. Step 3: Each sequence within the sliding window is passed through the filters utilizing the pre-defined filtering parameters (i.e. hairpin length, asymmetry). Step 4: For each sequence, the features used during training (sequence, structure and/or conservation) are generated according to the 16-letter key described earlier. This allows the simultaneous consideration of information for every nucleotide position in the genomic sequence. Step 5: The trained HMM is used to assign a likelihood score to each genomic sequence within the sliding window. The HMM score threshold can be selected by the user. It is usually defined as the score where sensitivity and specificity from the training/validation process were optimal. Step 6: Candidates that overlap by ≤50 nt were grouped and the candidate with the highest score is used to represent the cluster. Thereafter, the candidates are assessed according to their expression in HeLa or HepG2 cells using tiling array data.
Figure 3.
Flowchart of the scanning procedure.
RNA extraction and northern blot analysis
Total RNA was extracted from HeLa cells grown in culture using Trizol. Eighty micrograms of total RNA was analyzed on a 15% denaturing polyacrylamide gel containing 8 M urea and transferred to Nytran N membrane (Schleicher & Schuell, Germany). Membranes were probed with standard DNA oligonucleotides, complementary to both polarities. Due to the difficulty in predicting accurately the location of the mature on the pre-miRNA, we select both stem sequences (maximum size 50 nt) from the stem-loop structure of the miRNA gene candidates (Table 2). Ten picomoles of each oligonucleotide probe was end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Pre-hybridization of the filters was carried out in 7% SDS, 5× SSC, 1× Denhardt's solution and 0.02 M Na2HPO4 pH 7.2. Hybridizations were performed in the same solution at 50°C after the addition of the radiolabeled DNA oligonucleotide. Followed an overnight hybridization, the membranes were washed at 50°C in low stringency buffer [2× SSC, 0.3% SDS] twice for 30 min (37). The membranes were stripped by washing in a high stringency buffer (0.1× SSC and 0.5% SDS) for 30 min at 80°C and reprobed with the negative polarity oligonucleotides.
Table 2.
DNA oligonucleotides
| Oligo stem 1 | Oligo stem 2 | |
|---|---|---|
| Candidate 1 | 5′-ACCTCTCCCCCTGCCAGGTTCCACCAGGGGACACCGTGTGTGT-3′ | 5′-CGAGCAGGGCTCCCCCACCTGAGTACCTGACCATGGGCTTTGGAGAGGC-3′ |
| Candidate 2 | 5′-TACGCCCACAGCCCCCAGGCCCCCGAAGACAGGTGTCATGGA-3′ | 5′-TCCAAGAGCATCAAGCAGCAGGGGCTGGGGGAGCCAGCAGG-3′ |
| Candidate 3 | 5′-ATCTACCAGGTCCTGGGCTTCGGGCCGCGTTCCCAAGGCAAGC-3′ | 5′-ACCCGCGGCGAGGACACGGCCGACCGCCCGCCTGCGCCC-3′ |
| Candidate 4 | 5′-GCCCAGGAGGAGGTGGCACATCTGGGCTCCAGTCCTCGCAC-3′ | 5′-GTGCGGGCACCGCGCGAGCCTCGCCCCTTCCCACCTGCGC-3′ |
RESULTS
Learning characteristic features of human precursor miRNAs
Human miRNA precursors from the RNA registry version 12.0 were used to assess the performance accuracy of the SSCprofiler while over 35 000 negative miRNA sequences were used for evaluation purposes (see ‘Materials and Methods’ section). Negative sequences were hairpin structures derived from 3′-UTR regions. These regions were selected because they have not yet been documented to contain miRNA genes. To obtain a reliable control set, negative sequences were filtered according to free energy and conservation criteria (see ‘Materials and Methods’ section) to ensure their resemblance with true pre-miRNAs in both structure and conservation. Prior to training, all positive and negative examples were filtered as follows: initially, filtering exclusively by a minimum energy threshold of −25.44 kcal/mol resulted in 258 true miRNAs and ∼8000 negative sequences. Consequently, seven additional filtering parameters were used to further eliminate false positives. Figure 4 shows the histogram distributions of the sequences prior to filtering as generated by the SSCprofiler interface with respect to three filtering parameters: Hairpin Length, Asymmetry and Bulges-Loops. Similar distributions were generated for all eight filtering parameters in order to determine the respective cut-off values that were optimal for discriminating true miRNA genes from negative data. Sequences were only retained if they met the following criteria:
Hairpin = 1
Bulges ≤16
Loops ≤32
Asymmetry ≤13
Bulges-loops ≤37
Hairpin length ≤16
Folding min energy ≤−25.44 kcal/mol
Conservation ≥25% of nucleotides conserved
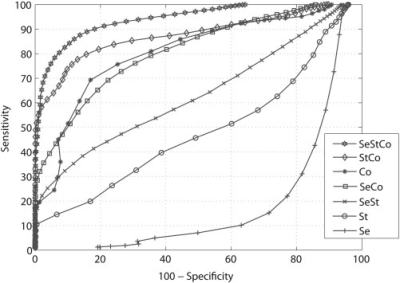
The above-mentioned filtering procedure resulted in 249 true miRNAs and 2330 negative sequences. Subsequently, HMMs were trained solely on the true miRNAs using a 5-fold (three-fifths for training, two-fifths for validation) boosting validation procedure (as described in the ‘Materials and Methods’ section). The procedure was repeated for different combinations of biological features and the HMMs average performance accuracy was reported for each case. ROC curves showing the average validation performance of HMMs that utilize all possible combinations of sequence, structure and conservation information are shown in Figure 5. There was a significant improvement in prediction accuracy for the validation set when certain features were combined, highlighting the importance of simultaneously incorporating additional biological information during the training procedure. The best results were obtained when all three features were used to train the HMMs, achieving on average 88.95% sensitivity and 84.16% specificity in the validation set for a score threshold of 3 (Figure 5 and Supplementary Figure S1). Once a good performance on the training/validation sets was achieved, all 249 true miRNA precursors were pooled together and used to train the final HMM taking into account the same feature combination and filtering parameters. This final model was used to build the scanning interface of the SSCprofiler.
Figure 5.
ROC curves for all possible combinations of sequence (Se), structure (St) and conservation (Co) features for the validation set averaged, over 100 repetitions. As evident from the figure, the area under the curve is maximized when all three features are combined. Note that conservation alone significantly outperforms sequence, structure and sequence + structure (SeSt).
To demonstrate the ability of SSCprofiler to generalize on unseen data we used 373 recently identified miRNAs from miRBase version 12.0 that were not contained in our training/validation sets. Of these, 219 precursors passed the SSCprofiler filters. Table 3 shows the classification performance obtained by SSCprofiler on the 249 training/validation and 219 unseen test precursors for different HMM thresholds. Classification of the 219 unseen precursors was performed using the scanning interface of SSCprofiler and precursors were considered as ‘identified’ by the model if a significant hit was observed at their respective genomic coordinates. For this reason we only report prediction accuracy for the test set. As evident from the table, generalization performance is maximum for an HMM threshold of 1.
Table 3.
SSCprofiler prediction accuracy on validation and blind test sets for three different thresholds
| Threshold | Validation |
Test | |
|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Pred. Acc (%) | |
| 3 | 88.95 | 84.16 | 72.15 |
| 2 | 90.07 | 81.77 | 78.08 |
| 1 | 91.3 | 78.9 | 85.84 |
Predicting miRNA genes in cancer-associated genomic regions (CAGRs)
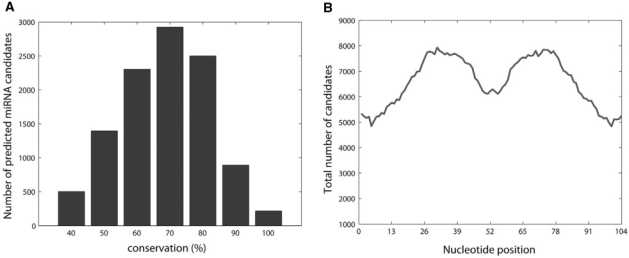
According to Calin et al. (32), there is a large probability that cancer-associated genomic regions contain miRNA genes. This hypothesis is based on the finding that at least 98 known miRNA genes reside in CAGRs, including 80 miRNAs that are located exactly in minimal regions of LOH or minimal regions of amplification described in a variety of tumors such as lung, breast, ovarian, colon, gastric and hepatocellular carcinoma, as well as leukemias and lymphomas. To investigate this hypothesis, we used the final trained SSCprofiler to search for novel miRNA candidates within these regions. Both positive and negative DNA strands were scanned for a number of regions which represent over 350 MB of the human genome and are known to be deleted or amplified in over 20 different types of cancers (Supplementary Table S1). Filtering parameters and conservation retrieval were the same as described in the previous section. The scanning procedure (see Materials and Methods section, Figure 3) lasted ∼8 days (real-time) using a parallel PC cluster with 10 dual opteron processors. An example of the SSCprofiler output for this scanning is shown in Supplementary Figure S2. Figure 6 shows the conservation of all predicted miRNA candidates for an HMM threshold of 3. As shown in Figure 6A, the majority of predicted miRNA candidates had a high degree of conservation (over 50%) across the seven different species. Moreover, the conservation for each nucleotide position along the 104–nt long predicted sequence dropped significantly near the loop.
Figure 6.
Conservation of all 10‱511 predicted miRNA candidates (HMM threshold = 3) across seven species. (A) Histogram of conserved miRNA candidates. As evident from the figure, the majority of candidates are more than 50% conserved across the seven species. (B) Distribution of conserved nucleodites for all candidates along the 104-nt positions of the scanning window. As evident from the figure, there is a large drop in conservation near the loop (middle area) while positions around the loop are higly conserved in a symmetrical way.
Identification of the 98 known miRNAs in the CAGRs regions that were scanned was assessed as a function of the HMM score as shown in Table 4. As expected, the number of miRNA gene candidates decreases with increasing HMM score. Consequently, as the HMM score becomes larger, the sensitivity drops while the specificity increases. According to the training and testing procedures discussed previously, the HMM score threshold for which both sensitivity and specificity values were maximized ranged between 1 and 3 (Table 3). However, when scanning large genomic sequences multiple false positives tend to accumulate, even for an average specificity value of ∼85% (threshold of 3). Since experimental verification is an expensive and time consuming process, we chose candidates attaining a significantly higher HMM score in order to obtain the most probable miRNA gene candidate.
Table 4.
Predicted miRNA genes as a function of the HMM score
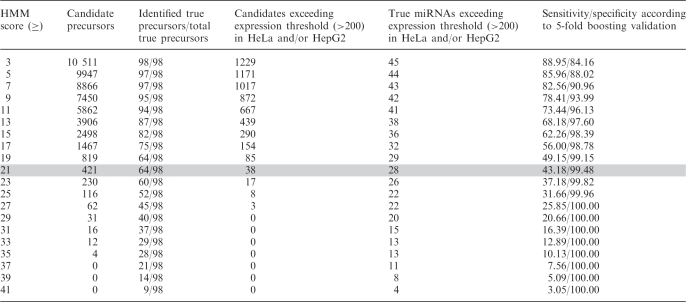 |
For each HMM score in the range of 3–41 (first column), the table shows: (a) the number of predicted miRNA precursors (second column); (b) the number of true precursors included in the predicted list as a function of all true precursors within CAGRs (third column); (c) the number of predicted candidates (fourth column) and true miRNAs (fifth column) that passed the 200 expression threshold in HeLa and/or HepG2 cells and (d) the respective sensitivity and specificity values (sixth column). The 421 sequences that were predicted for an HMM threshold of 21 were selected for further processing, which is tinted grey in the table.
Experimental verification of top scoring candidates
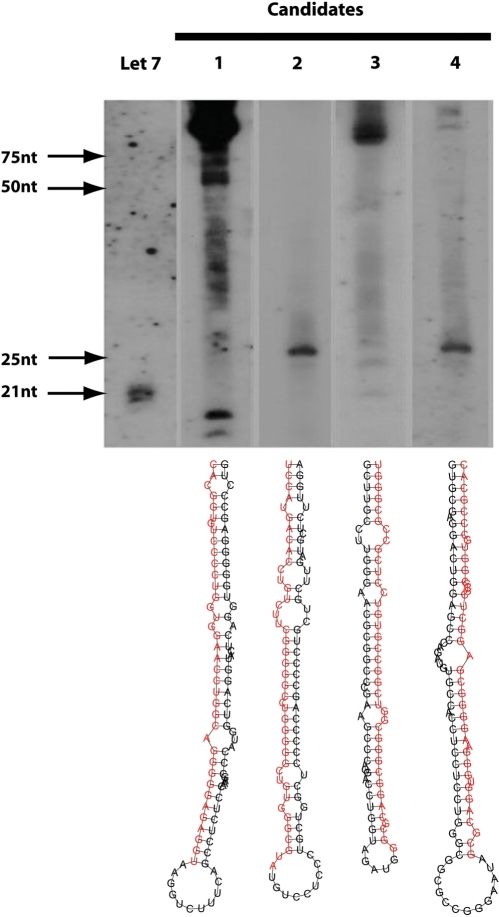
According to sensitivity and specificity measures, the prediction accuracy of SSCprofiler with respect to the identification of novel miRNA genes is very high. However, these statistical evaluation criteria depend highly on the specific data sets used to train and evaluate the computational model. Experimental verification of predicted miRNA genes is the optimal way to assess the model's performance. Towards this goal, we experimentally validated a few of our top scoring precursor candidates. The HMM threshold score for selecting these candidates was set according to the following criteria: (i) high enough to decrease the number of false positives and at the same time and (ii) low enough to capture many of the true miRNAs. A threshold of 21 was finally selected, at which 421 candidates were predicted with 65.31% (64/98) accuracy for the true miRNAs. At this threshold, the predicted list included candidates with only partial conservation when compared to higher scoring candidates. The expression of all 421 candidates in HeLa and HepG2 cells was assessed using recent data from a full genome tiling array (21) which provides a small-RNA expression map. A total of 38 candidates whose expression at the stem region was above a threshold of 200 were retained (see tinted grey in Table 4). Of these, only 20 were expressed in HeLa cells. The top four of these 20 candidates, according to their expression value (listed in Table 5), were tested experimentally using northern blot analysis on cultured HeLa cells (see ‘Materials and methods’ section for details). Northern blot analysis concurred with the tiling array expression data in all four of the candidates tested. As shown in Figure 7, all candidates produced specific signals whose size is within the mature miRNA range (19–27 nt) while, in some cases, the pre-miRNA was also detected. Moreover, we found that in all four candidates only one strand of the predicted precursor produced a specific signal, further suggesting that our candidates are likely to be true miRNA precursors.
Table 5.
Candidate miRNA genes verified by northern blot analysis located in minimal deleted regions involved in human cancers
| Candidate | Candidate Informationa | CAGR | Type of cancer | Closest miRNA | Expression in HeLa |
|---|---|---|---|---|---|
| 1 | chr9:123327358-123327460 st− | chr9:121153509-128793509 | Bladder cancer | miR-181a; miR-199b | 1667.5 |
| 2 | chr5:148958951-148959053 st− | chr5:144121683-156051683 | Prostate cancer aggressiveness | miR-145/miR-143 | 363.5 |
| 2 | chr5:148958951-148959053 st− | chr5:148181683-151101683 | Myelodysplastic syndrome | miR-145/miR-143 | 363.5 |
| 3 | chr22:40863894-40863996 st+ | chr22:31530000-43583971 | Colorectal cancer | miR-33a | 345.0 |
| 3 | chr22:40863894-40863996 st+ | chr22:31530000-42193557 | Astrocytomas | miR-33a | 345.0 |
| 4 | chr5:149984684-149984786 st− | chr5:144121683-156051683 | Prostate cancer aggressiveness | miR-145/miR-143 | 264.0 |
| 4 | chr5:149984684-149984786 st− | chr5:148181683-151101683 | Myelodysplastic syndrome | miR-145/miR-143 | 264.0 |
Positions are according to build 35 (hg17) version of the Human Genome at http://genome.ucsc.edu/
aChromosomal location and strand (st+ or st−).
Figure 7.
Northern blot analysis shows a specific signal for all 4 miRNA gene candidates. The let7 probe hybridizes on multiple members of the let7 miRNA gene family, accounting for the three bands shown on the reference membrane. The membranes labeled 1–4 represent the miRNA gene candidates predicted by SSCprofiler in the same order as shown in Table 5. The band patterns of four miRNA gene candidates resemble those of known miRNAs; displaying a band at the range of 19–27 nt. The higher signals represent the unprocessed precursors (range ∼70 nt). Structures for the four miRNA gene candidates as predicted by RNAfold are also shown. The strand on each precursor that produces a signal is shown in red.
Blat analysis (38) against the human genome provided additional supporting evidence for our four miRNA gene candidates. We found that all candidates are more than 45% conserved across eight other organisms and are located within expressed intergenic regions, consistent with the majority of miRNAs (39,40). Moreover, Blat search produced 100% identity hits at the level of 20–26 nt in other regions of the genome and secondary structure prediction for the genomic sequences flanking these regions resembled that of true miRNA precursors in five out of nine cases. Since the mature miRNA is the main unit of regulation for the miRNA gene it may be more conserved than the rest of the precursor, suggesting the existence of additional miRNA genes. Blast analysis of the oligonucleotides concurs with the results from the Blat search and reveals significant hits in other regions of the human genome.
Tool comparison
Finally, to assess the prediction accuracy of our tool compared to existing algorithms, we used the four verified candidates (the predicted precursors) as a query set in four existing miRNA gene prediction tools. Interestingly, all of these tools failed to identify our candidates as likely miRNA genes. MiRRim (19), ProMir II (41) (HMM algorithms) and BayesMiRNAfind (12) (Bayes classifier) were not able to identify any of our four candidates while TripletSVM (17) (SVM classifier) predicted one out of four candidates (candidate 1). It is important to note that all of these tools are considered as highly accurate with respect to traditional sensitivity/specificity measures, which in some cases outperform that of SSCprofiler (24). However, only for one tool (ProMir II) the authors performed experimental testing of their predicted miRNA gene candidates.
The most recent of these tools [MiRRim (19)] is similar to SSCprofiler as it also uses an HMM algorithm that considers structure and conservation features for predicting novel miRNA genes. The main conceptual differences between the two tools include: (1) the selection of negative data, (2) the size of the sliding window and (3) the ability of the user to take into account information from large scale tiling arrays. A detailed comparison between the two tools is provided in Supplementary Table S2. Briefly, for SSCprofiler both positive and negative sequences are similar in their structure and degree of conservation while for miRRim negative data are filtered according to conservation alone. The latter may bias results and make the discrimination between the two classes an easier task, resulting in higher sensitivity/specificity measures on the training and validation sets but not necessarily on a blind test set. Moreover, differences in the scanning procedure utilized by each tool can affect the total number of predicted miRNA candidates. A larger window such as the one used in miRRim, translates into a substantially smaller search space and consequently a smaller number of genomic regions that could be identified as potential miRNA genes. Thus, the number of predicted candidates is not directly comparable between the two tools. An important advantage of SSCprofiler is that it allows the user to further filter resulting candidates using a large scale tilling array data set (21). Such a feature is not provided in miRRim. Overall, we believe that the differences described above, together with the sequential consideration of sequence, structure and conservation at the nucleotide level offer an important advantage to our tool compared to existing ones. It is the combination of all of these factors that allowed SSCprofiler to identify four novel miRNA gene candidates, which were experimentally verified but could not be identified by four other prediction tools. We firmly believe that for a computational tool to prove its value, especially for the biology oriented user group it is designed for, it must provide a full prediction pipeline from computational identification to experimental verification for at least a few top scoring candidates.
DISCUSSION
In this study we introduced an efficient miRNA gene prediction tool (SSCprofiler) which is based on Profile HMMs and evaluated its performance against a blind set of recently identified miRNAs as well as via the experimental verification of four top scoring candidates. Our tool is provided both as a user friendly trainable interface and a web-based scanning application which can be used for querying genomic regions. In both cases, the user has a large degree of flexibility in terms of dataset specification and parameter tuning.
Our algorithm works by combining sequence, structure and conservation information taken at the nucleotide level throughout the length of miRNA precursors. We show that multiple feature integration is advantageous with respect to prediction accuracy and argue that this type of combination is more effective than other approaches. Incorporation of expression information for predicted candidates is another important advantage of our tool. The use of full genome tiling array data (21), which provide a small-RNA expression map in HeLa and HepG2 cells at 5-nt resolution, increases the reliability of model predictions and can be extremely useful when selecting miRNA gene candidates for experimental verification due to the tissue specific expression of miRNA genes.
The effectiveness of SSCprofiler in recognizing human miRNA genes was demonstrated using a blind set of 219 recently identified human miRNAs from the latest version of miRBase (version 12). For an HMM threshold of 1, the method reached a prediction accuracy of 85.84% similar to its training/validation performance (91.3% sensitivity, 78.9% specificity). The tool's ability to identify novel miRNA genes located within 350 MB of human cancer-associated genomic regions (32) was also assessed. For an HMM threshold of 11 (73.44% sensitivity, 96.13% specificity) SSCprofiler predicted a total of 5862 novel miRNA candidates within these regions. Assuming an analogy between CAGRs and the whole human genome, SSCprofiler would predict approximately 58 000 new miRNA genes, in agreement with a recent estimate provided by the study of Miranda et al. (40). However, it should be noted that CAGRs are known to contain a disproportionably large number of miRNAs (over 20% of all miRNAs in miRBase 12.0); therefore an analogy between those regions and the entire human genome might not be valid. Taking into account the high costs of reagents and the time consuming nature of experimental procedures we decided to select candidates that were more likely to be successful. For this reason we used a higher threshold (HMM score of 21) for which fewer candidates were predicted. Of the 421 predicted candidates only 20 were highly expressed in HeLa cells. Northern blot analysis of the top four of these candidates verified the presence of a specific RNA molecule at the miRNA range (19–27) which strongly suggests the presence of a small noncoding RNA. In future efforts, additional predicted candidates for lower HMM thresholds should be analyzed experimentally in order to obtain a more accurate cut-off value for reliably predicting novel miRNAs.
Our findings regarding miRNA gene identification are in accordance to the uniform system of miRNA annotation (42). The miRNA biogenesis criterion is satisfied by the prediction of a potential fold-back precursor structure that contains the ∼22-nt miRNA sequence within one arm of the hairpin. The hairpin displays a very low free energy, as predicted by RNAfold and only one stem of the precursor shows a northern blot signal. Our candidates do not contain large internal loops or bulges, particularly large asymmetric bulges and all fall within the miRNA precursor range of ∼60–100 nt reported in animals. Phylogenetic conservation of the whole miRNA precursor sequence for all four candidates across seven other organisms is also observed. The miRNA expression criterion is also met by our candidates. A distinct ∼22-nt RNA transcript is detected by hybridization to a size-fractionated RNA sample by northern blot analysis for all four candidates. In addition, expression of ∼22-nt RNA transcripts from the active stem region of the candidates is observed in HeLa cells using tiling arrays. These criteria provide strong evidence that our top scoring candidates are likely to be true miRNA precursors and consequently, that SSCprofiler is a reliable and efficient tool for predicting novel miRNA genes. Interestingly, a comparison study with other miRNA gene prediction tools reveals that three out of four of our verified miRNA gene candidates are not predicted by four other published tools (12,17,19,41). Only one out of four tools predicted one-fourth of our verified miRNA candidates (17). This finding highlights the superior prediction capacity of SSCprofiler and further substantiates its significance as a miRNA gene prediction tool. The tool's availability in both a trainable and a web-based scanning version can further facilitate its use as a part of a prediction pipeline for novel miRNA genes, hence allowing for minimization of time, cost and effort.
An important finding of this work is the identification of four novel miRNA gene candidates residing within genomic regions which are implicated in numerous cancers (CAGRs). Although a detailed experimental characterization of the mature miRNA function is pending, these molecules are likely to play an important role in regulating carcinogenesis, possibly by acting as ‘oncogenes’ or ‘tumor suppressors’. The CAGRs corresponding to each miRNA candidate are commonly deleted in various types of cancers (Table 5). Deletion of a region containing a miRNA gene prohibits the expression of the functional miRNA. As a result, gene(s) regulated by this miRNA will function uncontrollably, a process which may result in a cascade of events that triggers oncogenesis. Since the mode of action of mature miRNAs usually results in downregulation of targeted genes, one possibility is that our candidates play a tumor suppressor role perhaps by stopping a major tumorigenic turning point in the cell. However, since cumulating data support both a negative and a positive regulatory role for miRNAs (43) it is hard to predict the effect of these potential miRNAs on their target genes. Target prediction programs can provide a starting point for identifying possible target genes for these candidates, thus providing more insights into their potential role in specific types of cancer. Towards this goal, an RNA–RNA duplex prediction algorithm will be incorporated in future versions of the SSCprofiler. This can be achieved using programs such as RNAcofold (35) which calculate secondary structures of two RNA sequences in the form of a hybrid duplex. This extension will enable the user to train profile HMMs that can recognize the pairing rules between two hybrid RNA molecules, thus allowing the prediction of new miRNA–mRNA interactions that obey similar rules. Our ultimate goal is to develop a stand alone application which will be able to predict novel miRNA genes as well as their probable targets. Such a tool may provide a more concise biological picture of the pathways and genes regulated by our four novel miRNA gene candidates. Experimental verification will ultimately be needed to characterize the production of a mature miRNA, show that predicted interactions take place in the system of interest and that functional interactions are strongly associated with the emergence of a cancerous phenotype.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online
FUNDING
Action 8.3.1 (Reinforcement Pro-gram of Human Research Manpower—‘PENED 2003’, [03EΔ842]) of the operational program ‘competitiveness’of the Greek General Secretariat for Research and Technology; INFOBIOMED NoE [FP6-IST-2002-507‱585] EU funded project; Marie Curie Outgoing Fellowship [PIOF-GA-2008-219‱622] of the European Commission. Funding for open access charge: PENED 2003 [03EΔ842].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Prof. Angelos Bilas for the use of his PC cluster.
RERERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Fantom C. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 8.Helvik SA, Snove O, Jr., Saetrom P. Reliable prediction of Drosha processing sites improves microRNA gene prediction. Bioinformatics. 2006;23:142–149. doi: 10.1093/bioinformatics/btl570. [DOI] [PubMed] [Google Scholar]
- 9.Hertel J, Stadler PF. Hairpins in a haystack: recognizing microRNA precursors in comparative genomics data. Bioinformatics. 2006;22:e197–e202. doi: 10.1093/bioinformatics/btl257. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003b;16:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267–281. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yousef M, Nebozhyn M, Shatkay H, Kanterakis S, Showe LC, Showe MK. Combining multi-species genomic data for microRNA identification using a Naive Bayes classifier. Bioinformatics. 2006;22:1325–1334. doi: 10.1093/bioinformatics/btl094. [DOI] [PubMed] [Google Scholar]
- 13.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42–R61. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber MJ. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 15.Legendre M, Lambert A, Gautheret D. Profile-based detection of microRNA precursors in animal genomes. Bioinformatics. 2004;21:841–845. doi: 10.1093/bioinformatics/bti073. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Zhang. J, Li. F, Gu. J, He. T, Zhang. X, Li. Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- 17.Xue C, Li F, He T, Liu GP, Li Y, Zhang X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310–316. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam JW, Shin KR, Han J, Lee Y, Kim VN, Zhang BT. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acid Res. 2005;33:3570–3581. doi: 10.1093/nar/gki668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terai G, Komori T, Asai K, Kin T. miRRim: a novel system to find conserved miRNAs with high sensitivity and specificity. RNA. 2007;13:2081–2090. doi: 10.1261/rna.655107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 21.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 22.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452:1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oulas A, Reczko M, Poirazi P. MicroRNAs and cancer—the search begins! IEEE Trans. Inf. Technol. Biomed. 2008;13:67–77. doi: 10.1109/TITB.2008.2007086. [DOI] [PubMed] [Google Scholar]
- 25.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific MicroRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 28.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 29.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 31.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High Expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2003;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 32.Calin GA, Sevignani. C, Dumitru. CD, Hyslop. T, Noch. E, Yendamuri. S, Shimizu. M, Rattan. S, Bullrich. F, Negrini. M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. PNAS. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 37.Koscianska E, Baev V, Skreka K, Oikonomaki K, Rusinov V, Tabler M, Kalantidis K. Prediction and preliminary validation of oncogene regulation by miRNAs. BMC Mol. Biol. 2007;8:79. doi: 10.1186/1471-2199-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 40.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 41.Nam JW, Kim J, Kim SK, Zhang BT. ProMiR II: a web server for the probabilistic prediction of clustered, nonclustered, conserved and nonconserved microRNAs. Nucleic Acids Res. 2006;34:W455–W458. doi: 10.1093/nar/gkl321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.