Abstract
Repair of single-stranded DNA breaks before DNA replication is critical in maintaining genomic stability; however, how cells deal with these lesions during S phase is not clear. Using combined approaches of proteomics and in vitro and in vivo protein–protein interaction, we identified the p58 subunit of DNA Pol α-primase as a new binding partner of XRCC1, a key protein of the single strand break repair (SSBR) complex. In vitro experiments reveal that the binding of poly(ADP-ribose) to p58 inhibits primase activity by competition with its DNA binding property. Overexpression of the XRCC1-BRCT1 domain in HeLa cells induces poly(ADP-ribose) synthesis, PARP-1 and XRCC1-BRCT1 poly(ADP-ribosyl)ation and a strong S phase delay in the presence of DNA damage. Addition of recombinant XRCC1-BRCT1 to Xenopus egg extracts slows down DNA synthesis and inhibits the binding of PCNA, but not MCM2 to alkylated chromatin, thus indicating interference with the assembly of functional replication forks. Altogether these results suggest a critical role for XRCC1 in connecting the SSBR machinery with the replication fork to halt DNA synthesis in response to DNA damage.
INTRODUCTION
The cellular response to DNA damage produced by environmental agents or generated by the cellular metabolism involves the coordinated activation of various enzymatic activities aimed at detecting, signaling and resolving faithfully genomic discontinuities. XRCC1 plays a crucial role in the coordination of two overlapping repair pathways, base excision repair (BER) and single strand break repair (SSBR), through the association with and stimulation of several key enzymes involved at different steps of these pathways [reviewed in (1,2)]. The two BRCT domains (BRCT1, from amino acids 314 to 403; and BRCT2, from amino acids 538 to 633) of XRCC1 mediate a network of protein–protein interactions with these repair factors. The BRCT1 domain is the most evolutionarily conserved and is required for survival after methylation damage (3,4). It interacts with PARP-1 and PARP-2, and contains a binding site for poly (ADP-ribose) (PAR) mediating the rapid recruitment of XRCC1 at the site of DNA damage (5–9). The BRCT2 domain of XRCC1 binds to and stabilizes DNA ligase III (Lig III) (10).
Several observations suggest that the hypersensitivity of XRCC1-mutant cell lines to monofunctional alkylating agents results from the persistence of unrepaired single strand breaks (SSBs) that are encountered by the DNA replication fork during S phase. The XRCC1 deficient EM9 cell line exhibits an increased doubling time and an elevated level of sister-chromatid exchange (SCE) (11,12). Kubota and Horiuchi (4) found that a mutant in the BRCT1 domain of XRCC1 is defective in the restart of DNA replication following methyl-methane sulfonate (MMS) treatment, while this mutant is proficient in DNA repair. Recently, Lan et al. (13) showed that suppressing XRCC1 expression by RNA interference decreased PCNA accumulation on SSBs induced by laser irradiation and Fan et al. (14) reported a direct interaction between XRCC1 and PCNA in vitro and in vivo in S phase. Altogether, these results further extended a possible link between the SSBR machinery and the replicative apparatus.
The formation of functional DNA replication forks occurs by the sequential assembly of large multiprotein complexes at DNA replication origins [reviewed in (15,16)]. The origin recognition complex (ORC1-6) together with the Cdc6 and Cdt1 proteins, catalyze the formation of pre-replicative complexes (pre-RCs), namely the assembly of the MCM2-7 helicase complex. Activation of pre-RCs during S phase allows the recruitment of additional replication factors to form pre-initiation complexes (pre-ICs) that can support DNA unwinding and recruit the DNA polymerases and other factors required to promote DNA synthesis. To begin DNA synthesis, an initial RNA primer is synthesized by the DNA primase, a heterodimer of two subunits, p48 and p58. This short RNA primer is then extended by DNA Pol α and marks the formation of initiation complexes (ICs). Then replication factor C (RFC) binds to the primer template junction and catalyzes the loading of the ring-shaped replication factor PCNA that encircles DNA and associates with the replicative polymerases Pol δ or -ϵ, taking over DNA synthesis from Pol α (elongation step).
Here, we show that the BRCT1 domain of XRCC1 specifically interacts in vitro and in vivo with the p58 subunit of DNA Pol α-primase in HeLa cells. p58 also interacts with PAR resulting in the inhibition of the p48–p58 primase activity in vitro. Consistent with these findings, the expression of the BRCT1 domain of XRCC1 in HeLa cells or in Xenopus extracts interferes with ongoing DNA synthesis in the presence of DNA damage in a PAR-dependent manner. These results suggest that the BRCT1 domain of XRCC1 plays a central role in regulating DNA replication across SSBs during S phase.
MATERIALS AND METHODS
Construction of XRCC1 and p58 expression vectors
From the human DNA primase p48-His-tagged-p58 and p58 C-terminus cloned in pET11 (17), we amplified the DNA sequence encoding p58 by PCR (amino acids 1–266) and cloned it in the NdeI and BamHI restriction sites of the pET 15b vector (Novagen).
Vectors allowing the expression in mammalian cells of GST-tagged fragments of human hXRCC1 are described in (18). PCR amplified fragment encoding Xenopus xXRCC1-BRCT1 (amino acids 307–414) was subcloned into the EcoRI and XhoI of pGEX 4T vector allowing overproduction of the GST fused proteins in Escherichia coli.
Antibodies
The following antibodies were used: rat monoclonal anti-p58 antibody (19), mouse monoclonal anti-GST antibody (kindly given by M. Oulad, IGBMC, Illkirch), rabbit polyclonal anti-RPA antibody (D. Maiorano, IGH, Montpellier), rabbit polyclonal anti-ORC1 antibody (20), rabbit polyclonal anti-MCM2 antibody (AbCam), mouse monoclonal anti-PARP-1 (EGT69), rabbit polyclonal anti-GST, mouse monoclonal anti-PCNA antibody (PC10, Sigma-Aldrich), goat polyclonal anti-XRCC1 antibody (D-18 sc-5902, Santa Cruz Biotechnology Inc.), rabbit polyclonal anti-XRCC1 (Alexis), rabbit polyclonal anti-PAR, and mouse monoclonal anti-BrdU antibodies (Becton Dickinson and Company), and rabbit polyclonal anti-phospho Chk1 (S345) antibody (Cell Signalling). Secondary antibodies, from Molecular Probes, are either goat anti-mouse, donkey anti rat or goat anti-rabbit antibodies, peroxidase-conjugated or alexa 488/alexa 568-conjugated.
Protein expression, purification, GST pull-down assays and immunoprecipitation
Fusion proteins were expressed in E. coli (BRCT1: amino acids 282–428 or BRCT2: 538–633 domains) and purified using the affinity of either GST for glutathione-coupled beads (GE Healthcare, Amersham Biosciences) or His-tag for Ni2+ ions immobilized on a silica-based resin (ProtinoR Ni, Machery-Nagel) as recommended by manufacturers.
GST pull-down and immunoprecipitation experiments were carried as described in (18). When indicated, cells were treated with aphidicolin (A, 5 μg/ml for 16 h) followed or not by hydroxyurea treatment (HU, 4 mM for 4 h) or by MMS (2.5 mM for 30 min) with or without PARP [Poly (ADP-ribose) Polymerase] inhibitor (Ku-0058948, 100 nM).
Immunofluorescence
Cells (105) grown on glass cover slips, in DMEM medium containing 10% fetal bovine serum and 0.5% gentamicin were treated or not with HU (4 mM for 4 h at 37°C) and processed further as described in (18).
Mass spectrometry
GST or GST-hXRCC1-BRCT1 (amino acids 282–428) produced in E. coli, fixed on glutathione-coupled beads were used to purify interacting proteins from HeLa cell extracts (18). Proteins were separated on SDS–Poly Acylamide Gel Electrophoresis (PAGE) gel, stained with SYPRO-ruby and gel slices containing protein bands of interest were excised and processed for mass spectrometry. In-gel digestion was performed with an automated protein digestion system, MassPREP Station (Waters, Milford, MA, USA). The resulting peptide extracts were directly injected for nanoscale capillary LC-MS/MS (nano-LC-MS/MS) analysis (21). Mass data acquisitions were piloted by MassLynx software (Waters) using automatic switching between MS and MS/MS modes as described previously (22).
PAR binding, DNA binding and far-western blotting
Proteins were separated by SDS–PAGE (2 μg) or dot-blotted directly on membrane (1 or 2 μg, as indicated). Polyacrylamide gels were incubated for 1 h at room temperature in a 50 mM Tris pH 8, 30% glycerol buffer, and proteins were transferred on nitrocellulose membranes for PAR and DNA binding or on Polyvinylidene Fluoride (PVDF) membranes for far-western blotting. Proteins were then re-naturated overnight at 4°C in a 50 mM Tris pH 8, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 mM EDTA, 0.3% Tween 20 and 5% non-fat dried milk buffer. In vitro PAR synthesis was performed as in (23). For PAR binding either anti-PAR immunostaining was performed or, as for DNA binding, membranes were incubated for 1 h at 4°C with 32P radiolabeled PAR or DNA, washed three times in PBS and submitted to autoradiography. Far-western blotting was done as described in (18).
Band shift assay
Electrophoretic mobility shift assays (EMSAs) were carried out to analyze primase binding with PhiX174-ssDNA as previously described (24). Briefly, 2 µg of primase (p48/p58) were incubated with 70 ng PhiX174-ssDNA and increasing amounts (0, 63, 127 and 255 ng) of PAR in binding buffer (final concentration: 10 mM Tris/HCl pH 7.5, 5 mM EDTA, 50 mM NaCl and 1 µg Bovin Serum Albumin (BSA)) at 25°C for 30 min. The protein–DNA complexes were separated by 0.8% agarose gel electrophoresis with 60 mA for 4 h. The DNA was then stained with ethidium bromide and ssDNA and protein–DNA complexes were determined using a fluoroimager FLA-5100 and the software ImageGauge version 4.2.3 (Fuji Europe, Düsseldorf, Germany).
Primase activity
One unit of DNA primase (p48–p58) was incubated with 0.1 mM (nucleotide concentration) oligo (dT)20, 500 μM ATP and 10 µCi [α32P]ATP in a 10 mM Tris Ac pH 7.3, 10 mM MgAc, 1 mM DTT and 0.1 mg/ml BSA buffer. After 15 min at 37°C, the reactions were spotted on DE81 filters (Whatman). Filters were washed four times in 0.4 M ammonium bicarbonate, 1% PPi rinsed twice in H2O, dried, and submitted to scintillation counting in Ultima Gold scintillation liquid (Packard).
Cell cycle experiments
HeLa cells (106) grown in 10-cm Petri dish were transiently transfected with the pBC or pBC-hXRCC1-fragments vectors using JetPEI transfection reagent (Polyplus Transfection). When indicated, cells were incubated for 30 min at 37°C with 1 mM MNU (Sigma), washed and grown in fresh culture medium for 20 h. Cells were pulse labeled for 30 min at 37°C in DMEM medium supplemented with 5 µM BrdU (Sigma). After two PBS washes, cells were detached by trypsination, washed in PBS buffer Glucose EDTA (PGE) (PBS 1×, 1 g/l glucose, 1 mM EDTA) buffer and fixed for 30 min on ice in 70% EtOH in PGE. Cells were centrifuged and incubated for 4 h at 4°C in 5 ml PGE for rehydration. Cells were treated for 15 min at room temperature in 2N HCl in PGE, collected by centrifugation, and resuspended in 2 ml neutralization PBS buffer Sodium tetraborate Tween BSA (PTTB) buffer (PBS 1×, 0.5% Tween 20, 0.5% BSA, 0.1 M sodium tetraborate) and washed twice in PTB buffer (PBS 1×, 0.5% Tween 20, 0.5% BSA). Cells were then incubated for 1 h at room temperature in mouse monoclonal anti-BrdU antibodies (Becton Dickinson) diluted to 1:3 ratio and rabbit polyclonal anti-GST antibodies (Sigma) diluted to 1:2000 ratio. After two washes in PTB buffer, cells were incubated for 1 h at room temperature in Alexa 488 conjugated goat anti-mouse secondary antibodies (Invitrogen) at 1:500 ratio and PE conjugated donkey anti-rabbit secondary antibodies (Jackson Immuno Research) at 1:200 ratio. After two washes in PTB buffer, cells were counterstained with 10 µg/ml 7-aminoactinomycin-D (7-AAD, Sigma) in PGE. Flow cytometry analysis was performed using a FACS Calibur and the Cell Quest software (Becton Dickinson).
Xenopus extracts and DNA replication assay
Sperm nucleus and egg extracts were prepared as described previously (25). Upon thawing, extracts were supplemented with cycloheximide (250 µg/ml) and an energy regeneration system (10 µg/ml creatine kinase, 10 mM creatine phosphate, 1 mM ATP, 1 mM MgCl2). MMS treatment (either 50 or 75 mM depending upon egg extracts) of sperm nuclei was carried out as previously described (26). When recombinant proteins were added, these were incubated in egg extracts for 10 min on ice before addition of sperm chromatin. To follow DNA replication by incorporation of radiolabeled nucleotide into newly replicated DNA, 1 µl of α-[32P] dATP or dCTP (3000 Ci/mmol) was added to a standard reaction of 50 µl and the amount of newly synthesized DNA was determined by TCA precipitation on GF/C glass fibers filters followed by scintillation counting. Ku-0058948 was used as PARP inhibitor (27). Alkaline gel electrophoresis of DNAs and chromatin isolation were performed as previously described in (28) and (29), respectively.
RESULTS
hXRCC1 interacts with the p58 subunit of DNA Pol α-primase
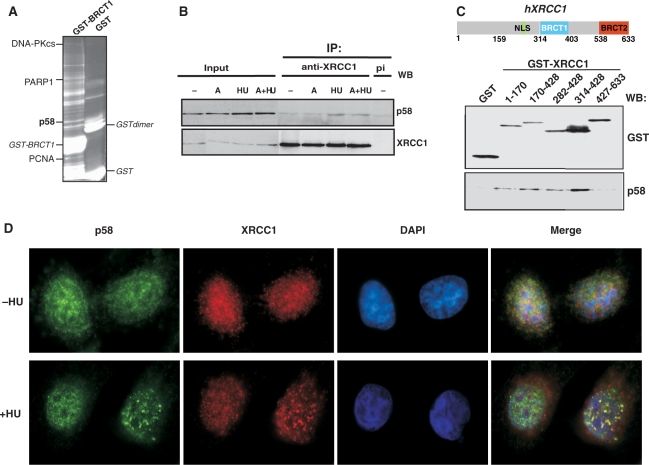
To identify new proteins interacting with the BRCT1 domain of hXRCC1, we used recombinant GST-tagged hXRCC1 fragment (amino acids 282–428) to pull down protein partners from HeLa cell extracts. Co-purified proteins were separated by SDS–PAGE and analyzed by mass spectrometry. The identification of proteins already known to interact with XRCC1 such as PARP-1, PCNA and DNA-PKcs validated our analysis (5,14,18). In addition, we identified the p58 subunit of the DNA Pol α-primase as a novel hXRCC1-BRCT1 interacting protein (Figure 1A).
Figure 1.
Interaction between XRCC1 and p58 subunit of Pol α-primase. (A) Identification of proteins interacting with GST-XRCC1-BRCT1 by mass spectrometry. Sypro ruby stained gel after GST pull-down of HeLa cell extracts expressing either GST or GST-hXRCC1-BRCT1 fused proteins. (B) Identification of XRCC1 associated p58 by immunoprecipitation of XRCC1 from extracts of HeLa cells treated or not with aphidicolin (A: 5 μg/ml, 16 h) and with or without HU (4 mM, 4 h). Co-purifying p58 and immunoprecipitated XRCC1 were identified by western blot. Inputs represent 5% of the total proteins used in immunoprecipitation (pi: pre immune serum). (C) GST pull-down of p58 with different GST-fused XRCC1 domains overexpressed in HeLa cells. A schematic drawing of XRCC1 domain structure is shown. (D) Indirect immunofluorescence microscopy showing co-localization of XRCC1 and p58 in HeLa cells treated (+ HU) or not (−HU) with 4 mM HU for 4 h.
To test whether endogenous p58 and XRCC1 coexist in a common protein complex, immunoprecipitation experiments were performed using extracts from HeLa cells treated or not with aphidicolin (A), with HU or with both drugs to block DNA replication. As shown in Figure 1B, p58 was weakly but reproducibly co-immunoprecipitated with XRCC1, specifically after HU treatment. These results suggest a preferential interaction between XRCC1 and DNA primase when DNA replication forks are stalled.
In order to map the p58 interaction domain(s) within XRCC1, GST fusion proteins that encompass truncated versions of human XRCC1 were generated (Figure 1C). These fusion proteins were expressed in HeLa cells and GST pull-down experiments were performed, followed by western blot analysis. The endogenous p58 subunit efficiently co-purified with polypeptides carrying either the N-terminal part of XRCC1 (amino acids 1–170) or the BRCT1 domain but not the BRCT2 domain (Figure 1C).
To further characterize the interaction between XRCC1 and p58, we analyzed the localization of both endogenous proteins by immunofluorescence in asynchronous cells treated or not with HU. No or very few XRCC1 and p58 foci co-localized in untreated cells; whereas after HU treatment, p58 and XRCC1 foci number increased and significant co-localization was observed between the two proteins (Figure 1D).
Taken together, these results suggest that hXRCC1 and DNA primase, via its p58 subunit, can associate in vivo in response to stalled replication forks.
In vitro association of XRCC1 with p58-Nter
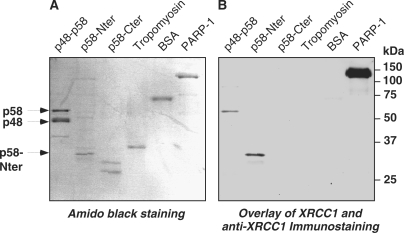
Far-western blot analyses were performed to assess whether the interaction between p58 and hXRCC1 was direct. The p48-His-tagged p58 complex and the N- (amino acids 1–266) or C-terminal (amino acids 267–509) part of p58 were independently expressed in E. coli, purified by affinity chromatography and separated by SDS–PAGE along with negative (tropomyosine, BSA) and positive (PARP-1) controls. Proteins transferred to PVDF membranes were re-naturated prior incubation with purified hXRCC1 protein and immunodetected with anti-XRCC1 antibody (Figure 2B). XRCC1 interacted only with p58 and its N-terminus fragment, interacted neither with p48 nor tropomyosine or BSA. These results demonstrated a direct contact between XRCC1 and the N-terminus domain of p58.
Figure 2.
In vitro association of XRCC1 with the N-terminal domain of p58 assessed by far-western blot. Purified p48-His-tagged-p58, His-tagged p58 N-terminal (Nter) or C-terminal (Cter) domains expressed in E. coli were separated by SDS–PAGE and transferred onto a nitrocellulose membrane. Tropomyosin, BSA and PARP-1 were also included as internal negative and positive controls, respectively (A). The membrane was stained with amido black or incubated with purified XRCC1 and revealed with an anti-XRCC1 antibody (B). p58 domains were determined by partial tryptic digests and peptide analysis (Schlott and Nasheuer, unpublished data).
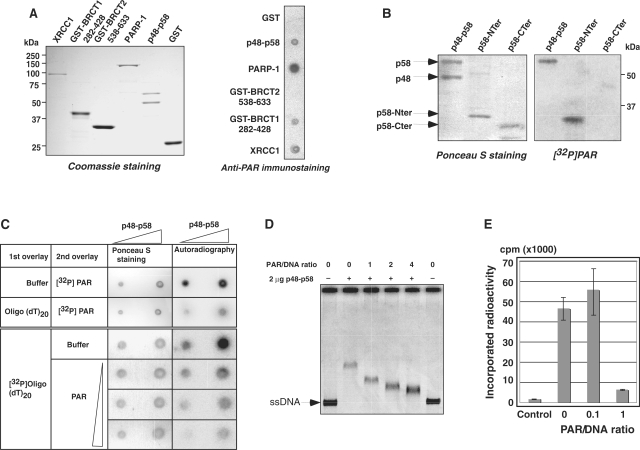
PAR binds p58 and strongly inhibits DNA primase activity of the p48–p58 complex
Since XRCC1-BRCT1 has strong affinity for PAR, we investigated whether p58 could also bind PAR. A search for putative PAR binding motifs in the sequence of p48 and p58 identified two potential PAR binding sites in p58, one in the N-terminal part (amino acids 101–111) and one in the C-terminal part (amino acids 286–296) of the protein. To determine whether these sites were functional, binding of DNA primase to PAR was first analyzed by dot blot. As expected, PAR bound to PARP-1, XRCC1 and the BRCT1 domain of XRCC1 but not to the BRCT2 domain of XRCC1 or to GST. Under the same conditions, the p48–p58 complex interacted with PAR (Figure 3A). To identify precisely the domain of the DNA primase responsible for PAR binding, we separated the p48–p58 subunits along with the N-terminal (amino acids 1–266) and C-terminal (amino acids 266–509) part of p58 by SDS–PAGE and analyzed by radiolabeled PAR binding on proteins blotted on membrane. Results showed that PAR only bound to the N-terminal fragment of p58 (Figure 3B).
Figure 3.
The p58 subunit of DNA primase binds to PAR leading to its inactivation. (A) The indicated purified proteins (1 μg each) were dot-blotted onto nitrocellulose and incubated with PAR. Bound PAR was immunodetected with anti-PAR antibody. Loading of proteins was quantified on a Coomassie stained SDS–PAGE. (B) The indicated purified proteins (2 μg) were separated on SDS–PAGE, transfered on nitrocellulose membranes and incubated with 32P-labeled PAR. Visualization of the loaded proteins was performed by Ponceau S staining of the membrane. (C) Increasing amounts of purified p48–p58 (1 and 2 μg) were dot blotted on nitrocellulose membrane and a first overlay (buffer, radioactively labeled or unlabeled oligo(dT)20, as indicated) was followed by a second incubation (radioactively labeled or unlabeled PAR or buffer) to analyze the competition between PAR and DNA for p58 binding. (D) Band shift assay to analyze complex formation of p48–p58 and ssDNA (ssDNA: PhiX174 single-stranded DNA). Two micrograms of purified p48–p58 was incubated with ssDNA (70 ng) and PAR was added in increasing concentration to displace the p48–p58–ssDNA complex. PAR to DNA ratio varied from 0 to 1–4 excess of PAR. (E) Addition of increasing amounts of PAR to primase assay (ratio 0, 1:10, 1:1) significantly inhibited primase activity.
Next, we wanted to determine whether PAR binding could affect the ability of primase to bind DNA. Purified p48–p58 DNA primase (1 and 2 µg) was dotted on nitrocellulose and incubated successively with oligo(dT)20 and radioactively labeled PAR. PAR was able to bind to p48–p58 in the absence of pre-incubation with DNA; whereas, incubation with unlabeled oligo(dT)20 prior to PAR addition dramatically decreased the efficiency of PAR binding (Figure 3C). When the membranes were first incubated with radioactively labeled DNA oligo(dT)20 and then with increasing concentrations of unlabeled PAR, we observed a decrease in the intensity of the radioactive signal retained onto the membrane indicating that the PAR can dislodge DNA from p48–p58 complex (Figure 3C). This observation was confirmed by band-shift assays performed with purified recombinant p48–p58 and single-stranded DNA. Adding increasing concentrations of PAR (PAR/DNA ratio: 0, 1–4) to the reaction strongly decreased the binding of p48–p58 to DNA (Figure 3D).
To evaluate the effect of PAR on primase activity, we monitored the incorporation of radiolabeled ribonucleotide on an oligo(dT)20 DNA substrate in the presence of increasing PAR concentrations. Low PAR/template DNA ratio had no effect on primase activity; whereas, equimolar conditions of PAR and DNA in the reaction strongly inhibited the radioactive nucleotide incorporation (Figure 3E). Altogether these results indicate that PAR competes with DNA for p58 binding to inhibit DNA primase activity.
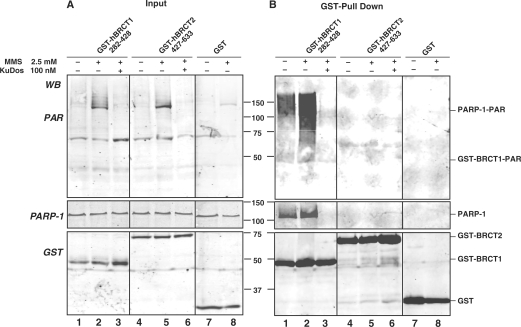
XRCC1-BRCT1 stimulates PARP-1 activity and is poly(ADP-ribosyl)ated in response to DNA damage
The fact that PAR affects primase activity and hXRCC1-BRCT1 interacts with p58 prompted us to examine whether hXRCC1-BRCT1 could be poly(ADP-ribosyl)ated in vivo. GST-tagged hXRCC1-BRCT1 overexpressed in undamaged HeLa cells was slightly poly(ADP-ribosyl)ated, in contrast to GST alone or GST-hXRCC1-BRCT2 which were not poly(ADP-rybosyl)ated (Figure 4B, compare lane 1 with lanes 4 and 7). Treatment of cells with 2.5 mM MMS for 30 min triggered PARP-1 activation and PAR synthesis, and cells overexpressing GST-hXRCC1-BRCT1 displayed high levels of automodified PARP-1 suggesting that hXRCC1-BRCT1 overexpression stimulates PARP-1 activity (Figure 4B, lanes 2, 5 and 8). In addition, GST-hXRCC1-BRCT1 was poly(ADP-ribosyl)ated in MMS-treated cells and this modification decreased in the presence of the PARP inhibitor Ku-0058948 (Figure 4B, lanes 2 and 3). The signals recognized by the PAR antibody with a molecular weight higher than 116 kDa, co-purifying with GST-hXRCC1-BRCT1 correspond to automodified PARP-1 which strongly interacts with the BRCT1 domain of XRCC1 (5). The observation that GST-hXRCC1-BRCT1 is poly(ADP-ribosyl)ated in vivo confirmed previous in vitro data showing that this domain could be covalently poly(ADP-ribosyl)ated by PARP-1 (and PARP-2) in addition to its non-covalent binding to PAR (6). The increased PAR level observed in undamaged GST-hXRCC1-BRCT1 expressing cells was confirmed by immunofluorescence microscopy. PAR synthesis was detected in HeLa cells overexpressing GFP-hXRCC1-BRCT1 but not GFP-hXRCC1-BRCT2 (Supplementary Figure 1). Treatment of cells with hydrogen peroxide triggered higher amount of PAR produced in GFP-hXRCC1-BRCT1 expressing cells compared to untransfected or GFP-hXRCC1-BRCT2 expressing cells (Supplementary Figure 1). In addition, some PAR foci colocalized with GFP-hBRCT1 but not with GFP-hBRCT2.
Figure 4.
GST-XRCC1-BRCT1 overexpressed in HeLa cells triggers PAR synthesis and is poly(ADPribosyl)ated. HeLa cells overexpressing GST-hXRCC1-BRCT1 (amino acids 282–428, lanes 1–3), GST-hXRCC1-BRCT2 (amino acids 427–633, lanes 4–6) or GST (lanes 7, 8) were either untreated (lanes 1, 4 and 7) or treated with 2.5 mM MMS for 30 min (lanes 2, 5 and 8). Where indicated, cells were pre-incubated for 2 h with 100 nM of the PARP inhibitor Ku-0058948 (lanes 3 and 6). (A) Inputs corresponding to 2% of each lysates. Proteins were analyzed by GST pull down and western blot (B) using successively the anti-PAR, anti-PARP-1 and anti-GST antibodies.
Altogether, these results reveal that the overexpression of hXRCC1-BRCT1 domain in HeLa cells stimulates PAR synthesis in damaged cells which leads to its poly(ADP-ribosyl)ation.
XRCC1-BRCT1 overexpressing cells accumulate in S phase following DNA damage
The poly(ADP-ribosyl)ation of hXRCC1-BRCT1 in response to DNA damage, the interaction of p58 with PAR and hXRCC1-BRCT1 and the inhibition of DNA primase activity by PAR lead us to hypothesize that the overexpression of the BRCT1 domain of XRCC1 would have the potential to halt DNA replication of damaged DNA.
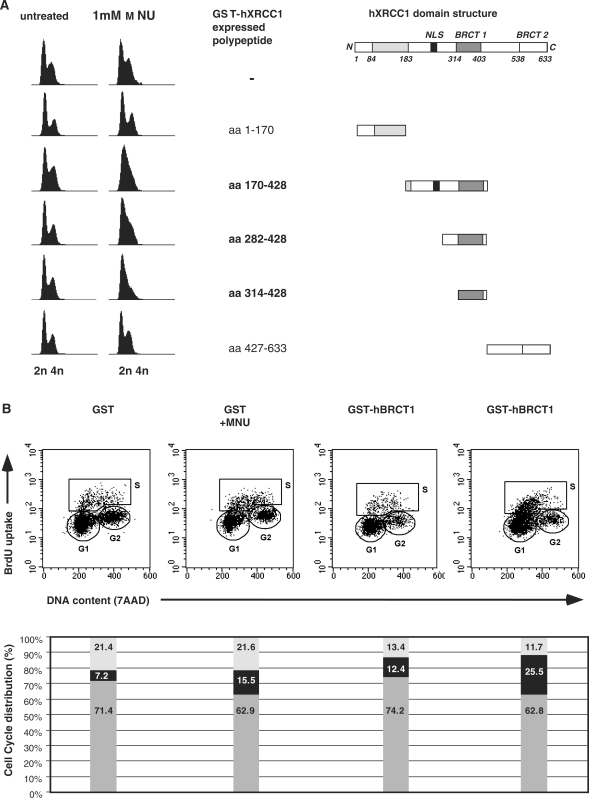
To test this possibility, truncated fragments of hXRCC1 fused to GST were overexpressed in HeLa cells and the progression through the cell cycle was analyzed by flow cytometry (FACS). The cell cycle of untreated transfected cells was not affected by the overexpression of any of the hXRCC1 fusion proteins (Figure 5A). In contrast, 20 h following treatment with the alkylating agent methyl nitrosourea (MNU), cells expressing hXRCC1 fragments containing the BRCT1 domain (amino acids 170–428, 282–428 and 314–428) showed a strong accumulation in S phase leading to a decreased G2/M phase (Figure 5A). Quantification of the DNA synthesis was performed by monitoring BrdU incorporation in cells overexpressing either the hBRCT1 (amino acids 282–428) fused to GST or GST alone, 20 h after treatment with 1 mM MNU (Figure 5B). MNU treatment lead to an increased number of BrdU positive cells that was higher for GST-hXRCC1-BRCT1 than GST overexpression (25% versus 15.5%, respectively, Figure 5B). However, HeLa cells expressing GST were evenly distributed throughout the S phase; whereas, GST-hXRCC1-BRCT1-expressing cells were enriched in the early S phase just after the G1 exit.
Figure 5.
GST-hXRCC1-BRCT1 overexpressing cells accumulate in S phase following DNA damage. (A) Cell cycle analysis (right) of HeLa cells overexpressing GST-tagged fragments of hXRCC1 (schematically represented on the right), treated by 1 mM MNU and analyzed 20 h after. (B) Flow cytometry profiles of BrdU incorporation (DNA synthesis) versus DNA content (7AAD fluorescence) of HeLa cells overexpressing either GST alone or GST-hXRCC1-BRCT1 20 h after treatment or not with 1 mM MNU.
Altogether, these results indicate that the overexpression of the BRCT1 domain of hXRCC1 leads to a strong accumulation of MNU-treated human cells in early S phase. This effect is specific for the BRCT1 domain, since overexpression of GST-hBRCT2 had no particular effect on cell cycle progression whether the cells were treated with alkylating agents (Figure 5A and data not shown).
XRCC1-BRTC1 slows down DNA synthesis in Xenopus egg extracts
To gain further insight into the biological significance of the interaction between XRCC1, PAR and the p58 subunit of Pol α-primase during the replication of damaged DNA, we used Xenopus laevis egg extracts. The cell-free aspect of this system allows the detailed analysis at the molecular level of the different steps of DNA synthesis and its biochemical manipulation, by the addition of either purified proteins to assess for a dominant effect or pharmacological drugs to inhibit specific activities. Introduction of Xenopus sperm chromatin in such extracts results in the assembly of a nuclear membrane around DNA and the execution of a single complete round of semiconservative replication (30). To this end, sperm chromatin treated or not with MMS (‘Materials and Methods’ Section) was introduced into egg extracts synchronized in early S phase and DNA replication was monitored by following the incorporation of a radiolabeled nucleotide precursor. Either GST or GST-xXRCC1-BRCT1 (amino acids 307–414 from X. laevis XRCC1) was added to egg extracts prior to the initiation of DNA synthesis.
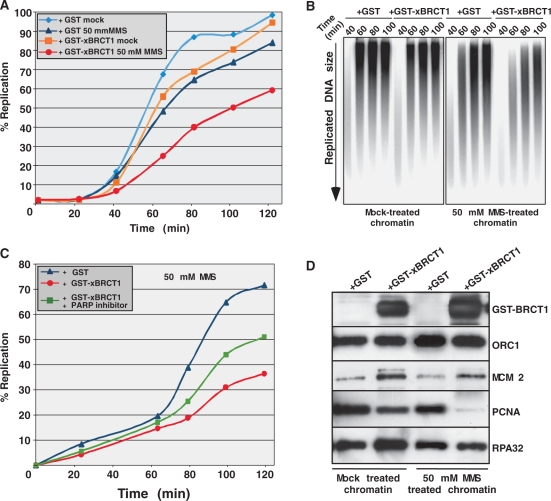
As shown in Figure 6A, replication of sperm chromatin treated with low concentrations of MMS was slightly impeded in extracts supplemented with GST as control (∼ 85% of replication compared to the 100% in the untreated reaction), confirming that replication of alkylated DNA is slowed down in this system (31). Consistent with what was observed in HeLa cells (Figure 5), replication of MMS-treated chromatin was strongly slowed down when egg extracts were supplemented with GST-xXRCC1-BRCT1 (>40% reduction of replication, Figure 6A). To further characterize the defect in DNA synthesis induced by the xXRCC1-BRCT1 domain, replication intermediates were analyzed by alkaline gel electrophoresis. In this assay, nascent DNA is detected as a smear corresponding to growing DNA chains, while fully replicated DNA is visible as high molecular weight species. As expected, nascent DNA accumulated in the presence of MMS compared to the mocked-treated reaction (Figure 6B, compare right to left panel), which is due to a delay in ongoing DNA synthesis. Upon MMS treatment, the addition of GST-xXRCC1-BRCT1 resulted in a much stronger accumulation of nascent DNA compared to the GST control (Figure 6B, right panel). After 100 min of incubation in the presence of GST-XRCC1-BRCT1, very little fully replicated DNA was synthesized, while by this time the replication of mock-treated chromatin was complete. Moreover, the abundance of early replication intermediates (within 40 min) decreased in the presence of GST-xXRCC1-BRCT1 suggesting inhibition of both the initiation and elongation steps of DNA synthesis.
Figure 6.
xXRCC1-BRCT1 inhibits the initiation of DNA synthesis in Xenopus egg extracts. (A) Kinetics of replication of either mock- or MMS-treated sperm chromatin after the addition of either purified GST or GST-xXRCC1-BRCT1 to egg extracts. (B) Size of replication products synthesized in (A) at different times as determined by alkaline gel electrophoresis and autoradiography. (C) Replication reactions were performed as in (A) in the presence of MMS or MMS and PARP inhibitor Ku-0058948 (green curve). (D) Western blot of chromatin-associated proteins after 60 min incubation in Xenopus extracts treated or not with MMS, in the presence of either GST or GST-xBRCT1.
We examined whether PAR production could be observed on MMS-treated sperm chromatin in Xenopus extracts supplemented with GST-xXRCC1-BRCT1. Indeed, PAR production, which was detectable in nuclei treated with MMS, was dramatically increased by the addition of GST-xXRCC1-BRCT1 (data not shown) as observed in HeLa cells. Owing to the inhibitory effect of PAR on primase activity, we hypothesized that the massive PAR production caused by the presence of GST-xXRCC1-BRCT1 could be responsible for the inhibition of replication and that this effect could be reversed by addition of a PARP inhibitor. Results shown in Figure 6C confirmed this hypothesis, since pre-incubation of Xenopus extracts with the PARP inhibitor rescued the DNA synthesis delay induced by the addition of GST-xXRCC1-BRCT1, whereas this inhibitor had no effect on replication of MMS-treated chromatin in the presence of GST (Supplementary Figure 2A). Taken together, these results confirm that the BRCT1 domain of XRCC1 inhibits replication of damaged DNA in Xenopus egg extracts.
XRCC1-BRTC1 binds to chromatin and interferes with formation of functional replication forks
To characterize in more detail the DNA synthesis defect induced by xXRCC1-BRCT1 in the presence of DNA damage, we analyzed the recruitment to chromatin of replication factors specific of the different steps of DNA synthesis. Hence, sperm chromatin was exposed to MMS and incubated in egg extracts supplemented with the indicated proteins, as described in Figure 6A. Chromatin fractions were analyzed by western blot for the binding of the indicated proteins. The GST-xXRCC1-BRCT1 domain bound Xenopus chromatin, and its chromatin association was enhanced by MMS treatment (Figure 6D). Addition of the xXRCC1-BRCT1 domain did not induce phosphorylation of the Chk1 protein kinase in the absence of MMS, demonstrating that the inhibition of DNA synthesis observed when xXRCC1-BRCT1 is added to MMS-treated chromatin is not due to a direct activation of the DNA damage checkpoint (Supplementary Figure 2B). The chromatin binding of ORC1, a subunit of the ORC essential for the assembly of preRCs, and that of RPA32, a component of pre-ICs that binds to single stranded DNA at replication forks, were not significantly affected by the addition of GST-xXRCC1-BRCT1, indicating that pre-RCs and pre-ICs assembled normally. Consistent with this conclusion, the chromatin binding of the pre-RC component MCM2 was not affected, but even it increased following addition of GST-xXRCC1-BRCT1. In contrast, PCNA binding to MMS-treated sperm chromatin was abolished in the presence of GST-xXRCC1-BRCT1 (Figure 6D), while this inhibition was significantly rescued in the presence of PARP inhibitor (Supplementary Figure 2C).
Since PCNA requires the activity of DNA Pol α-primase to bind to chromatin and stimulates replication elongation (32,33), these results strongly argue that the BRCT1 domain of xXRCC1, by interfering with the activity of DNA Pol α -primase, regulates the progression of replication forks through damaged DNA.
DISCUSSION
Several reports have put forward the idea that XRCC1 could play a role in the control of replication fork progression when DNA is damaged. The first indication came from the observation that XRCC1 deficient EM9 cells display high levels of SCEs, reflecting the accumulation of unrepaired SSB converted to DBS at collapsed replication forks (2). Kubota and Horiuchi (4) showed that complementation of EM9 cells with XRCC1 point mutated in the BRCT1 domain could not restore nascent DNA replication after MMS treatment. While the BRCT2 domain of XRCC1 is only required for BER/SSBR during G1, the BRCT1 domain was shown to be critical for efficient repair during G1 but also S/G2 phase of the cell cycle (3). More recently, Brem and Hall (34) found that lowering XRCC1 levels by RNA interference led to a significant delay in S-phase progression after exposure to MMS. XRCC1 was shown to interact with PCNA and both proteins displayed co-localization during S phase of undamaged cells (14). In addition, Parlanti et al. (35) demonstrated the existence of a multiprotein complex containing the DNA replicative polymerases Pol α-δ-ϵ the replication protein MCM7, BER/SSBR components including XRCC1 and Pol β and the cell cycle regulatory protein cyclin A. They proposed that XRCC1 could act as an early effector of cellular response to DNA breaks at stalled replication. All these studies clearly identified XRCC1 as a critical factor acting when replication forks encounter damaged DNA. The idea most commonly advanced was that BER/SSBR machineries are associated to the replication machinery in order to coordinate the repair of DNA lesions with replication fork progression, thus avoiding the conversion of unrepaired damaged bases or single-strand breaks to mutations or highly toxic DSBs during replication. However, the precise role of XRCC1 in this process remained an open question.
In this study, we identified the p58 subunit of the Pol α-primase complex as a novel important element that links XRCC1 to the replication apparatus. We demonstrate an interaction between the BRCT1 domain of XRCC1 and the N-terminal part of the p58 and observed co-localization between XRCC1 and p58 in damaged cell nuclei. In addition, we found that the N-terminal domain of p58 bound PAR, leading to inhibition of primase activity. XRCC1-BRCT1 domain overexpressed in HeLa cells is poly(ADP-ribosyl)ated following DNA damage and led to the accumulation of MNU-treated HeLa cells in early S phase. The inhibition of replication of damaged DNA was also observed in Xenopus egg extracts, suggesting that this regulation is conserved in vertebrates. Looking more deeply into the molecular mechanism, we found that in the presence of DNA damage, the BRCT1 domain of Xenopus XRCC1 did not interfere with the formation of both pre-RCs and pre-ICs, but strongly inhibited the association of PCNA with replicating chromatin. Thus, our results suggest that overexpression of XRCC1-BRCT1 could interfere either with SSBR, as a dominant negative factor, or with the establishment of functional replication forks and/or ongoing DNA synthesis. But the observed interaction of XRCC1-BRCT1 with p58, the distribution of cells in S phase in HeLa XRCC1-BRCT1 and the inhibition of chromatin association of PCNA with MMS-treated chromatin lead us to preferentially propose a role for XRCC1 in the coordination of DNA repair and replication during S phase.
The inhibition of p58 primase activity by PAR in the presence of damaged DNA is strikingly reminiscent of the mechanism recently described in Bacillus subtilis by which guanosine penta- and tetraphosphate [(p)ppGpp] were synthetized by RelA in response to nutritional stress, directly inhibits primase activity to stop ongoing DNA replication (36). It was proposed that this regulation might avoid replication fork collapse during nutrient deprivation and therefore maintain genome integrity. In eukaryotes, PAR may play a similar role in response to DNA damage during S phase. The implication of PARP-1 and poly(ADP-ribosyl)ation in the replication arrest in response to DNA damage has been previously documented. PARP-1 activity was shown to inhibit the replicative polymerase activities of DNA Pol α and -δ and the DNA repair polymerase activity of Pol β in vitro (37,38). We and others previously reported the physical interaction between PARP-1 and the 180 kDa catalytic subunit of DNA Pol α-primase, as well as the impaired S-phase DNA synthesis in PARP-1-deficient or depleted mouse embryonic fibroblasts (39,40). In contrast, our experiments presented here reveal that HeLa cells overexpressing GST-hXRCC1-BRCT1 displayed an increased PAR synthesis under damage conditions and cells accumulated in the early S phase. In addition, the delayed replication of MMS-treated chromatin from Xenopus egg extracts upon addition of GST-xXRCC1-BRCT1 was alleviated in the presence of a PARP inhibitor, thus highlighting the role of PAR in the replication arrest of damaged chromatin. The inhibition of primase activity in vitro by PAR that competes with template DNA points to a critical step subjected to regulation by poly(ADP-ribosyl)ation. Our results shed light onto previous observations by Yoshihara et al. (38) who reported that PARP-1, in the presence of NAD+, could inhibit, in vitro, the DNA Pol α and the primase activities, which are restored by a PARP inhibitor, suggesting that both the primase and polymerase are directly controlled by PAR. Very recently, PARP-1 was also shown to slow down fork progression in response to damage induced by the topoisomerase I inhibitor camptothecin (41).
This points out the need of an appropriate PAR bearing protein, such as XRCC1 through its BRCT1 domain, and suggests that poly(ADP-ribosyl)ated XRCC1 could be a factor that targets p58 to inhibit replication on a damaged template.
Interaction between XRCC1 and primase could also help to stabilize stalled replication forks preventing them from collapsing. Subsequently, degradation of PAR could help to stabilize the replication machinery and promotes its reactivation. This hypothesis refers to the work of Maruta et al. (42) who proposed that PAR degradation by PARG could be a source of ATP required for replisome stability. If replication fork arrest fails when a SSB is present and leads to the generation of a DSB, then the activation of double-strand end-joining repair will be turned on by another function of XRCC1, through its interaction and stimulation of the NHEJ factor DNA-PK (18). In a recent paper, Brem et al. showed that unresolved BER intermediates give rise to lesions activating both ATM and ATR and leading to S-phase delay. This delay is required to repair damages through a recombinational repair pathway involving Rad51 (43).
These findings raise the question about the relevance of the XRCC1-PAR-p58 interaction. Inhibiting primase activity by PAR, which is facilitated by the interactions of XRCC1 with p58 and PARP-1, may give cells more time to repair DNA lesions prior to resume the replication process and thus avoid conversion of SSBs into DSBs. A role for DNA primase, delaying the progression of replication when DNA needs first to be mended, is consistent with its role as a molecular brake in DNA replication when leading strand synthesis is prevented from outspacing lagging strand synthesis, as reported for bacteria and T7 bacteriophage DNA replication (44,45). As the Pol α-primase complex is required for both the initiation of DNA replication and the lagging strand DNA synthesis, it is therefore a likely target for coupling DNA replication to the DNA damage response (46).
We propose that XRCC1 plays a central role in the coordination of DNA repair and replication during S phase. In response to DNA damage, poly(ADP-ribosyl)ated XRCC1 slows down replication via its functional interaction with the p58 subunit of the Pol α-primase complex in order to allow damage repair to take place or acting, if necessary, as a molecular switch between SSBR and Double Strand Break Repair (DSBR).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; Association pour la Recherche contre le Cancer; Electricité de France; Ligue contre le Cancer Comité du Bas-Rhin; Commissariat à l’Energie Atomique and Agence Nationale pour la Recherche; Science Foundation Ireland; Health Research Board, Ireland; INTAS (Brussels, Belgium). Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wished to thank all the members of the team Poly(ADP-ribosyl)ation et intégrité du génome, especially F. Dantzer, for fruitful discussions. Special thanks are due to C. Bouakaze, a summer student from the ESBS, for IP experiments. They also wish to thank M. Mechali for putting them in touch with D. Maiorano and allowing N. Levy to perform experiments in Montpellier's laboratory.
REFERENCES
- 1.Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutat. Res. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 2.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 3.Taylor RM, Thistlethwaite A, Caldecott KW. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol. Cell Biol. 2002;22:2556–2563. doi: 10.1128/MCB.22.8.2556-2563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubota Y, Horiuchi S. Independent roles of XRCC1’s two BRCT motifs in recovery from methylation damage. DNA Repair. 2003;2:407–415. doi: 10.1016/s1568-7864(02)00242-2. [DOI] [PubMed] [Google Scholar]
- 5.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 7.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor RM, Wickstead B, Cronin S, Caldecott KW. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr. Biol. 1998;8:877–880. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 11.Dillehay LE, Thompson LH, Minkler JL, Carrano AV. The relationship between sister-chromatid exchange and perturbations in DNA replication in mutant EM9 and normal CHO cells. Mutat. Res. 1983;109:283–296. doi: 10.1016/0027-5107(83)90053-2. [DOI] [PubMed] [Google Scholar]
- 12.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., III XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiorano D, Lutzmann M, Mechali M. MCM proteins and DNA replication. Curr. Opin. Cell Biol. 2006;18:130–136. doi: 10.1016/j.ceb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 2007 doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider A, Smith RW, Kautz AR, Weisshart K, Grosse F, Nasheuer HP. Primase activity of human DNA polymerase alpha-primase. Divalent cations stabilize the enzyme activity of the p48 subunit. J. Biol. Chem. 1998;273:21608–21615. doi: 10.1074/jbc.273.34.21608. [DOI] [PubMed] [Google Scholar]
- 18.Levy N, Martz A, Bresson A, Spenlehauer C, de Murcia G, Menissier-de Murcia J. XRCC1 is phosphorylated by DNA-dependent protein kinase in response to DNA damage. Nucleic Acids Res. 2006;34:32–41. doi: 10.1093/nar/gkj409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisshart K, Forster H, Kremmer E, Schlott B, Grosse F, Nasheuer HP. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 2000;275:17328–17337. doi: 10.1074/jbc.M000717200. [DOI] [PubMed] [Google Scholar]
- 20.Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 21.Brizard JP, Carapito C, Delalande F, Van Dorsselaer A, Brugidou C. Proteome analysis of plant-virus interactome: comprehensive data for virus multiplication inside their hosts. Mol. Cell Proteomics. 2006;5:2279–2297. doi: 10.1074/mcp.M600173-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Richert S, Luche S, Chevallet M, Van Dorsselaer A, Leize-Wagner E, Rabilloud T. About the mechanism of interference of silver staining with peptide mass spectrometry. Proteomics. 2004;4:909–916. doi: 10.1002/pmic.200300642. [DOI] [PubMed] [Google Scholar]
- 23.Dantzer F, Giraud-Panis MJ, Jaco I, Ame JC, Schultz I, Blasco M, Koering CE, Gilson E, Menissier-de Murcia J, de Murcia G, et al. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol. Cell Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosse F, Nasheuer HP, Scholtissek S, Schomburg U. Lactate dehydrogenase and glyceraldehyde-phosphate dehydrogenase are single-stranded DNA-binding proteins that affect the DNA-polymerase-alpha-primase complex. Eur. J. Biochem. 1986;160:459–467. doi: 10.1111/j.1432-1033.1986.tb10062.x. [DOI] [PubMed] [Google Scholar]
- 25.Menut S, Lemaitre JM, Hair A, Méchali M. DNA replication and chromatin assembly using Xenopus egg extracts. In: Richter JD, editor. Advances in Molecular Biology: A Comparative Methods Approach to the Study of Oocytes and Embryos. New York, USA: Oxford University Press; 1988. [Google Scholar]
- 26.Stokes MP, Michael WM. DNA damage-induced replication arrest in Xenopus egg extracts. J. Cell Biol. 2003;163:245–255. doi: 10.1083/jcb.200306006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 28.Maiorano D, Cuvier O, Danis E, Mechali M. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell. 2005;120:315–328. doi: 10.1016/j.cell.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Maiorano D, Lemaitre JM, Mechali M. Stepwise regulated chromatin assembly of MCM2-7 proteins. J. Biol. Chem. 2000;275:8426–8431. doi: 10.1074/jbc.275.12.8426. [DOI] [PubMed] [Google Scholar]
- 30.Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 31.Stokes MP, Van Hatten R, Lindsay HD, Michael WM. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 2002;158:863–872. doi: 10.1083/jcb.200204127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005);40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 33.Philpott A, Yew PR. The Xenopus cell cycle: an overview. Mol. Biotechnol. 2008;39:9–19. doi: 10.1007/s12033-008-9033-z. [DOI] [PubMed] [Google Scholar]
- 34.Brem R, Hall J. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res. 2005;33:2512–2520. doi: 10.1093/nar/gki543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parlanti E, Locatelli G, Maga G, Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eki T. Poly (ADP-ribose) polymerase inhibits DNA replication by human replicative DNA polymerase alpha, delta and epsilon in vitro. FEBS Lett. 1994;356:261–266. doi: 10.1016/0014-5793(94)01280-6. [DOI] [PubMed] [Google Scholar]
- 38.Yoshihara K, Tanaka Y, Itaya A, Kamiya T, Hironaka T, Minaga T, Koide SS. In vitro evidence for poly(ADP-ribosyl)ation of DNA polymerasea-primase, & phosphorylation of poly(ADP-ribose) synthetase by protein kinase C. In: Jacobson MK, Jacobson EL, editors. I. ADP-ribose transfer reactions. Mechanisms and biological significance. Springer-Verlag; 1989. pp. 39–46. [Google Scholar]
- 39.Dantzer F, Nasheuer HP, Vonesch JL, de Murcia G, Menissier-de Murcia J. Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: a link between DNA strand break detection and DNA replication. Nucleic Acids Res. 1998;26:1891–1898. doi: 10.1093/nar/26.8.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simbulan-Rosenthal CM, Rosenthal DS, Iyer S, Boulares H, Smulson ME. Involvement of PARP and poly(ADP-ribosyl)ation in the early stages of apoptosis and DNA replication. Mol. Cell Biochem. 1999;193:137–148. [PubMed] [Google Scholar]
- 41.Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol. 2008;183:1203–1212. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maruta H, Okita N, Takasawa R, Uchiumi F, Hatano T, Tanuma S. The involvement of ATP produced via (ADP-Ribose)n in the maintenance of DNA replication apparatus during DNA repair. Biol. Pharm. Bull. 2007;30:447–450. doi: 10.1248/bpb.30.447. [DOI] [PubMed] [Google Scholar]
- 43.Brem R, Fernet M, Chapot B, Hall J. The methyl methanesulfonate induced S-phase delay in XRCC1-deficient cells requires ATM and ATR. DNA Repair. 2008;7:849–857. doi: 10.1016/j.dnarep.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 45.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 46.Foiani M, Lucchini G, Plevani P. The DNA polymerase alpha-primase complex couples DNA replication, cell-cycle progression and DNA-damage response. Trends Biochem. Sci. 1997;22:424–427. doi: 10.1016/s0968-0004(97)01109-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.