Abstract
SeqA is a negative regulator of DNA replication in Escherichia coli and related bacteria that functions by sequestering the origin of replication and facilitating its resetting after every initiation event. Inactivation of the seqA gene leads to unsynchronized rounds of replication, abnormal localization of nucleoids and increased negative superhelicity. Excess SeqA also disrupts replication synchrony and affects cell division. SeqA exerts its functions by binding clusters of transiently hemimethylated GATC sequences generated during replication. However, the molecular mechanisms that trigger formation and disassembly of such complex are unclear. We present here the crystal structure of a dimeric mutant of SeqA [SeqAΔ(41–59)-A25R] bound to tandem hemimethylated GATC sites. The structure delineates how SeqA forms a high-affinity complex with DNA and it suggests why SeqA only recognizes GATC sites at certain spacings. The SeqA–DNA complex also unveils additional protein–protein interaction surfaces that mediate the formation of higher ordered complexes upon binding to newly replicated DNA. Based on this data, we propose a model describing how SeqA interacts with newly replicated DNA within the origin of replication and at the replication forks.
INTRODUCTION
Initiation of replication is the cascade of events that causes unwinding of DNA at an origin of replication. In Escherichia coli, regulation of replication initiation ensures that the chromosome is replicated once, but only once, during the cell cycle. Three main processes control timing and synchrony of replication initiation: regulatory inhibition of the initiator protein DnaA (RIDA), titration of free DnaA and sequestration by SeqA (1–3). The initiator protein DnaA forms two different complexes with the origin of replication (oriC). These are analogous to the eukaryotic origin recognition and the pre-priming complexes (4). The origin recognition complex is formed when DnaA binds to the three high-affinity DnaA boxes within oriC. This complex persists throughout most of the cell cycle, whereas assembly of a pre-priming complex only occurs at the time of initiation of DNA synthesis and requires binding of DnaA to high and low-affinity recognition sites in an ATP-dependent fashion (5). Upon initiation of replication, the SeqA protein forms a high-affinity complex with transiently hemimethylated GATC sites within oriC that partially overlap low-affinity DnaA boxes. This process, known as sequestration of oriC, represses the assembly of the pre-priming complex and, in turn, ensures that all origins are reset to form an origin recognition complex before a new round of replication starts (6,7). SeqA sequestration is also important for survival of replication fork damage because it prevents convergence of forks upon DNA damage (8).
Fluorescently labeled SeqA proteins form visible foci in the cells. These appear to be clusters of SeqA bound to newly replicated DNA at the replication forks rather than at the origin of replication (9–11). Formation of these foci depends on Dam methylation and ongoing DNA replication, but not on the presence of oriC (10,12,13). This suggests that SeqA binding to newly replicated DNA also plays a role in organization of the chromosome. Indeed, seqA null strains exhibit increased negative superhelicity and abnormal localization of nucleoids (14,15), and mutation of the condensin-like protein MukB, a known participant in DNA segregation, has a mutually suppressive effect (16). SeqA binding beyond the origin is also necessary for a full stringent response and cell cycle arrest (17). Excess SeqA also interferes with nucleoid segregation, causing a delay in cell division and affecting topoisomerase IV activities (18,19). These roles of SeqA have also been identified in bacteria bearing more complex genomes than E. coli such as Vibrio cholerae (20).
SeqA has two functional domains: an N-terminal oligomerization domain (residues 1–35), and a C-terminal DNA-binding domain (residues 64–181) (21). The C-terminal domain of SeqA (SeqA-C), binds specifically to hemimethylated GATC sequences (21), but it can also recognize other hemimethylated sequences to a lesser extent (22). The N-terminal domain mediates dimerization of SeqA. SeqA dimers further associate to form left-handed spiral linear polymers (23). While SeqA dimer formation is sufficient to form a high-affinity complex with DNA, filament formation is required for proper function in vivo. A molecular model based on the crystal structures of the N- and C-terminal domains of SeqA suggests that the SeqA foci at the replication forks could be SeqA polymers restraining negative supercoils on newly replicated DNA (23). In agreement with this, wild type SeqA restrains negative supercoils in DNA (24). Interestingly, SeqA mutants with impaired ability to form filaments have been shown to introduce positive supercoils in DNA (24). A wealth of information regarding how various mutations affect the activities of SeqA has been generated (23–26). However, the molecular interactions that trigger the formation of a high-affinity complex of SeqA with newly replicated DNA have remained unclear.
In an effort to understand the topological constraints imposed by the SeqA dimer on newly replicated DNA, we have solved the crystal structure of a SeqA mutant unable to form oligomers larger than dimers bound to a DNA duplex containing two adjacent hemimethylated GATC sequences. The structure reveals how SeqA forms a high-affinity complex with newly replicated DNA and why it prefers to bind to GATC sites that are on the same face of the DNA helix. The organization of the linkers connecting the dimerization and DNA-binding domains suggests that SeqA uses a stepwise unwinding mechanism to bind pairs of GATC sequences with different spacings. Moreover, the association of SeqA–DNA complexes in the crystal unveils additional protein–protein interaction surfaces within the DNA-binding domain of SeqA that do not play a role in replication synchrony but could affect chromosome organization at the replication fork. Based on these findings, we propose a model to explain the independent roles of SeqA at oriC and the replication forks.
MATERIALS AND METHODS
Crystallization and structure determination
SeqAΔ(41–59)-A25R was over-produced and purified as previously described (27). Oligonucleotides 5′GAGTCG(mA)TCGGCGGG(mA)TCCTTA3′ and 5′TCTAAGGATCCCGCCGATCGAC3′ were annealed to yield a 20-bp duplex encompassing two hemimethylated GATC sites. Crystallization and data collection details of SeqAΔ(41–59)-A25R bound to DNA were described elsewhere (27). The initial phases were determined by molecular replacement using PHASER (28) and the structures of the N- and C-terminal domains of SeqA as models (PDB ID: 1XRX and 1LRR). The structure was refined using standard protocols in REFMAC and PHENIX.REFINE (29,30).
SeqA mutants
All SeqA mutants were derived from a pET11a plasmid encoding wild-type SeqA (pSS1) using QuikChange site-directed mutagenesis kit (Stratagene). Sequences of the mutants were verified by DNA sequencing (MOBIX Laboratory at McMaster). Mutant SeqA proteins were over-produced and purified as described elsewhere (23).
Electromobility shift assays
The same oligonucleotides used for crystallization were annealed and used for electromobility shift assays. Constant amounts of DNA duplex (80 nM) were incubated with increasing amounts of protein (15–250 nM) and incubated 15 min at room temperature followed by 30 min at 4°C. Samples were resolved on 10% native TBE gels and stained with SYBR Green (1:10 000) (Cambrex, Inc.). Bands were quantified using the ImageJ software (http://rsbweb.nih.gov/ij/index.html). Experiments were run in triplicate and the kD was estimated from the average plots.
DNA-binding specificity assays
The randomly-chosen 72-bp sequence and the design of hemimethylated DNA duplexes with two GATC sites at various spacings used for SeqA protein binding were as described earlier (9,31). The preparation and radioactive labeling of hemimethylated duplex DNA and the conditions for the SeqA binding electrophoretic mobility shift assay (EMSA) were done as described previously (31,32). Unless otherwise stated, SeqA protein binding assays were performed using 0.2 μM of either wild-type or mutant SeqA protein.
Flow cytometry assays
Host strain BL21DE3/pLysS was made ΔseqA::tet by P1 transduction with lysate from MM294 ΔseqA::tet (a kind gift from Dr Kleckner). This strain was supplemented with pET11a derivatives encoding: SeqA (pSS1), SeqA-A25R (pAG8015), SeqAΔ(41–59) (pAG8023), SeqAΔ(41–59)-A25R (pAG8033), SeqA-R70S-R73S (pAG8270), SeqA-A25R-R70S-R73S (pAG8268) and SeqAΔ(41–59)-A25R-R70S-R73S (pAG8269). In each case, the average number of origins per cell was determined by the flow cytometry ‘run-off’ method with modifications (33). Overnight cultures were grown in the absence or presence of 25 μM IPTG at 37°C in M63 minimal media with the appropriate antibiotics. The overnight cultures were diluted to an OD600 of 0.02 and grown to an OD600 ∼0.1 prior to incubation for 3 h with rifampicin (200 μg/ml) and cephalexin (36 μg/ml). After fixing with 77% ethanol, cells were analyzed in a Bryte SH flow cytometer (Biorad) using WinBryte software (Figure 1) and in an Apogee A40-Mini FCM flow cytometer (Apogee Flow Systems) using Apogee Histogram Software version 1.94 (Figure 4D).
Figure 1.
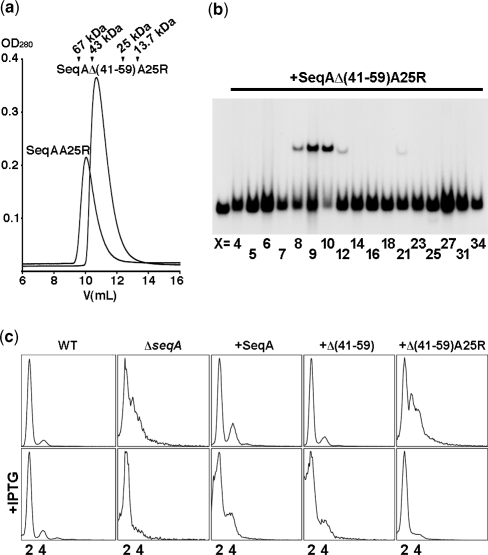
Characterization of the SeqAΔ(41–59)-A25R mutant. (a) Elution profiles of SeqA-A25R (20 315 Da) and SeqAΔ(41–59)-A25R (18 513 Da) over a Superdex-75 size exclusion chromatography column (GE Healthcare). Elution volumes of albumin (67 kDa), ovoalbumin (43 kDa), chymotrypsinogen A (25 kDa) and ribonuclease A (13.7 kDa) are indicated. (b) Electrophoretic mobility shift assay of SeqAΔ(41–59)-A25R with DNAs containing two hemimethylated GATC sequences separated by a variable number of base pairs (X). The left-most lane contains an equimolar mixture of DNAs with 5, 7, 12, 21, 25 and 34 base pairs between the two GATC sites but no protein. (c) From left to right, flow cytometry profiles of a wild-type strain, the ΔseqA::tet strain and the ΔseqA::tet strain transformed with pET11a plasmids encoding wild-type SeqA, SeqAΔ(41–59), and SeqAΔ(41–59)-A25R. Wild-type SeqA and SeqAΔ(41–59) restore replication synchrony, however synchrony is lost upon protein over-expression. Conversely, SeqAΔ(41–59)-A25R only restores replication synchrony upon protein overexpression by addition of 25 μM IPTG.
Figure 4.
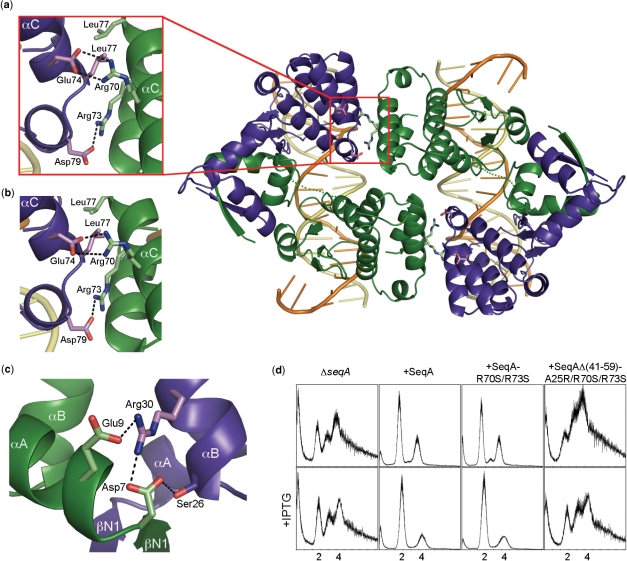
Additional aggregation surfaces in SeqA. (a) Ribbon diagram depicting the protein–protein interaction surfaces in the DNA-SeqAΔ(41–59)-A25R complex relating the two complexes of the asymmetric unit. Relevant residues involved in polar interactions are depicted as color-coded sticks. A detailed view of the interaction surface between the adjacent SeqAΔ(41–59)-A25R dimers with hydrogen bonds depicted as dashed lines is shown on the left-hand side. (b) Detail of the interface between the two SeqA-C molecules [PDB ID: 1LRR and (21)]. (c) Network of hydrogen bonds between Asp7, Glu9 and Arg30. (d) Flow cytometry profiles of the ΔseqA::tet strain and the ΔseqA::tet strain transformed with pET11a plasmids encoding wild-type SeqA, SeqA-R70S/R73S and SeqAΔ(41–59)-A25R/R70S/R73S in the absence (top) or presence of 25 μM IPTG (bottom). Panels (a–c) were prepared using PyMol (42).
RESULTS AND DISCUSSION
DNA-binding plasticity of SeqA
We had previously shown that the SeqA-A25R mutant disrupts the ability of SeqA to form filaments and hence causes loss of replication synchrony (23). However, replication synchrony of the SeqA-A25R mutant could be restored by protein over-expression suggesting that filament formation aids the assembly of a sequestered origin complex, but that the complex can still form in the presence of a high local concentration of SeqA. For this study, a double mutant of SeqA lacking residues 41–59 within the linker connecting the two functional domains and carrying the A25R point mutation was generated (27). Similarly to SeqA-A25R, the SeqAΔ(41–59)-A25R double mutant was stable at low ionic strength and eluted from a size-exclusion column at a volume consistent with the formation of a dimer (Figure 1A). Deletion of residues 41–59 abolished the ability of SeqA to bind hemimethylated GATC sequences separated by more than one helical turn (Figure 1B). This restricted DNA binding favored crystallization of SeqAΔ(41–59)-A25R bound to a tandem GATC repeat. The plasmid encoding SeqAΔ(41–59) restored replication synchrony of the ΔseqA::tet strain similarly to wild-type SeqA (Figure 1C), suggesting that filament formation can compensate the DNA-binding defects caused by the deletion of the linker region. However, the SeqAΔ(41–59)-A25R double mutant behaved similarly to SeqA-A25R (23) and only restored replication synchrony upon protein over-expression as seen by the even number of chromosome equivalents (Figure 1C).
Structure determination of SeqAΔ(41–59)-A25R bound to DNA
SeqAΔ(41–59)-A25R was crystallized in complex with a hemimethylated duplex containing two GATC sequences separated by 9 bp (27). The structure was solved by molecular replacement using the structures of the N- and C-terminal domains of SeqA (PDB codes 1XRX and 1LRR, respectively) and refined using standard protocols in REFMAC and PHENIX.REFINE (29,30). The asymmetric unit contains two identical protein–DNA complexes related by a 2-fold axis. The final model comprises two copies of protomer A (residues 1–40/60–181), two copies of protomer B (residues 1–35 and 60–181), two copies of the hemimethylated DNA duplex (with the exception of nucleotide Cyt2 from the unmethylated strands), 32 water molecules and four 2-methyl-2,4-pentanediol (MPD) molecules (Table 1 and Figure 2). Over 97% of the residues lie in the most favored regions of the Ramachandran plot, and none in disallowed regions.
Table 1.
Refinement statistics
| SeqAΔ(41–59)-A25R | |
|---|---|
| Refinement | |
| Resolution (Å) | 35–3.0 |
| No. reflections (work/test) | 30 285/1533 |
| Rwork/Rfree | 21.9/24.5 |
| No. atoms | |
| Protein | 5142 |
| DNA | 1758 |
| MPD | 4 |
| Water | 32 |
| B-factors | |
| Protein/DNA | 68.52 |
| Solvent | 46.59 |
| R.m.s.d | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.877 |
Figure 2.
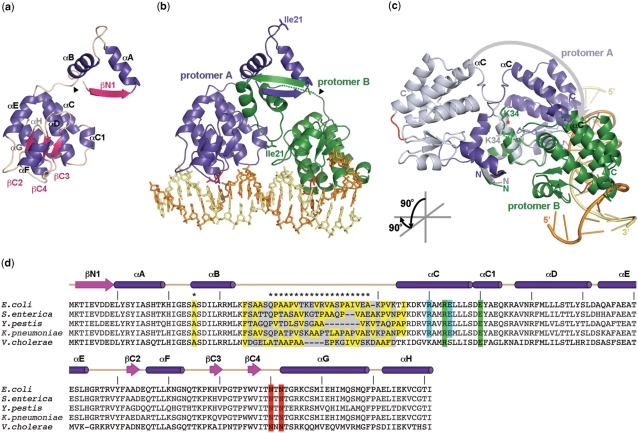
Structure of SeqAΔ(41–59)-A25R bound to a hemimethylated GATC repeat. (a) Ribbon diagram of the SeqAΔ(41–59)-A25R monomer with helices in purple and strands in pink. The junction between residues Gln40 and Lys60 is indicated with a black arrowhead. (b) SeqAΔ(41–59)-A25R dimer bound to DNA. Protomer A (encompassing residues 1–40/60–181) is shown in purple and protomer B (encompassing residues 1–35/60–181) is shown in green. The disordered linker in protomer B is shown as a green dotted line. DNA binding loops are shown in red with the side-chains of residues Asn150 and Asn152 as red sticks. Ile21 at the tip of the αA–αB loops are shown as sticks. The hemimethylated DNA is shown in orange (methylated strand) and light yellow (unmethylated strand) with the methylated adenine in brown. (c) Ribbon diagram depicting how the C-terminal domain breaks the two-fold symmetry of the N-terminal domain. The SeqAΔ(41–59)-A25R dimer is shown in purple (protomer A) and green (protomer B). Protomer A superimposed onto the N-terminus of protomer B is shown in light-grey. The first α-helix on the C-terminal domain (αC) is labeled for reference. The grey arrow indicates the rotation of the C-terminal domain around Lys34 (red asterisk and sticks). The cartoon illustrates the transformation from (b to c). (d) Sequence alignment of SeqA from Escherichia coli K-12, Salmonella enterica serovar Typhi str. CT18, Yersinia pestis CO92, Klebsiella pneumoniae MGH 78578 and Vibrio cholerae 623-39 (top to bottom). Secondary structure motifs from SeqAΔ(41–59)-A25R are shown. The deletion and point mutations in SeqAΔ(41–59)-A25R are marked with asterisks. Ala25 and hydrophobic residues within the linker region are highlighted in yellow. The variable linker between the two functional domains is shadowed in grey. DNA-binding residues are highlighted in red and those involved in reciprocal salt-bridges between neighbour C-terminal domains in green and blue. Residues (a–c) were prepared using PyMol (42).
Organization of SeqA functional domains
The SeqA monomer is organized into two domains, an N-terminal oligomerization domain (residues 1–33) and a C-terminal DNA-binding domain (residues 65–181) joined by a flexible linker (residues 34–64). Each SeqA monomer includes four β-strands (βN1 at the N-terminus, and βC2, βC3 and βC4 that define the small anti-parallel β-sheet within the C-terminal domain), and nine α-helices (αA and αB at the N-terminus and αC, αC1, αD, αE, αF, αG and αH forming the C-terminal domain) (Figure 2A and D). Formation of the SeqAΔ(41–59)-A25R dimer is mediated exclusively by the N-terminal domain, whereas the two DNA-binding domains of the dimer are unrelated to one another (Figure 2B and C). Notably, the A25R mutation did not change the extended conformation of the αA–αB loop (residues 18–24), leaving the side-chain of isoleucine 21 completely exposed to solvent (Figure 2B). Minor changes on the tracing of the main-chain at loop αA–αB were attributed to crystal-packing environment.
Although SeqA can recognize pairs of hemimethylated GATC sites with the methyl groups on the same or opposite DNA strands in vitro (31), newly replicated GATC sequences have all methyl groups on the template strand. Binding in this configuration forces the SeqA dimer to re-arrange its DNA-binding domains to recognize tandem hemimethylated GATC sites. Indeed, while the two N-terminal domains in the SeqA dimer are related by a 2-fold axis, the symmetry of SeqAΔ(41–59)-A25R is lost at the flexible linker joining the N- and C-terminal domains (Figure 2B and C). This linker mediates the 180°-rotation of one of the DNA-binding domains required to recognize a tandem GATC repeat (Figure 2C).
In protomer A, the linker is completely ordered and mainly helical, with helix αB encompassing residues Arg25 to Ser39 (Figure 2A–C). However, the electron density of this linker in protomer B was very weak. Consequently, residues Ser36 to Gln40 were not included in the final model (Figure 2B). Lys34 is the pivotal point that re-orients the C-terminal domains towards the target DNA. While in protomer A Lys34 is part of helix αB, Lys34 exchanges the orientations of its main and side chains in protomer B, breaking the 2-fold symmetry (Figures 2C and 3D). Note that the two protomers of the dimer cannot adopt the conformation seen in protomer A concurrently as this would cause steric hindrance at residues Ala37-Gln40 (Figure 2C). Therefore, Lys34 may be an intrinsically flexible point in the SeqA protein even in the absence of DNA.
Figure 3.
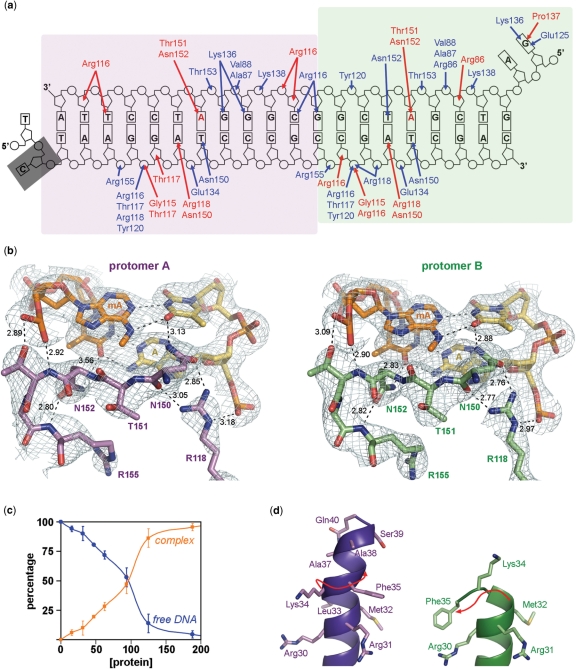
Recognition of hemimethylated DNA by SeqAΔ(41–59)-A25R. (a) Diagram of the protein–DNA interactions. The purple and green shadow boxes indicate the SeqAΔ(41–59)-A25R protomer that interacts with each GATC site with hydrogen bonds shown in blue and van der Waals interactions in red. The methylated adenines are labeled in red and the disordered Cyt2 is shadowed with a grey box. (b) Detail of the interaction between SeqAΔ(41–59)-A25R and the methylated and unmetylated A–T base pairs. The refined model is shown as sticks with protomer A shown in purple (left panel), protomer B in green (right panel), the methylated DNA strand in orange and the unmethylated DNA strand in yellow with the 2Fo−Fc electron density maps contoured at 1 σ. Hydrogen bonds are shown as black dashed lines with distances labeled. (c) Electrophoretic mobility shift assays of the oligonucleotide used for crystallization (80 nM) when incubated with increasing quantities of SeqAΔ(41–59)-A25R (nM). (d) Detail of helix αB in the two protomers of the dimer. Red arrows indicate the path of the main chain on each protomer. Panels (b and d) were prepared using PyMol (42).
DNA conformation
The 22-bp long oligonucleotides used for crystallization formed a 20-bp DNA duplex with two overhanging nucleotides at each 5′-end (Figure 3A). The overhanging dinucleotide on the methylated strand projects away from the duplex helical axis and interacts with the C-terminal domain of protomer B (Figure 2B and C). The overhanging dinucleotide on the unmethylated strand is more flexible and only the 5′ thymine, which stacks on top of adenine 22 from the methylated strand, could be fitted on the electron density maps (Figure 3A).
The duplex portion of the DNA molecule adopted an overall B-DNA conformation with local distortions around the methylated mA-T base pair (Table 2). The N6-methylated A (mA) maintains both canonical hydrogen bonds with its paired T, but the propeller twist between the bases is −19° for both methylated sites rather than the average −11.4° (34,35). This distortion is also present in the structure of the isolated C-terminal domain (SeqA-C) bound to a single hemimethylated GATC sequence, but not in the structures of free DNA encompassing hemi or unmethylated GATC sites. Similarly to SeqA, the mismatch repair protein MutH specifically recognizes hemimethylated GATC sequences. Binding of MutH to un- and hemimethylated GATC sites also imposes significant distortions on the DNA (36), however the nature of these distortions is different to those imposed by SeqAΔ(41–59)-A25R binding to DNA.
Table 2.
DNA parameters at the hemimethylated GATC sites
| Structure | PDB ID | GATC | GATC methylation | Propeller mAT | Opening mAT | Roll mA→T | Inclination mA→T | Helical twist |
||
|---|---|---|---|---|---|---|---|---|---|---|
| G→mA | mA→T | T→C | ||||||||
| SeqAΔ4-A25R/DNA | 3FMT | 2 | Hemimeth. | −17.0 | −5.7 | −14.3 | −21.1 | 43.0 | 40.6 | 29.8 |
| −19.3 | −5.9 | −13.6 | −21.2 | 45.2 | 38.4 | 33.3 | ||||
| SeqA-C/DNA | 1LRR | 1 | Hemimeth. | −21.2 | −9.7 | −10.3 | −17.0 | 46.1 | 35.9 | 31.5 |
| MutH/DNA | 2AOQ | 1 | Unmethylated | −6.5 | 0.5 | 0.9 | 1.7 | 40.1 | 31.7 | 20.0 |
| MutH/DNA | 2AOR | 2 | Hemimeth. | −6.3 | −0.7 | 1.8 | 3.3 | 40.4 | 31.8 | 20.3 |
| −5.8 | 0.6 | −0.7 | −1.2 | 40.5 | 32.0 | 20.5 | ||||
| GATC duplex | 1OPQ | 1 | Unmethylated | 11 | −8.7 | −12.3 | −19.4 | 38.1 | 37.7 | 32.6 |
| GATC duplex | 1OQ2 | 1 | Hemimeth. | −8.3 | 4.1 | −7.2 | −18.5 | 48.6 | 23.0 | 38.0 |
| 1DA3 | 2 | Hemimeth. | −9.3 | −3.7 | 4.2 | 8.0 | 36.5 | 30.5 | 32.5 | |
| −11.5 | −0.1 | −3.3 | −5.9 | 36.3 | 33.0 | 35.6 | ||||
| Average B-DNA (35) | −11.4 | 0.6 | 0.6 | 2.1 | 36.5 | |||||
Other significant distortions (namely roll and inclination) were also seen at the mA–T→T–A base-pair junction (Table 2). However, they are not discussed further because similar distortions are found in the NMR structures of free DNA encompassing a single GATC site (37), and hence cannot be caused by protein binding. Interestingly, a recent structural study on a fully methylated GATC sequence reveals that methylation of the second strand compensates the base-opening distortions introduced by the first methyl group (38), suggesting that distortions at a hemimethylated GATC sequence are both a requisite and a consequence of specific binding of SeqA and MutH.
Interestingly, the DNA duplex is significantly overwound at the base-pair junctions between G–C→mA–T and mA–T→T–A. This overwinding is partially compensated by the underwinding at the T–A→C–G junction of the GATC site (Table 2). It is tempting to speculate that duplex overwinding is the mechanism used by dimeric SeqA to introduce positive supercoils into DNA (24). However, the minor overwinding detected in our structure (11° over 19-bp steps) could also be related to the overall DNA sequence. In fact, the G–C→mA–T base-pair junction is also significantly overwound in the crystal structures of MutH bound to DNA, as well as, the NMR structures of free unmethylated and hemimethylated DNA (Table 2). Conversely, the high-resolution crystal structure of a hemimethylated GATC site does not show this distortion (39), suggesting that the GATC sequence is intrinsically dynamic and that protein binding may stabilize a specific DNA conformation.
DNA-SeqAΔ(41–59)-A25R interactions
Although it had been predicted that the dimerization domain of SeqA would introduce restraints in the complex forcing the interaction with the 4 bp of the GATC site (21,22), the structure of SeqA(Δ41-59)-A25R bound to DNA exhibited the same sequence-specific interactions seen on the structures of the isolated C-terminal domain bound to a single GATC site (Figure 3A–B). Hence, the presence of the N-terminal domain does not seem to modulate the specificity of the interaction with hemimethylated GATC sites. However, the binding of a second GATC site significantly increases the stability of the DNA-SeqAΔ(41–59)-A25R complex (kD ∼90 nM), suggesting that the binding cooperativity of the SeqA dimer might be related to fulfilling its valence (Figure 3C).
Protomer B binds the GATC sequence very similarly to the previous structures; however, protomer A is only bound loosely to its GATC site (Figure 3B). Conceivably, the tighter binding of protomer B could be due to the additional interactions with the overhanging 5′GA dinucleotide. However, the isolated C-terminal domain of SeqA (SeqA–C) achieves a similar interaction with the GATC site in the absence of any additional contacts with DNA (21,22). The linker region, connecting the oligomerization and DNA-binding domains, is ordered and mostly helical (αB) in protomer A, while it is flexible and unstructured in protomer B (Figures 2 and 3D). Unwinding of a helical turn on αB may allow the apparent tighter DNA binding displayed by protomer B. Conversely, the longer αB helix in protomer A may prevent its tight binding with DNA. Interestingly, the last helical turn on αB (Ser36-Gln40) from protomer A no longer maintains the hydrogen bonding pattern expected for an alpha helix, suggesting that this helix is also unwinding to reach its GATC site (Figures 3D). Therefore, the αB of protomer A in the SeqAΔ(41–59)-A25R crystal structure may represent an intermediate state on the formation of a high-affinity complex with DNA.
The linker region (residues Lys34-Lys63) is chiefly hydrophobic, with 18 residues out of 30 being Ala, Ile, Phe, Pro or Val. While the length and sequence of the linker varies among different organisms, its hydrophobic character is conserved (Figure 2D). Some of these residues would be shielded if the linker region forms an amphipathic α-helix running along the surface of the dimer. The presence of several proline residues within the linker would aid on kinking the helix along the surface. Indeed, the C-terminal domains of the SeqAΔ(41–59)-A25R dimer fold against one side of the dimerization domain (Figure 2C) rather than adopting the extended conformation predicted previously (23). This organization only allows interactions with GATC sites that reside approximately on the same face of the DNA and at a very restricted spacing (Figure 1B). Assuming that the full-length SeqA dimer adopts a similar organization, binding of GATC repeats separated by more than one helical turn could be mediated by further stepwise unwindings of the helical linker.
Alternate surfaces facilitate DNA organization beyond oriC
The two SeqA–DNA complexes in the asymmetric unit interact through a reciprocal network of hydrogen bonds and hydrophobic interactions between residues Glu74, Asp79 and Leu77 from protomer A on one dimer and Arg70, Arg73 and Leu77 from protomer B on the adjacent dimer (Figure 4A). Arg70 forms a bi-dentate salt bridge with Glu74, while Arg73 is hydrogen-bonded to Asp79 (Figure 4A). This interaction was also found in the structure of SeqA-C encompassing residues 50–181 bound to DNA (Figure 4B), but not in the structures of shorter SeqA-C mutants lacking Arg70 (22). This interface is relatively small, only ∼400 Å2 of surface is excluded by the pair-wise interactions. However, the residues involved in this interaction are well-conserved (Figure 2D) and their presence in several crystal structures suggests that this surface could contribute to the multimerization properties of SeqA. Indeed, a previous study had already shown that mutation of Lys66 and Arg70 did not affect DNA binding by SeqA but abrogated protein aggregation in vitro and foci formation in vivo (25). Furthermore, conservation of Arg70 and Arg73 is correlated with conservation of Glu74 and Asp79, respectively even in most divergent species such as V. cholerae (Figure 2D). Interestingly, protein–protein association through αC seems to be promoted by DNA binding since SeqA-A25R and SeqAΔ(41–59)-A25R do not form higher order species in solution [Figure 1A and (23)].
To further explore the role of this surface on the function of SeqA, we mutated residues Arg70 and Arg73 and analyzed the ability of the SeqA-R70S/R73S mutant to restore synchrony of replication using flow cytometry. Although the R70S/R73S double mutation did not affect synchrony of replication (Figure 4D), ΔseqA cells transformed with plasmids encoding SeqA-R70S/R73S or SeqAΔ(41–59)-A25R/R70S/R73S grew much slower than ΔseqA cells transformed with plasmids encoding other SeqA mutants, reinforcing the idea that Arg70 and Arg73 are important for the function of SeqA in vivo as it had been previously proposed (25). Therefore, we concluded that the SeqA interaction mediated by Arg70 and Arg73 is not required to sequester or reset oriC. However, it may contribute to additional functions of SeqA in fork management or chromosome segregation.
Conceivably, these weak interactions between αC helices from adjacent SeqA dimers may assist on compacting the SeqA-DNA filament (Figure 5). Interactions through αC may cross-link dimers, either bound to different DNA duplexes (intermolecular cross-link), or bound to the same DNA duplex (intramolecular cross-link). We favor the latter because previous studies of complexes with duplex containing multiple GATC sites do not show the presence of intermolecular cross-linked complexes (40).
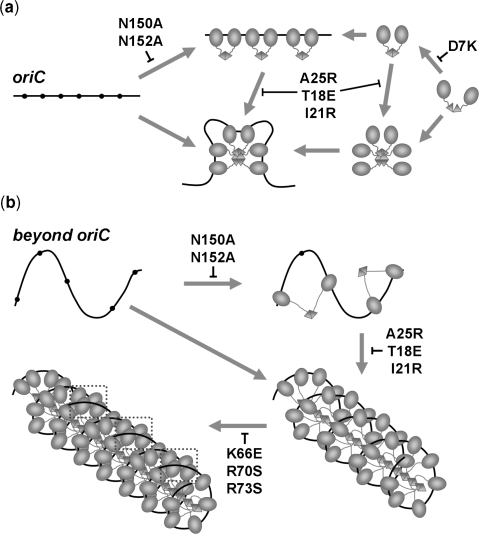
Figure 5.
Modes of operation of SeqA at oriC and the replication forks. (a) Reduced spacing between GATC sites (shown as black dots) at oriC permits sequestration with only few SeqA molecules. Sequestration of the origin is abolished by lack of DNA binding (SeqA-N150A/N152A) and weakened by lack of SeqA oligomerization (SeqA-A25R, SeqA-T18E, SeqA-I21R). (b) SeqA works similarly at the replication forks. However, formation of a left handed SeqA-DNA filament at the forks compensates for the increased spacing between GATC sites. SeqA-DNA filaments are further stabilized by protein–protein interactions through their DNA-binding domains (marked with boxes on the bottom left panel). Mutations in this surface (SeqA-K66E/R70E) affect SeqA-induced DNA aggregation. Mutations destabilizing the dimerization domain (SeqA-D7K, SeqA-E9A) affect the functions of the SeqA dimer but not those of the SeqA filament and, hence, they likely have a more deleterious effect at the oriC.
Distinct roles of SeqA at oriC and the replication forks
Apart from residues Asn150 and Asn152 in the DNA-binding loop (21,22), other key residues have been identified for the proper function of SeqA. Mutants that disrupt filament formation (A25R, T18E or I21R) retain binding to pairs of hemimethylated sites [Figure 5 and (23)], but lose cooperative binding to additional GATC sites (25). SeqA mutants that are unable to form filaments cause replication asynchrony (23). However, synchrony can be restored by protein overexpression, suggesting that the role of SeqA oligomerization is to facilitate saturation of linked sites rather than to remodel the origin of replication (Figure 5A).
Mutants affecting SeqA oligomerization may have a more deleterious effect at the replication forks where the GATC sites are spaced further apart and GATC-binding of few protomers along the SeqA filament might be the only mechanism to form a high-affinity complex with DNA [Figure 5 and (23)]. Thus, the SeqA–DNA complex will likely weaken as the forks enter regions of the chromosome with low GATC content. Indeed, the original study by Campbell and Kleckner already revealed that the rate of adenine methylation by dam methylase was inversely proportional to the distance from oriC with the exception of regions containing GATC clusters like the recB gene (41). In this context, additional weak interactions between neighbouring DNA-binding sites mediated by helix αC may aid in extending the half-life of the SeqA–DNA complex (Figure 5B) and, in turn, assist in organizing newly replicated DNA.
Additional residues within the dimerization domain of SeqA also modulate its interaction with DNA. SeqA-D7K or SeqA-E9K do not bind pairs of GATC sites but can form larger complexes when more than two GATC sites are present (26). Close inspection of the SeqA(Δ41–59)-A25R structure reveals that Asp7 stabilizes the dimerization domain in two ways (Figure 4C). On one hand, it caps and hence stabilizes α-helix αA. On the other hand, it interacts with the side chain of Arg30 from the neighboring protomer, stabilizing the dimerization interface. Similarly, Glu9 is also hydrogen-bonded to Arg30. Mutation of Glu9 to an amino acid bearing an opposite charge (Glu→Arg) disrupts replication synchrony, but mutations removing the net charge of the side chain do not (23). Conceivably, introducing a positive charge (E9R or E9K) in close vicinity to Arg30 may be more disruptive than simply removing the side chain of Glu9 (E9A), thus explaining the different phenotypes observed in these mutants (23,26).
In the absence of the Asp7 or Glu9 anchors, the SeqA dimer probably loosens up, causing the loss of detectable DNA binding by the SeqA dimer (Figures 4C and 5A). However, filament formation would presumably compensate for the weaker dimerization surface, since these mutants display enhanced DNA binding when more than two GATC sites are present (26), suggesting once again that the SeqA filament can compensate the defects of the dimer. Mutations affecting the stability of the SeqA dimer are likely to have a significant impact in oriC sequestration where filament formation plays a mere supportive role (Figure 5A).
In conclusion, the structure of SeqA(Δ41–59)-A25R bound to a pair of hemimethylated GATC sites reveals how SeqA forms a high affinity complex with DNA and starts to unravel how different interaction surfaces contribute either to origin sequestration or DNA aggregation at the replication forks.
COORDINATES
Atomic coordinates and structure factors have been deposited with the Protein Data Bank (accession code 3FMT).
FUNDING
Canadian Institutes of Health Research (MOP-67189 to A.G.); Intramural Research Program of the National Institutes of Health (to S.A.) in part. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the PXRR staff at the NSLS (Brookhaven National Laboratory) for assistance during data collection and Monica Pillon for help with DNA purification.
REFERENCES
- 1.Camara JE, Skarstad K, Crooke E. Controlled initiation of chromosomal replication in Escherichia coli requires functional Hda protein. J. Bacteriol. 2003;185:3244–3248. doi: 10.1128/JB.185.10.3244-3248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kitagawa R, Ozaki T, Moriya S, Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 4.Leonard AC, Grimwade JE. Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol. Microbiol. 2005;55:978–985. doi: 10.1111/j.1365-2958.2004.04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan VT, Grimwade JE, Camara JE, Crooke E, Leonard AC. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol. Microbiol. 2004;51:1347–1359. doi: 10.1046/j.1365-2958.2003.03906.x. [DOI] [PubMed] [Google Scholar]
- 6.Nievera C, Torgue JJ, Grimwade JE, Leonard AC. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell. 2006;24:581–592. doi: 10.1016/j.molcel.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boye E, Stokke T, Kleckner N, Skarstad K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl Acad. Sci. USA. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutera VA, Jr., Lovett ST. The role of replication initiation control in promoting survival of replication fork damage. Mol. Microbiol. 2006;60:229–239. doi: 10.1111/j.1365-2958.2006.05093.x. [DOI] [PubMed] [Google Scholar]
- 9.Brendler T, Sawitzke J, Sergueev K, Austin S. A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J. 2000;19:6249–6258. doi: 10.1093/emboj/19.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiraga S, Ichinose C, Onogi T, Niki H, Yamazoe M. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells. 2000;5:327–341. doi: 10.1046/j.1365-2443.2000.00334.x. [DOI] [PubMed] [Google Scholar]
- 11.Yamazoe M, Adachi S, Kanaya S, Ohsumi K, Hiraga S. Sequential binding of SeqA protein to nascent DNA segments at replication forks in synchronized cultures of Escherichia coli. Mol. Microbiol. 2005;55:289–298. doi: 10.1111/j.1365-2958.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 12.Niki H, Hiraga S. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onogi T, Niki H, Yamazoe M, Hiraga S. The assembly and migration of SeqA-Gfp fusion in living cells of Escherichia coli. Mol. Microbiol. 1999;31:1775–1782. doi: 10.1046/j.1365-2958.1999.01313.x. [DOI] [PubMed] [Google Scholar]
- 14.Bahloul A, Meury J, Kern R, Garwood J, Guha S, Kohiyama M. Co-ordination between membrane oriC sequestration factors and a chromosome partitioning protein, TolC (MukA) Mol. Microbiol. 1996;22:275–282. doi: 10.1046/j.1365-2958.1996.00106.x. [DOI] [PubMed] [Google Scholar]
- 15.Weitao T, Nordstrom K, Dasgupta S. Escherichia coli cell cycle control genes affect chromosome superhelicity. EMBO Rep. 2000;1:494–499. doi: 10.1093/embo-reports/kvd106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weitao T, Nordstrom K, Dasgupta S. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol. 1999;34:157–168. doi: 10.1046/j.1365-2958.1999.01589.x. [DOI] [PubMed] [Google Scholar]
- 17.Ferullo DJ, Lovett ST. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bach T, Krekling MA, Skarstad K. Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division. EMBO J. 2003;22:315–323. doi: 10.1093/emboj/cdg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang S, Han JS, Park JH, Skarstad K, Hwang DS. SeqA protein stimulates the relaxing and decatenating activities of topoisomerase IV. J. Biol. Chem. 2003;278:48779–48785. doi: 10.1074/jbc.M308843200. [DOI] [PubMed] [Google Scholar]
- 20.Saint-Dic D, Kehrl J, Frushour B, Kahng LS. Excess SeqA leads to replication arrest and a cell division defect in Vibrio cholerae. J. Bacteriol. 2008;190:5870–5878. doi: 10.1128/JB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guarné A, Zhao Q, Ghirlando R, Yang W. Insights into negative modulation of E. coli replication initiation from the structure of SeqA-hemimethylated DNA complex. Nat. Struct. Biol. 2002;9:839–843. doi: 10.1038/nsb857. [DOI] [PubMed] [Google Scholar]
- 22.Fujikawa N, Kurumizaka H, Nureki O, Tanaka Y, Yamazoe M, Hiraga S, Yokoyama S. Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein. Nucleic Acids Res. 2004;32:82–92. doi: 10.1093/nar/gkh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarné A, Brendler T, Zhao Q, Ghirlando R, Austin S, Yang W. Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization. EMBO J. 2005;24:1502–1511. doi: 10.1038/sj.emboj.7600634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odsbu I, Klungsoyr HK, Fossum S, Skarstad K. Specific N-terminal interactions of the Escherichia coli SeqA protein are required to form multimers that restrain negative supercoils and form foci. Genes Cells. 2005;10:1039–1049. doi: 10.1111/j.1365-2443.2005.00898.x. [DOI] [PubMed] [Google Scholar]
- 25.Kang S, Han JS, Kim SH, Park JH, Hwang DS. Aggregation of SeqA protein requires positively charged amino acids in the hinge region. Biochem. Biophys. Res. Commun. 2007;360:63–69. doi: 10.1016/j.bbrc.2007.05.225. [DOI] [PubMed] [Google Scholar]
- 26.Kang S, Han JS, Kim KP, Yang HY, Lee KY, Hong CB, Hwang DS. Dimeric configuration of SeqA protein bound to a pair of hemi-methylated GATC sequences. Nucleic Acids Res. 2005;33:1524–1531. doi: 10.1093/nar/gki289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung YS, Guarne A. Crystallization and preliminary X-ray diffraction analysis of SeqA bound to a pair of hemimethylated GATC sites. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008;64:567–571. doi: 10.1107/S1744309108014851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 30.Afonine PV, Grosse-Kunstleve RW, Adams PD. phenix.refine. CCP4 Newsletter. 2005;42 contribution 8. [Google Scholar]
- 31.Brendler T, Austin S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 1999;18:2304–2310. doi: 10.1093/emboj/18.8.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brendler T, Abeles A, Austin S. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 1995;14:4083–4089. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skarstad K, Bernander R, Boye E. Analysis of DNA replication in vivo by flow cytometry. Methods Enzymol. 1995;262:604–613. doi: 10.1016/0076-6879(95)62047-8. [DOI] [PubMed] [Google Scholar]
- 34.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protocol. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu XJ, Neidle S, Shakked Z, et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 36.Lee JY, Chang J, Joseph N, Ghirlando R, Rao DN, Yang W. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell. 2005;20:155–166. doi: 10.1016/j.molcel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Bae SH, Cheong HK, Cheong C, Kang S, Hwang DS, Choi BS. Structure and dynamics of hemimethylated GATC sites: Implications for DNA-SeqA recognition. J. Biol. Chem. 2003;278:45987–45993. doi: 10.1074/jbc.M306038200. [DOI] [PubMed] [Google Scholar]
- 38.Bang J, Bae SH, Park CJ, Lee JH, Choi BS. Structural and dynamics study of DNA dodecamer duplexes that contain un-, hemi-, or fully methylated GATC sites. J. Am. Chem. Soc. 2008;130:17688–17696. doi: 10.1021/ja8038272. [DOI] [PubMed] [Google Scholar]
- 39.Baikalov I, Grzeskowiak K, Yanagi K, Quintana J, Dickerson RE. The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn. J. Mol. Biol. 1993;231:768–784. doi: 10.1006/jmbi.1993.1325. [DOI] [PubMed] [Google Scholar]
- 40.Han JS, Kang S, Lee H, Kim HK, Hwang DS. Sequential binding of SeqA to paired hemi-methylated GATC sequences mediates formation of higher-order complexes. J. Biol. Chem. 2003;278:34983–34989. doi: 10.1074/jbc.M304923200. [DOI] [PubMed] [Google Scholar]
- 41.Campbell JL, Kleckner N. The rate of Dam-mediated DNA adenine methylation in Escherichia coli. Gene. 1988;74:189–190. doi: 10.1016/0378-1119(88)90283-1. [DOI] [PubMed] [Google Scholar]
- 42.DeLano WL. The PyMOL Molecular Graphic Systems. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]