Abstract
Despite being plagued by heavily degraded DNA in palaeontological remains, most studies addressing the state of DNA degradation have been limited to types of damage which do not pose a hindrance to Taq polymerase during PCR. Application of serial qPCR to the two fractions obtained during extraction (demineralization and protein digest) from six permafrost mammoth bones and one partially degraded modern elephant bone has enabled further insight into the changes which endogenous DNA is subjected to during diagenesis. We show here that both fractions exhibit individual qualities in terms of the prevailing type of DNA (i.e. mitochondrial versus nuclear DNA) as well as the extent of damage, and in addition observed a highly variable ratio of mitochondrial to nuclear DNA among the six mammoth samples. While there is evidence suggesting that mitochondrial DNA is better preserved than nuclear DNA in ancient permafrost samples, we find the initial DNA concentration in the bone tissue to be as relevant for the total accessible mitochondrial DNA as the extent of DNA degradation post-mortem. We also evaluate the general applicability of indirect measures of preservation such as amino-acid racemization, bone crystallinity index and thermal age to these exceptionally well-preserved samples.
INTRODUCTION
An integral aspect of any ancient DNA (aDNA) work is to deal with inevitably aged and degraded specimens, most commonly bones or teeth (1). Despite countless samples being available, only few are suitable for aDNA studies as little or no DNA has survived over time (2). In the attempt to facilitate the identification of samples with adequate DNA preservation, indirect measures of preservation have been established (3–6), under the assumption that these parameters correlate with degradation processes to which DNA is subjected during diagenesis. Although many aspects of DNA degradation have been meticulously studied in vitro (7–11), the direct application of this knowledge to the aDNA context remains difficult because of numerous factors contributing to the complexity of DNA break-up in tissues post-mortem (2,12).
While some research has been done on DNA damage since the application of PCR to the field of aDNA, most studies have focused on polymerase-bypassable damage (13–15), analyzed as by-products inherent in the process of sequencing aDNA amplification and cloning products. The analysis of these bypassable damages is crucial in order to distinguish miscoding lesions from real substitutions and thus prevent artificial sequence reconstruction (2). In contrast to this, non-bypassable DNA damage has received far less attention (16,17). The assessment of these forms of damage in aDNA extracts has typically relied on the inverse correlation between amplicon size and the success of PCR amplification simply measured by presence or absence of a PCR product visualized via gel electrophoresis (18,19). The implementation of qPCR has greatly improved the practice of ancient DNA by adding a quantitative component to a successful PCR while at the same time providing insight into sensitivity and efficiency of the reaction itself (20). Despite these obvious advantages however, qPCR is still only marginally applied in aDNA studies. The serial qPCR approach (i.e. the utilization of various amplicon sizes spanning the same locus) enables characterization of aDNA extracts on a much finer scale. It provides a qualitative assessment of samples with respect to DNA content and degradation (1,20), elucidates the differential preservation of endogenous and exogenous DNA (21), and allows for the modeling of the distribution of non-bypassable damage over DNA molecules in an extract (22). Here, we extend the serial qPCR approach to a cross-comparison between samples and extraction fractions, i.e. decalcification product (demineralized supernatant) versus protein digest, in order to characterize the preservation of endogenous aDNA from both mitochondrial and nuclear genomes in bone extracts. To further address the possible effects of DNA degradation over different time scales, we compare both quantitative and qualitative results obtained from permafrost preserved mammoth samples ranging between 12 and >48 ky BP to a year-old ‘modern’ elephant sample. Finally we review the generality and applicability of the repeatedly purported ‘1000 to 1 ratio of mitochondrial to nuclear DNA’ (20,23–25) and address the implications of our results for large-scale palaeogenomic studies.
MATERIALS AND METHODS
Samples
Six permafrost mammoth bone samples along with one modern Asian elephant bone sample have been used in this study (Table 1). Five of the six mammoth samples were obtained from the Cerpolex/Mammuthus collection (Khatanga, Taimyr Peninsula, Siberia) where they have been kept at subzero temperatures since recovered from the permafrost while Ber12 was obtained from a museum collection (Zoological Institute, St. Petersburg, Russia), where it has been stored at low ambient temperature for about 30 years (Table 1). Immediately upon receipt, all mammoth samples were stored at –20°C in a dedicated clean room facility, which is physically separated from post PCR labs (http://socserv.mcmaster.ca/adna/labs.htm). We chose these six samples to compile a set representing a range form fair to excellent in terms of DNA yield, based on previously obtained qPCR results targeting mitochondrial DNA (26).
Table 1.
Bone samples used in this study
| Sample ID | Species | Skeletal element | 14C date | Geographic origin | Latitude/Longitude | Excavated | Storage temperature since excavation |
|---|---|---|---|---|---|---|---|
| Ber12 | M. primigenius | Left femur | 12 350 ± 35 | Berelekh | 70° 24′N/143° 57′E | 1971–73 | Low ambient temperature |
| 2002/473 | M. primigenius | Left femur | 46 700 ± 2800 | Taimyr, Arilakh | 74° 25′N/107° 45′E | 2002 | Below 0°C |
| 2000/173 | M. primigenius | Right ulna | 11 900 ± 40 | Taimyr, Arilakh | 74° 25′N/107° 45′E | 2000 | Below 0°C |
| 2002/472 | M. primigenius | Right ulna | >48 000 | Taimyr, Arilakh | 74° 25′N/107° 45′E | 2002 | Below 0°C |
| 2005/917 | M. primigenius | Vertebra | 35 380 ± 550 | Taimyr, Baikura-Turku | 73° 45′N/102° 00′E | 2005 | Below 0°C |
| 2005/915 | M. primigenius | Mandible | 27 740 ± 220 | Taimyr, Baikura-Turku | 73° 45′N/102° 00′E | 2005 | Below 0°C |
| BMV | E. maximus | Ulna | ∼1 year | Bowmanville Zoo, Bowmanville, Ontario | 44° 48′N/78° 42′W | April 2007 | Below 0°C |
Bone samples with their associated radiocarbon dates and geographic origin, 14C dates are given in Year Before Present (YBP).
The modern elephant bone was sampled from the carcass of an Asian elephant buried outside of the Bowmanville Zoo (Bowmanville, Ontario) in late summer 2006. Since excavation and sampling in April 2007 it has been kept at –20°C.
DNA extraction
Extractions of mammoth samples as well as setup of quantitative PCRs were performed in a dedicated clean room facility following strict procedures required for studies of ancient DNA [(20,26); http://socserv.mcmaster.ca/adna/labs.htm].
Bones were chopped into small pieces using hammer and chisel. The maximum dimension of the chopped bone material was ∼3–4 mm.
Approximately 150 mg of each sample along with one extraction blank were incubated in 1.5 ml 0.5 M EDTA pH 8 for 3 days at room temperature and very gentle agitation (recent extractions have shown that an extended decalcification time is often required to make sure that the bone material is completely dissolved after the PK digest, data not shown). Samples were briefly spun down and the EDTA supernatants (SNs) were transferred to new tubes and processed separately. To the remaining decalcified bone material we added 1.5 ml of Proteinase K (PK) digestion buffer [10 mM Tris (EMD Chemicals Inc.) at pH 8.5, 0.5% N-Lauroylsarcosine (Sigma), 250 μg/ml Proteinase K (Fisher Biotech), 5 mM CaCl2 (EMD Chemicals, Inc.), 50 mM DTT (EMD Chemicals Inc.), 1% PVP (EMD Chemicals Inc.), 2.5 mM PTB (Prime Organics, Inc.)]. Demineralized bone pellets (herein referred to as the PLT fraction) were incubated at 55°C under gentle agitation. After 6 h the bone material was completely digested (further incubation will not dissipate the remaining, barely visible, fibrous material, data not shown).
To purify the raw extracts (i.e. SN and PLT fractions, separately), samples were organically extracted first using 0.5 ml of phenol/chloroform/isoamylalcohol (25/24/1, pH 8, Fluka) and then 0.5 ml of chloroform (Fluka). Samples were further ultra filtrated through a 30K membrane (microcon, Millipore) until there was no remaining liquid on the filter followed by three (SN) or two (PLT) washing steps with 300 μl 0.1× TE (10 mM Tris, 0.1 mM EDTA, pH 8.5), each time spinning to dryness again. Finally DNA was recovered by adding 200 μl of 0.1× TE, incubating for 5 min at 1000 r.p.m. (Eppendorf thermomixer) and an upside-down spin into collection tubes.
The extraction of the modern Asian elephant followed the same protocol as used for the mammoth samples (starting from ∼150 mg of bone) but was performed in a dedicated ‘semi’-clean room, which is located in the vicinity of the post PCR labs. In this ‘semi’-clean room similar but less stringent procedures are followed as compared to the clean room. No modern elephant DNA was ever processed in the clean room where the mammoth samples were processed.
Quantitative PCR
The numbers of amplifiable mitochondrial DNA (mtDNA) fragments were determined by serial quantitative PCR (qPCR) using one forward primer and six different reverse primers amplifying a part of the cytochrome B gene 84, 151, 279, 490, 677 and 921 bp in length, respectively (20).
For the quantitation of nuclear DNA (nucDNA) a similar set of one forward and five different reverse primers was designed to amplify fragments of 67, 112, 189, 360 and 632 bp of the nuclear (single-copy) von-Willebrand-Factor gene (see Figure S1 in Supplementary Data). The primers were designed to amplify the African elephant (Loxodonta africana, Genbank accession number U31615) and the Asian elephant (Elephas maximus, Genbank accession number U31611) while excluding human. They perfectly match these two sequences with the exception that the very 3′-end of vWF_R62–83 shows an A to T mismatch to the Asian elephant sequence. However, both available vWF sequences of the woolly mammoth (Genbank accession numbers AF154873, AF154874) spanning this primer binding site suggest that this position is conserved among African elephants and mammoths. To confirm the primer binding site of vWF_R62–83 in all three species, PCR products were generated from African and Asian elephant genomic DNA as well as from a former DNA extract of mammoth 2005/915 using vWF_F17–35 and vWF_R182–205. After cloning (TOPO TA kit, Invitrogen) and sequencing these PCR products, it was noted that all three species match the 3′-end of vWF_62–83 perfectly; only the 5′-end of this primer has a T to C mismatch for all three species. This is expected to have no influence on the quantitation as the unknown sample and the standard both contain the same sequence at this position.
Template DNA for the qPCR-standard was generated using primers vWF_F17–35 and vWF_R629–648 from Loxodonta africana whole genomic DNA. The obtained product was cloned (TOPO TA kit, Invitrogen) and re-amplified via colony PCR using M13for and M13rev primers.
Standard curves were generated for each of the five primer combinations by amplifying a serial dilution with the following copy numbers: 5, 50, 500, 5000 and 50 000 generated from the purified colony PCR product of known concentration (measured via UV spectrophotometry). Sensitivity of the five primer combinations was tested using a different standard serial dilution (5 × 0.01, 3 × 0.1, 3 × 1, 1 × 10, 1 × 100, 1 × 1000 copies), all five primer pairs amplify at least 10 copies under the reaction conditions specified below (for all primer pairs: 7 out of a total of 15 reactions starting from one copy and 1 out of 15 reactions starting from 0.1 copy yielded a specific product). Blasting the elephant vWF sequence yielded only one single match in the African elephant whole genome shotgun sequence (accession number: AAGU01197285, 2× coverage) suggesting that no vWF pseudo genes exist that might interfere with the quantitation of nuclear DNA. To check whether contaminating DNA in higher concentrations would result in non-specific amplification products or potentially interfere with the quantitation of low mammoth DNA concentrations in our samples, we spiked a 25 copy standard as well as the no template controls (NTC) with 10 ng of human genomic DNA (∼3000 nuclear genomic copies). In all cases the primers did not amplify the vWF gene target from human DNA (spiked NTCs) and quantitation of the 25 copies of standard DNA spiked with 3000 human nuclear DNA copies consistently yielded ∼25 amplifiable molecules when compared to the standard dilution series.
Quantitation of mitochondrial and nuclear DNA
For both standard series as well as for samples of unknown concentration, quantitations were done in duplicate (plus ∼1 NTC per 10 reactions). The standard dilution series used for the mtDNA assay spanned 5–500 000 copies, for the nucDNA 5–50 000 copies (both in 10-fold increments). For reaction composition and thermal cycling conditions using the CytB primers see (20). The reaction targeting the vWF gene contained 1× PCR Buffer II (Applied Biosystems), 2.5 mM (R62–83, R108–128) or 2.0 mM (R182–205, R355–376, R629–648) MgCl2 (Applied Biosystems), 250 μM dNTPs (each, Amersham), 0.75 mg/ml BSA (Sigma), 250 nM of each primer (vWF_F17–35 and the reverse primer, IDT), 30 nM ref dye (Stratagene), 0.333 × SYBR Green (Invitrogen), 1 unit AmpliTaq Gold (Applied Biosystems) and 5 μl of DNA template (standard, extract or water). Reaction volume was 20 μl. The temperature profile included an initial activation of the enzyme at 95°C for 7 min, followed by 45 cycles of 95°C for 30 s, 66°C for 30 s and 72°C for 90 s, and a final extension at 72°C for 3 min. The dissociation curves were generated using the following thermal profile: 95°C for 1 min, 55°C for 30 s and 95°C for 30 s. Optical data for the amplification was acquired following each extension step and for the dissociation curve during the 55–95°C ramp, using a Mx3000P cycler (Stratagene). The cycling conditions were the same for all five primer pair combinations.
Efficiencies of the six CytB assays were between 90% and 101% as calculated from the slope of the standard curve, with R2-values, a measure for the quality of the regression fit, between 0.996 and 1.000. Efficiencies of the five vWF assays were between 90% and 104%, with R2-values between 0.997 and 0.999.
To check for inhibition, a quantitation of all our extracts at a 1:10 dilution was performed, using primers CytB_F111–131 and CytB_R171–194 (84 bp mt fragment).
Calculation of inhibition in the quantitative PCRs.
For example, if the copy number (straight) is 10 times the copy number (at 1:10), inhibition is 0%; if the copy number (straight) is 5 times the copy number (at 1:10), inhibition is 50%; if the copy number (straight) is the same as the copy number (at 1:10), inhibition is 90%.
Cloning and sequencing of qPCR products
For the mitochondrial DNA the longest CytB qPCR product obtained for each sample and fraction was cloned using the TOPO TA cloning kit (Invitrogen, at one quarter reaction scale). Sixteen white colonies were lysed (95°C for 5 min) in 50 μl of 1× TE (10 mM Tris, 1 mM EDTA, pH 8). Two microlitres of lysate were used in a 30 μl colony PCR using M13 general primers and the following cycling conditions: 95°C for 5 min, 35 cycles of 95°C for 30, 58°C for 30 and 72°C for 90 s, a final extension of 72°C for 10 min. Colony PCR products were visualized on 1.5% agarose gels and purified over 30K AcroPrep plates (Pall). Eight colony-PCR products per cloning reaction were sequenced with the M13 forward primer; in case full coverage could not be reached some of the colony-PCR products were re-sequenced using the M13 reverse primer. For the nuclear DNA the longest obtained vWF qPCR product for each sample and fraction was sequenced directly after purification over 30K AcroPrep plates (Pall). Sequencing was done on an ABI 3130 using BDT v1.1 chemistry and the KB basecaller (version 1.2, Applied Biosystems) for data analysis at an N threshold setting of QV20 (i.e. bases below quality value = 20 were called N). Sequence data were aligned using BioEdit (ver. 7.0.5.2, Hall T.A. 1999) and consensus sequences were generated (see SOM for details). All sequences have been deposited at Genbank under the following accession numbers FJ753551–FJ753566.
DNA degradation analysis
The model described by Deagle et al. (22) has been applied to analyse the obtained qPCR results. It targets non-bypassable DNA damage and is based on the assumption that damage occurs randomly, following a Poisson distribution of parameter lambda. Briefly, log-transformed copy numbers for the different amplicon sizes are plotted against their respective sizes. Provided this results in a relation that can be described by a linear regression, the resulting slope (i.e. lambda values) reflects the damage frequency (and the reciprocal, 1/lambda, describes the mean amplifiable fragment length), whereas the Y intercept (i.e. parameter N) is a general measure of total amplifiable DNA. All data points below five copies were excluded from this analysis to minimize erroneous deviation at the low end of the copy number range. Due to the few data points obtained for nucDNA in the mammoth extracts, these data have not been used to generate estimates of lambda or N.
Mitochondrial to nuclear DNA ratio
MtDNA to nucDNA ratios were determined using qPCR copy numbers for the 112 bp nucDNA amplicon and size corrected copy numbers for a 112 bp mtDNA amplicon, based on the model of random degradation fitted for each individual sample (Figure 3). In addition to the qPCR results, 454 shotgun sequence data were available for two of the six mammoths: 915 (20) and 917 (unpublished data), which were analyzed for their number of nuclear and mitochondrial DNA hits found in the endogenous portion of the sequence data.
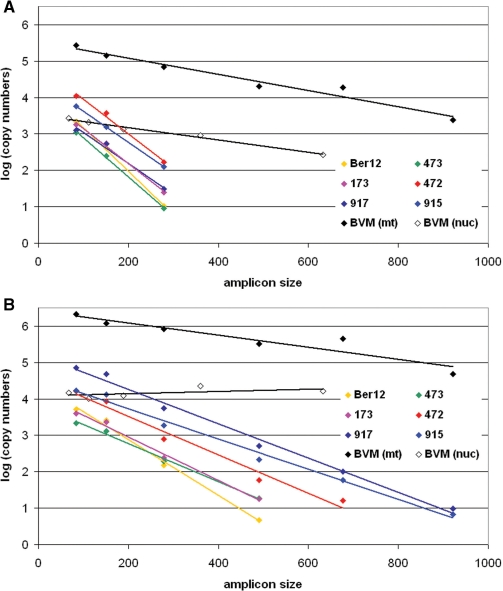
Figure 3.
Linear regression for the modeling of non-bypassable damage, according to Deagle et al. for the six mammoths (only mitochondrial DNA) and the modern elephant (mitochondrial and nuclear DNA) extracts. x-axis is the amplicon size in bp, y-axis is the log(10) transformed copy number as in Table S1. The corresponding three regression parameters slope ( = lambda value, i.e. damage frequency), Y-intercept (N parameter, i.e. number of DNA copies of the length zero) and RSq are given in Table 2. (A) shows the regression for the SN fractions, (B) the regression for the PLT fractions.
Cytosine deamination analysis
For each position in the CytB DNA sequence carrying a C or G we divided the number of clones revealing a substitution (C to T; G to A) by the total number of clones sequenced, yielding λI for that particular position. To take into account the length and the base composition of the sequence we divided the sum of all λI per sample/fraction by the number of C and G positions in the sequence (i.e. all potentially affected positions), yielding λ′, the average deamination rate over all positions (13).
Dating of samples, total collagen and stable isotope analysis
All data related to radiocarbon dating and isotope analysis of the samples in this study were originally reported in Debruyne et al. (26).
Thermal age calculations
Thermal age calculations were kindly provided by David Harker [http://drrc.oneiria.co.uk/, (6)]. Besides the given geographic location and sample date the following parameters were assumed for all samples (as no further information is available): soil type fresh/sandy, burial depth 2 m, no vegetation cover.
Amino-acid racemization analysis
Total hydrolyzable amino-acid (THAA) analyses were performed at Scripps Institution of Oceanography. Samples were weighed out into sterilized test tubes (16 × 150 mm) to target masses ∼20 mg. One milliliter of doubly distilled 6 N HCl was added to each sample, flame-sealed after evacuation with N2, and exposed to 100°C for 24 h. After removal from heat, the 1 ml HCl fractions were transferred into small test tubes (10 × 75 mm) and brought to dryness on a vacuum centrifuge system. These residues were transferred onto equilibrated columns of AG 50W-X8 resin and desalted according to the procedures of (27). The desalted extracts (3 ml ∼2 M NH4OH) were brought to dryness on a vacuum centrifuge and reconstituted in 100 μl of doubly distilled water (ddH2O). Amino acids were quantified using reverse-phase high performance liquid chromatography (RP–HPLC) after precolumn derivatization with o-phthaldialdehyde-N-acetyl-l-cysteine (OPA-NAC) according to the procedures of (28). In order to fall in the detector signal range, a dilution of 10× was necessary for sample 173, 100× for samples 472 and 473, while dilutions of 1000× were necessary for samples BER12, 915 and 917. Ten microliters of each diluted sample extract was added to an equal volume of 0.4 M sodium borate buffer. Five microliters of 10–2 M OPA-NAC was added to each sample and allowed to react for 1 min. The reaction was quenched with 475 μl of sodium acetate buffer (pH adjusted to 5.5) and 50 μl was injected into the HPLC sample loop. RP–HPLC separations were achieved using a Phenomenex Luna C18(II) 250 × 4.6 mm column with a stationary phase of 50 mM sodium acetate buffer (8% methanol) and a mobile phase of pure methanol. Amino-acid abundances and chirality were determined by comparison against racemic standards of known concentration (asp, glu, ser, gly, ala, val). Fluorescent detection was monitored using a Shimadzu RF-535 fluorescent detector at an excitation wavelength of 340 nm and an emission wavelength of 450 nm.
Crystallinity Index (CI) analysis
Bone from all seven samples (six mammoths, one modern elephant) was powdered in a freezermill (Spex model 6750) under liquid nitrogen for Fourier transform infrared (FTIR) spectroscopy. The particle size of the ground bone was less than 10 μm. An agate mortar and pestle were used to grind ∼200 mg of dried KBr and 2 mg of bone to a fine powder. The powder was then pressed at 10 000 PSI to create a 13 mm diameter pellet.
A BioRad FTS-40 FTIR Spectrometer with a sample chamber flushed with dry nitrogen was used to scan the samples 16 times at a resolution of 4 cm−1 over the frequency range 400–4000 cm–1. After correcting the resulting spectra with a KBr baseline, the peak absorbance at 565 and 605 cm–1, representing the absorption of the ν4 PO4 vibrations, was measured. The minimum absorbance around 595 cm–1 was also measured to allow calculation of the CI by:
where Ax is the absorbance at frequency x [3].
Principal component analysis (PCA)
A pair of PCA of the data were performed using Minitab (ver. 14) to evaluate the level of correlation between the actual preservation of DNA (both qualitatively and quantitatively) as measured by qPCR results (i.e. copy numbers, N and lambda), and the indirect factors often used for determining the state of DNA preservation (Table 2): amino-acid racemization (putative antagonist to the DNA preservation) and concentration (agonist), collagen concentration (agonist), C > T deamination rate (per SN or PLT fraction; antagonists), CI (antagonist), and thermal age (antagonist). The analyses were both performed on the correlation matrix to standardize the data, and the four first components (sufficient to explain >90% of the total variance) were computed each time.
Table 2.
Results obtained from the bone samples used in this study
| Sample ID | Total mtDNA (84 bp) | Total nucDNA (112 bp) | Modeling damage of mtDNA in the SN |
Modeling damage of mtDNA in the PLT |
Rate of deamination in mtDNA |
Amino acid concentration (p.p.m.) | D/L Asp | Collagen (mg/g) | Thermal age | Crystallinity Index | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lambda | N | RSq | Lambda | N | RSq | SN | PLT | ||||||||
| Ber12 | 7247 | 19 | 0.0120 | 24 082 | 0.9984 | 0.0077 | 26 767 | 0.9931 | 0.018 | 0.002 | 1966 | 0.058 | 182 | 300 + 80 (−8.2, + 15) | 2.99 |
| 2002/473 | 3241 | 2 | 0.0107 | 9224 | 0.9984 | 0.0052 | 6566 | 0.9938 | 0.001 | 0.000 | 611 | 0.067 | 240 | 1090 (−8.5) | 2.99 |
| 2000/173 | 5762 | 7 | 0.0097 | 13 425 | 0.9942 | 0.0060 | 14 028 | 0.9912 | 0.016 | 0.006 | 71 | 0.060 | 107 | 280 (−8.5) | 2.85 |
| 2002/472 | 27 230 | 9 | 0.0095 | 80 631 | 0.9914 | 0.0053 | 37 359 | 0.9766 | 0.011 | 0.002 | 541 | 0.056 | 224 | > 1120 (−8.5) | 2.93 |
| 2005/917 | 72 002 | 3 | 0.0085 | 7898 | 0.9861 | 0.0047 | 162 405 | 0.9911 | 0.000 | 0.000 | 5859 | 0.058 | 147 | 790 (−8.7) | 2.96 |
| 2005/915 | 22 858 | 40 | 0.0085 | 30 158 | 1.0000 | 0.0041 | 35 950 | 0.9893 | 0.012 | 0.000 | 9088 | 0.035 | 262 | 620 (−8.7) | 3.01 |
| BMV | 2 434 600 | 12 489 | 0.0022 | 343 321 | 0.9613 | 0.0017 | 2 633 298 | 0.8875 | 0.000 | 0.000 | ND | ND | ND | ND | 2.82 |
| BMV, nucDNA | 0.0017 | 3270 | 0.9859 | 0 (−0.0003) | 12 067 | 0.3128 | |||||||||
Copy numbers for mt and nucDNA are the sum of SN and PLT. The values for lambda, N and RSq are obtained from the linear regression using Deagle et al.'s model (see ‘Materials and Methods’ section). D/L Asp is the extent of racemization of aspartic acid. Thermal ages are given in years with the effective depurination temperature in degree Celsius in parentheses. The 80 years in the thermal age field for Ber12 reflects the 30 years period the sample was stored at low ambient temperature of ∼15°C. ND stands for not determined.
In the first analysis, the indirect factors were computed together with total nucDNA copy numbers for the 112 bp fragment and the 84 bp output for mtDNA (seven variables total). In the second analysis, only the mitochondrial modelized outputs N (putative agonist of quantitative preservation) and Lambda (putative antagonist of qualitative preservation) were segregated by fraction (SN versus PLT) and analyzed together with the indirect factors (11 variables total).
RESULTS
qPCR data
Copy numbers for all mitochondrial DNA (mtDNA) and nuclear DNA (nucDNA) fragments, found within the supernatant (SN) and pellet (PLT) fractions of the extracts are presented in Table S1 and plotted in Figures 1 and 2. It is worth noting that as ancient DNA molecules are often damaged and these damages most likely affect the efficiency of amplification during the early cycles of PCR, we presumably underestimate the number of DNA molecules present in an ancient extract. However the similar geographic origin and the comparable preservational history of our permafrost mammoth samples nevertheless warranted comparability as can be seen from the lambda values discussed later in the manuscript.
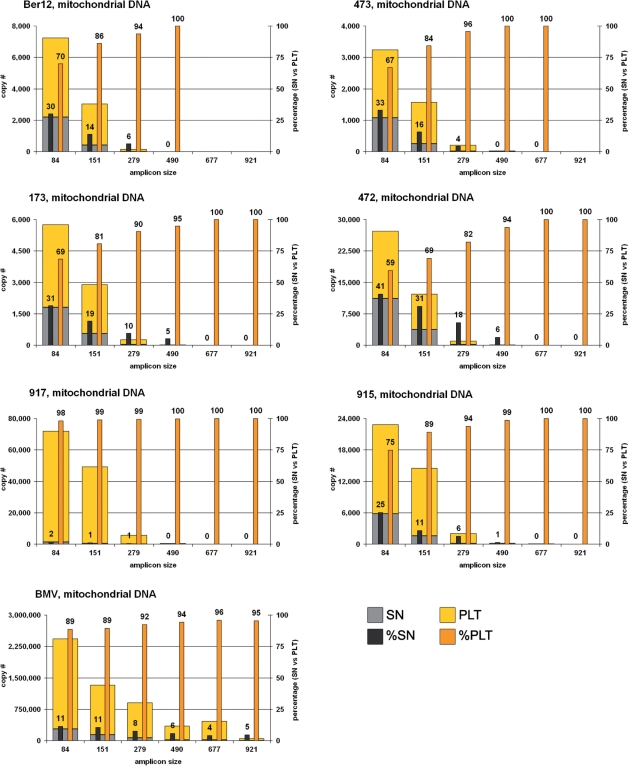
Figure 1.
Mitochondrial DNA yields of the six mammoths and the modern elephant extracts for the six amplicon sizes, in absolute copy numbers as given in Table S1 and relative contributions of the two fractions SN and PLT.
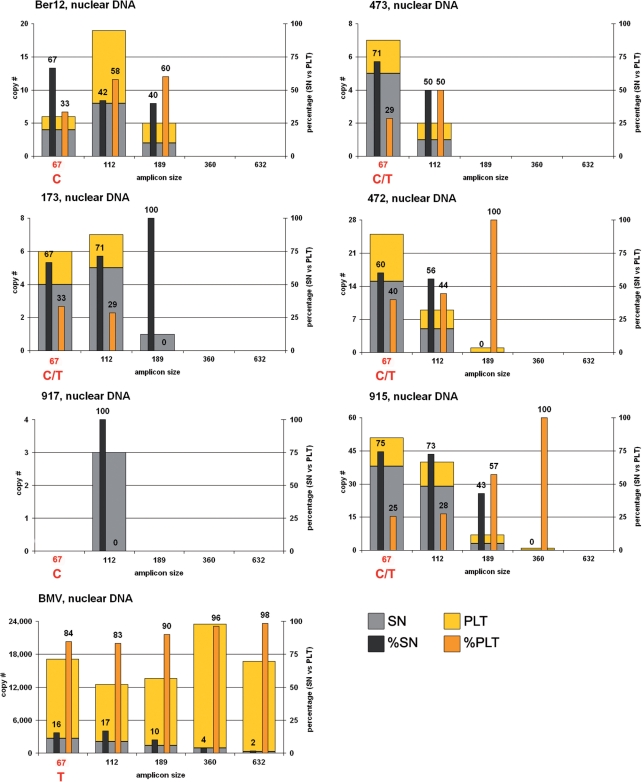
Figure 2.
Nuclear DNA yields of the six mammoths and the modern elephant extracts for the five amplicon sizes, in absolute copy numbers as given in Table S1 and relative contributions of the two fractions SN and PLT. See text and Figure S2 for information concerning a variable position for the six mammoths within the primer binding site of the reverse primer of the 67 bp amplicon.
For the sake of clarity the results are developed in two general sections addressing mtDNA and nucDNA, respectively.
Inhibition
The level of inhibition found in the mammoth extracts was on average 49% and showed little variation among samples (SD 9%). Inhibition was only slightly higher in the SN fractions (54%, SD 9%) when compared to the PLT fractions (44%, SD 6%). Due to its consistency across samples and extracts, inhibition is unlikely to bias the pattern of quantitative and qualitative preservation of DNA described below.
Mitochondrial DNA, mammoth extracts
For the shortest mtDNA fragment (84 bp), total copy numbers (SN and PLT fractions pooled) for the six mammoth extracts fall within the range of 3000–70 000 (per 5 μl of each extract fraction or per ∼3.75 mg of bone material). The relative proportion of available copy numbers for that amplicon in the SN and PLT fractions ranged from 2% to 41% and 59% to 98%, respectively. Whereas the sample 917 represents the most extreme situation in that almost all the amplifiable DNA is recovered from the PLT fraction, the remaining five samples showed only a slight prevalence of the PLT (59–75%) over the SN (25–41%).
With increasing amplicon length, total copy numbers decreased and the proportion shifted toward the PLT fraction. Likewise, no amplifiable DNA could be retrieved for the longest mtDNA amplicon (921 bp) from any of the six SN fractions, whereas five of the six PLT fractions yielded the expected product based on 10 or less initial copies.
Mitochondrial DNA, modern elephant extract
The modern elephant bone (BMV) yielded a substantially higher amount of mtDNA (almost two orders of magnitude more for the SN fraction and one order of magnitude more for the PLT fraction), yet shows a similar pattern of decreasing copy numbers with increasing amplicon length. The relative proportion of mtDNA in the SN and PLT fractions is 11% and 89%, respectively, and thus more shifted towards the PLT fraction than it is in the majority of the mammoths. While the SN fraction still provides template molecules for the longest amplicon (921bp), the relative yield of mtDNA in the PLT fraction increases with increasing amplicon length, depicting the same trend as for the mammoths (Figure 1).
Nuclear DNA, mammoth extracts
Total copy numbers for the nucDNA are, not surprisingly, much lower than those for mtDNA, with less than 50 copies for the 67 bp as well as for the 112 bp products. The 189 bp product could only be amplified from four of the six mammoth samples (starting from less than seven copies), while for the next amplicon length (360 bp) only the PLT fraction of sample 915 yielded a specific product starting from a single copy. No attempt has been made to amplify the longest product (632 bp) from any of the mammoth extracts.
Unexpectedly, copy numbers for the 67 bp amplicon were in some cases lower than for the 112 bp amplicon. Further investigation and sequencing of the primer binding site of vWF_R62–83 revealed that the eighth base position from the 5′-end was variable within mammoths (see Supplementary Data, Figure S2). In the modern elephant (BMV) this position was consistently a T while the mammoths were either C (Ber12, 917) or C/T (473, 173, 472, 915). While this finding is consistent with the reduced copy numbers for the 67 bp product for some of the samples (Figure 2), the obtained copy numbers for this amplicon length cannot be trusted and were therefore excluded from further analysis of the mammoth samples.
Contrary to the mitochondrial DNA results, the analysis of the 112 bp nucDNA data shows that a substantial amount of short fragments of nucDNA can be amplified from the SN fractions. Nevertheless, in the three samples (Ber12, 472, 915) which provided the highest (hence most significant) copy numbers for nucDNA, the ratio shifts towards the PLT fraction with increasing amplicon length consistent with the findings for mtDNA (Figures 1 and 2).
Nuclear DNA, modern elephant extract
The BMV sample yielded a higher total amount of amplifiable nucDNA than the six mammoth bones (over two orders of magnitude more for the SN fraction and over three orders of magnitude more for the PLT fraction). However the PLT fraction does not show a reduction in copy numbers with increasing amplicon length, as does the SN fraction and is generally seen for mtDNA in both the BMV and all mammoth extracts.
Regarding the relative contribution of the SN and PLT fractions the BMV sample shows a similar picture for nucDNA as for mtDNA since the majority of DNA has been found in the PLT fraction. The fact that shorter nucDNA is less abundant in the SN is apparently a distinctive feature between the modern elephant and the six ancient mammoth samples.
DNA degradation analysis
In order to study the rate of degradation in all seven elephantid bone samples, we analyzed our qPCR data using Deagle et al.'s model (22). Plots of the copy numbers in logarithmic scale are shown in Figure 3; values for lambda and N obtained from the linear regression as well as the corresponding R2-values are detailed in Table 2.
For the six mammoth samples the slopes for the SN fraction range from –0.0085 to –0.0121, whereas for the PLT fraction they were between –0.0040 and –0.0080, indicating a generally higher frequency of non-bypassable damage in the SN fractions (Table 2, Figure 3).
The BMV sample sets itself apart from the six mammoth samples with distinctly lower slopes for both mtDNA (–0.0022, SN and –0.0017, PLT) and nucDNA (–0.0017, SN), suggesting (i) a far better overall DNA preservation compared to the mammoth bones and (ii) that nucDNA is better preserved than mtDNA. For the nucDNA from the PLT fraction, some variation was observed although without any clear pattern of decrease for copy numbers with increasing fragment length. The least-square R2 of 0.31 results from the almost null slope (+0.0003) of the linear regression, indicating a spurious linear fit of the data.
The total amount of DNA, in this model reflected by N, was generally similar in the two fractions or slightly higher in the SN fraction for all mammoth samples except 917, where N(PLT) was approximately 20 times higher than N(SN). For the BMV extract, the ratio of N(PLT)/N(SN) was 7.7 for mtDNA and 3.7 for nucDNA.
Mt/nuc DNA ratios
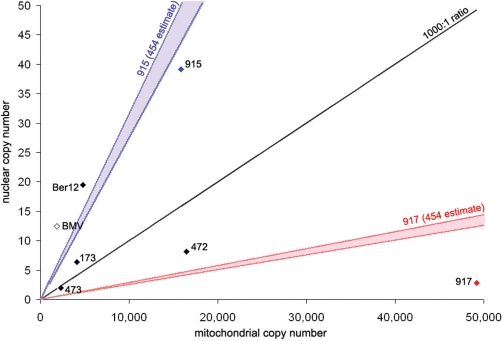
Based on the data obtained for the 112 bp nucDNA qPCR assay and the size corrected mtDNA results, shown in Table 3, the mitochondrial to nuclear DNA ratios range from 245:1 to 17 369:1 for the six mammoth samples. The modern elephant sample shows a ratio of 152:1 clearly outside the range for all mammoth samples. The analysis of the 454 data for sample 915 reveals a mtDNA to nucDNA ratio ranging from 317:1 to 362:1 (see table S3 in supplementary information) compared to a ratio of 405:1 based upon the qPCR results. The 454 data for sample 917 (unpublished material) shows a ratio spanning 3469–3965:1 compared with 17 369:1 from the qPCR.
Table 3.
Mitochondrial to nuclear DNA ratios for the SN and PLT fractions and the pool of both (amplicon size 112 bp)
| Sample ID | Ber12 |
473 |
173 |
472 |
917 |
915 |
BMV |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fraction | SN | PLT | SN | PLT | SN | PLT | SN | PLT | SN | PLT | SN | PLT | SN | PLT |
| mtDNA copy number | 1091 | 3675 | 584 | 1718 | 1100 | 2985 | 6958 | 9524 | 882 | 48 328 | 3368 | 12 488 | 194 670 | 1 698 635 |
| nucDNA copy number | 8 | 11 | 1 | 1 | 5 | 2 | 5 | 4 | 3 | 0 | 29 | 11 | 2089 | 10 400 |
| mt/nucDNA ratio | 129 | 334 | 1168 | 1209 | 235 | 1793 | 1504 | 2664 | 328 | n.a. | 118 | 1181 | 93 | 163 |
| mt/nucDNA ratio, pooled data | 245 | 1198 | 643 | 2010 | 17 480 | 405 | 152 | |||||||
mtDNA copy numbers were calculated for 112 bp using Deagle et al.'s model. The mt/nucDNA ratio for the pooled data was calculated as [mtDNA copy number, SN + mtDNA copy number, PLT] divided by [nucDNA copy number, SN + nucDNA copy number, PLT].
Cytosine deamination
For all seven extracts analysed (modern and ancient) the rate of cytosine deamination found in mtDNA was never higher in the PLT when compared to the SN; for two samples, 917 and BMV, it was 0.0000 in both fractions. The average rate of deamination in the SN fraction (λ′ave,SN) was 0.0083 (0.0097: for the six mammoths only), whereas in the PLT fraction (λ′ave,PLT) it was 0.0014 (0.0017: for the six mammoths only); the ratio of these two averages (λ′ave,SN/λ′ave,PLT) is 5.8, suggesting that DNA in the SN fraction is clearly more prone to deamination than DNA in the PLT fraction.
External criteria for estimating levels of DNA preservation
Protein related analyses
Total collagen content of the mammoth samples spanned a 2.5-fold range from 107 to 262 mg/g and does not show a clear correlation with sample age nor with DNA yield. The level of amino-acid racemization of aspartic acid (D/L asp) for all six mammoth bones lies below 0.08, deeming them all to be potentially positive for DNA retrieval.
CI
CI has been widely used as an indicator of the state of preservation of bones to determine their suitability for isotopic and other analyses (29,30). While some questions have been raised as to the effect of sample preparation on determination of CI (31) the procedures used here were controlled by comparison with modern, unaltered bone and should provide adequate indication of the degree of recrystallization of the hydroxyapatite in these samples.
The CI values suggest that little to no recrystallization has taken place in all bone samples. The overall variation of the CI values is low and there seems to be no correlation with either the age or the DNA yield of the samples. The slightly lower CI value observed in the BMV sample is within the error of the analysis or could be due to the fact that CI values can vary between species.
Thermal age analysis
The calculated thermal ages for DNA and the corresponding effective temperatures for depurination for the six mammoth bones and the BMV sample are given in Table 2. The thermal ages correlate almost linearly with the real ages, as one would expect, since many of the parameters used for the calculations, i.e. soil type, burial depth and coverage by vegetation, for which no information was available, are the same for all samples. The ratios of thermal age to real age varies for the six mammoth samples from 17 (Ber12) to 32 (173, 472), with sample Ber12 being lowest due to its relatively lower effective depurination temperature.
PCA analysis
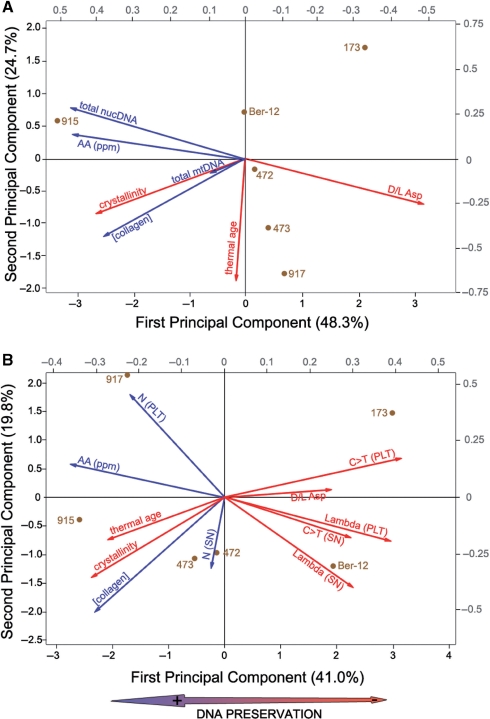
Both PCA analyses (with DNA factors split by nuc/mt and by SN/PLT fractions of the mtDNA, respectively, Figure 5) reveal relatively low levels of correlation between the variables analyzed: the first PC explains only 41% of the variance in the first analysis and 48% of variance in the second analysis, and the four first PCs showed eigenvalues >1 in each case (see supplementary information).
Figure 5.
Results of principal component analyses performed on seven variables (A) and 11 variables (B) (see ‘Materials and Methods’ section). Loading vectors (right and top gray scale) and specimens score (bottom and left black scales) plots are shown together. Vectors of putative preservation antagonists are shown in red while preservation agonists are in blue. The double arrow at the bottom indicates the main signal extracted from the first principal component and associated with general (both quantitative and qualitative) DNA preservation.
The two first principal components of the analysis which compares mtDNA and nucDNA yields with indirect factors of preservation (Figure 5A) depict vectors of mtDNA and nucDNA which have very different directions: nucDNA concentration is almost symmetrically opposed to amino-acid racemization whereas mtDNA concentration is rather positively correlated to increasing CI and collagen concentration. Amino-acid concentration, collagen content, CI and thermal ages show little correlations with each other.
The second analysis accounts for the relative degradation in the SN and PLT fractions, using both N parameters and lambda values from Deagle et al.'s model, correlated to the indirect factors (Figure 5B). This analysis thus distinguishes qualitative (lambda) and quantitative (N) preservation of DNA (although only for the mt genome) which do not show parallel vectors. Furthermore, the qualitative preservation of both fractions (lambda) is more strongly correlated than its quantitative counterpart (as measured by N). The C > T deamination rate seems to be a good proxy for qualitative degradation, but the other factors are quite widely distributed along the two first principal components. Once again, increasing thermal age and CI depict an unexpected behaviour along the first principal component and do not correlate strongly with the other preservation-antagonist variables.
DISCUSSION
DNA in the supernatant versus the pellet
Bone is a composite tissue comprised of both organic (mainly collagen) and inorganic matter (bioapatite) (32). Typically protocols designed for the extraction of DNA from bone aim at dissolving the mineral matrix (chelating the Ca2+ ions with EDTA) in order to make the organic matrix accessible to enzymatic digestion (using Proteinase K) through which the DNA is assumed to be released into solution. Depending on the protocol, demineralization and organic digestion are either performed as a single-step (13,15,33,34) or as a two-step (20,25,35) extraction.
In the single-step extraction protocol, the potential contribution of the demineralized fraction to the total DNA yield is confounded by the DNA from the organic fraction. In two-step approaches, the supernatant obtained after demineralization in EDTA is generally discarded or not further processed (25,35), under the assumption that it lacks significant amounts of DNA. However, our quantitative assessment of both SN and PLT fractions (SN = EDTA supernatant, associated with the mineral component and PLT = proteinase K digest on the remaining decalcified bone pellet, associated with the organic component) revealed that the SN can contain up to ∼40% of the 84 bp long mtDNA fragments recovered from mammoth samples. At first glance this result hints at a relatively large proportion of the amplifiable DNA recovered during bone extraction not being associated with the organic matrix, but rather resides within the mineral matrix.
Analyzing the difference in amplifiable length and amount of DNA molecules specifically obtained in each fraction, allowed us to observe two general trends. First, the SN fraction tends to be enriched in smaller DNA fragments compared with the PLT fraction of the same specimen. Thus, the proportion of SN-derived copies from the qPCR data generally decreases towards zero as the fragment length increases, to a point where only the PLT fraction provides template molecules for PCR (Figures 1 and 2). This pattern, although less pronounced, is seen in the modern elephant sample as well and could be explained by at least two non-exclusive processes. (i) The DNA associated with the mineral matrix is more prone to post-mortem degradation than the DNA associated with the organic matrix. (ii) A filtration effect, i.e. free-floating DNA fragments present within the collagen matrix are selectively retained in or released from the collagen matrix during demineralization depending on their size.
In addition to this fragment size shift, we observed another qualitative shift in the genomic composition of the two extract fractions. The SN fractions are typically enriched in nucDNA of short fragment size compared with the PLT fractions, with up to two thirds of the nuclear copies recovered from the SN fractions of the mammoths extracts. The apparent richness of SN fractions in nucDNA could be a by-product of a general enrichment in short fragments during bone extraction caused by the filtration effect mentioned above and insofar an artefact generated by the choice of the extraction method rather than a consequence of biochemical processes during diagenesis. NucDNA obtained from the mammoth bones, which seems to be of shorter molecular size than mtDNA for the mammoth samples, would be particularly affected. In fact, this hypothesis is consistent with the results derived from the BMV (modern elephant sample). It yielded, not surprisingly, more total DNA (in absolute copy numbers) and less degraded DNA (longer amplified fragments and lower lambda values) than any of the mammoth samples (Figures 1, 2 and 3). Although not a complete analog to the permafrost conditions of the mammoth samples, its relatively short time in the ground appears to have limited the accumulation of comparable DNA degradation. We therefore consider the BMV to be at an earlier stage of diagenesis. Contrary to the mammoth samples, the SN of the BMV is not particularly enriched in nucDNA: It contains between 2% and 17% of the amplifiable nucDNA for the different amplicon sizes, similar to the percentages of mtDNA (5–11%) for the same fraction. The relative paucity of nucDNA in the SN for the least diagenetically altered sample is surprising. It suggests a differential pattern of DNA degradation for mt- and nucDNA, something we address via direct comparisons of qPCR results later on.
Nuclear to mitochondrial DNA ratios
Most ancient DNA studies rely on a somewhat ideal mt:nuc ratio of 1000:1 copies of genome per cell, although the evidence for such a ratio in different tissue types and bone in particular is scarce (36). Comparison of copy numbers obtained for the 112 bp nuclear amplicon and a size adjusted amplicon for the mitochondrial locus (see ‘Materials and Methods’ section) yielded a mt:nuc ratio of 152:1 for the modern elephant bone which is at the low end of the typical range for different mammalian cell types [110:1 to 860:1 (37)]. In comparison, the six ancient samples returned higher ratios ranging from 245:1 to 17 369:1 (Figure 4). It is unclear how these results compare with the ratios reported by Robin and Wong, since in their study DNA is derived from cell lines and bone is not among the tissue types used. But the ratios derived from all mammoth samples are higher than the one from the modern bone, which shows a relatively similar richness of nucDNA in both SN and PLT fractions. This suggests a preferential preservation of mtDNA during diagenesis. To our knowledge this is the first empirical data indicating a preferential preservation of mitochondrial DNA. One possible mechanism causing this could be that mtDNA benefits from an additional protection against degradation by the double membrane of the mitochondrion. A less likely scenario would be different levels of endonuclease activities, to which the two types of DNA are exposed, as one would expect to see this effect, likely to happen at the very early stages of diagenesis, for the modern sample, too. Furthermore, Binladen et al. have suggested an interaction between chromosomal proteins and nucDNA (35), rendering it less accessible during PCR amplification or even causing its loss during DNA extraction.
Figure 4.
Ratio of mitochondrial to nuclear DNA in the six mammoths and the modern elephant sample. x-axis is the copy number for mitochondrial, y-axis the copy number for nuclear DNA with the black diagonal depicting the line for a mt:nuc ratio of 1000:1. The blue and red areas in the diagram show the mt:nuc ratio range derived from 454 data (for samples 915 and 917 only). See text for details.
Within the ancient samples, the mt:nuc ratios vary extensively, spanning some 60-fold difference. This indicates that the fate of either mt- or nucDNA is not necessarily the same for different bone specimens even when exposed to very similar environmental conditions, and thus questions the applicability of any generalized ratio, even to a specific tissue-type like compact mammalian bone.
For two of the six mammoth samples (namely 915 and 917), also 454 sequence data were available, allowing us to compare the mt:nuc ratios derived from both approaches. The accuracy of the 454 ratio estimate is limited by our current knowledge of the actual size of the elephant nuclear genome currently in the process of assembly, estimated at 4.2–4.8 Gb (38). Our 454-based calculations of the mt:nuc ratio for 917 yield a range of values consistently lower than the corresponding ratio based upon our qPCR results: 3469–3965:1 versus 17 369:1, although this might be the result of a quantitative bias of the qPCR approach for low nuclear copy numbers. Conversely, the qPCR results obtained for sample 915 agree quite well with the range derived from the 454 approach: 405:1 versus 317–362:1, respectively (Figure 4). Despite their difference, both estimates emphasize the same general pattern: 915 is, relatively speaking, almost as rich in nucDNA as the modern elephant bone, while 917 is extremely poor. This finding underscores the relevance of our qPCR framework to address the relative richness in nuc- and mtDNA for samples used for palaeogenomic approaches.
In addition, these results cast doubt on the usefulness of estimating the nuclear DNA content of a sample based on the quantitative assessment of its mitochondrial content (20,39). This is best exemplified by the sample 917. This specimen is distinct as it combines the second-lowest nucDNA yield with the highest mtDNA yield of all mammoths and emphasizing that a high level of mtDNA is no guarantee for a reciprocal high nuclear DNA content. The discrepancy between the mammoth samples clearly advocates for a specific nuclear quantitative assessment rather than an indirect mitochondrial approach to confirm the suitability of any ancient DNA extract in a whole-genome sequencing approach.
Modeling mtDNA damage
Non-bypassable DNA damages can a priori be classified into two main groups: damage creating new termini, i.e. strand breaks which limit the amplicon length to the actual size of the DNA molecule (40), and internal types of damage like AP-sites, base modifications or DNA–DNA or DNA–protein crosslinks (16,35), which can cause the Taq polymerase to stall and prevent the extension of the DNA molecule to its full length.
In Deagle et al.'s model (22) the average frequency at which a non-bypassable damage event occurs on any DNA strand is described by the parameter lambda. When applied to the mammoth mtDNA, lambda values are on average ∼2 times higher for the SN fraction than for the PLT fraction. Thus the mammoth DNA in the SN fractions appears to be twice as much prone to non-bypassable damage than that in the PLT fractions. This finding underlines the two different, non-exclusive mechanisms mentioned earlier: (i) post-mortem DNA degradation is not uniform across the bone but rather specific for (at least) the mineral and the organic matrix. (ii) the smaller and thus the more damaged the DNA fragments, the more likely they are flushed out of the organic matrix and into the SN fraction during the decalcification step.
Analysis of non-bypassable DNA damage using Deagle et al.'s model could not be achieved for the nucDNA of the mammoth samples due to an insufficient number of data points. However, this analysis was performed for the modern elephant bone, and the same trend (i.e. PLT fractions less degraded than SN fractions) was recovered with much lower lambda values, as expected from a less diagenetically altered bone. Interestingly, for both the SN and PLT, the nucDNA showed slightly lower lambda values than the mtDNA, which would suggest that nucDNA is possibly less prone to non-bypassable DNA damage during early diagenesis. This observation is at odds with the qPCR results from our mammoth data which suggests an apparently heavier degradation of the nucDNA compared to the mtDNA but might again point towards a differential susceptibility of DNA degradation during more advanced bone deterioration.
Although SN and PLT lambda values differ greatly for mtDNA within each mammoth, both sets of values appear to be consistent across the samples with almost parallel regression curves (Figure 3). The mean amplifiable length (estimated by 1/lambda) shows limited variation among the SN fractions (103.5 ± 13.5 bp) as well as the PLT fractions (189 ± 39 bp) in all mammoth samples. These results support the idea that, with regard to preservation, these samples are fairly comparable, possibly due to a leveling effect of the permafrost conditions against post-mortem DNA damage. Support for this view is strengthened by sample Ber12, which shows the highest lambda values (highest damage frequencies in both SN and PLT fraction) of all mammoth samples, suggesting that its storage at elevated temperatures during the last 30 years likely led to an accelerated DNA degradation after its excavation from the permafrost (the other five mammoths have been preserved in frozen conditions since their discovery less than 10 years ago). This emphasizes the need and usefulness of storing permafrost samples at below ambient temperatures after their excavation.
Whereas the lambda values derived from our samples apparently reflect the relative homogeneity of their preservation conditions in the permafrost, Deagle et al.'s other parameter N (22) describes yet another phenomenon. Parameter N, the Y-intercept of each degradation slope, can be related to the amount of starting molecules present in the qPCR reaction. Although the values for N are somehow theoretical, they can be analyzed relatively to each other between samples, and suggest that the richness in endogenous DNA may vary between samples by up to 10-fold in the SN and 25-fold in the PLT. Contrary to the uniform lambda, the N parameter thus reflects a wide variation in overall DNA content between the mammoth bones. Characteristics specific to each bone sample, such as the age of the individual (at the time of death), the thickness of the cortical bone, the difference in skeletal elements sampled or even the location of the sample within a specific skeletal element, could explain such differences.
When compared with other published serial qPCR data (see SOM) derived from degraded DNA it appears that the state of preservation of our mammoth samples (as modeled by lambda, mean = 0.0077 ± 0.0026) generally outperforms modern fecal samples [lambda = 0.0106–0.0116 on average; (22)], as well as recent archaeological remains spanning both the Neolithic and the Middle Age (21) not preserved in permafrost conditions (lambda = 0.0166 ± 0.0039). This is the first quantitative data supporting distinctly low DNA degradation rates due to unique preservation conditions in permafrost.
Cytosine deamination
Apart from non-bypassable damage, DNA extracted from ancient bone specimens is also subjected to bypassable damage mainly documented in the form of C to T deamination events (13), its most common form (15). Strikingly, the analysis of bypassable DNA damage follows the same trend as seen with non-bypassable damage: mtDNA in the SN appears to be consistently more prone to chemical modification as it contains on average 5.8 times more cytosine deamination events than mtDNA in the PLT. This result is supported by the multivariate analysis (Figure 5B) which shows that the vectors of C to T deaminations and lambda values are almost parallel for the SN and only slightly divergent along the second principal component for the PLT (in the latter case, the low total number of C to T deaminations recovered from the PLT fractions might have precluded its statistical significance compared with the SN).
Conversely, the amount of C to T deamination events does not appear to be related to the total amount of amplifiable DNA. Whereas the BMV and sample 917, which show the highest yields of mtDNA among our samples, both returned a rate of C to T of 0, the sample 473, which yields the lowest amount of both mt- and nucDNA, shows deamination rates almost identical to those of the BMV. These results underscore the absence of any correlation between the C to T rate and the parameter N which is also recovered from our multivariate analysis (Figure 5B). This outcome again emphasizes the usefulness of a qPCR approach with multiple amplicon sizes (compared to a single PCR or qPCR characterization) to assess the quality and quantity of DNA in extracts from ancient specimens.
In a previous report based on 454 sequence data, Gilbert et al. suggested deamination rates and hence degradation to be generally lower in ancient hair (average 0.47%, spanning 0.24–0.90%) than in comparably stored bone (1.70% for bone sample 2005/915 also used in the present study) (41). In contrast Debruyne et al. could not confirm these results and report a much lower deamination rate for another extract of the same sample 2005/915 (1). Our results based on an additional independent extraction of the very same sample yielded a rate of 0.55% for SN and PLT pooled (calculated as [rate(SN)*N(SN) + rate(PLT)*N(PLT)]/[N(SN) + N(PLT)]) comparable to the one reported by Debruyne et al. (0.48%).
Screening samples for DNA
Authenticity of ancient DNA remains an elusive goal, especially for ancient human DNA. As such, one is continuously striving to develop methods that facilitate an easy and accurate prediction of the presence or absence of (amplifiable) DNA of various fragment lengths in ancient samples. Those methods not only lend support to the authenticity of ancient DNA findings but can also be used as a screening tool in the situation when a large number of specimens is available/needs to be assessed (26). Chemical and biochemical characteristics of the bone have been utilized (3,4) under the assumption that they correlate with the overall preservation of DNA. Another approach currently employed is ‘thermal age’, where the age of a sample is calculated, as a function of its thermal history based upon the climatic conditions of the burial site, soil characteristics including thermal diffusivity and even shading due to possible vegetation cover (implicit to this model is the assumption that depurination is the only cause of DNA degradation) (6).
When compared to our qPCR results, the predictive power of these methods is questionable (Table 2; Figure 5). Some criteria apparently hint in the right direction but lack resolving power. Thus, the D/L ratios of aspartic acid certainly suggest that all mammoth samples are potentially good candidates for DNA retrieval, but they can not predict whether one of these samples will provide more or less total DNA. More specifically, although the PCA analysis suggests that this criterion might be the most reliable to estimate the nucDNA content (Figure 5A), it appears less strongly correlated to the mtDNA content (whether approached by lambda or N; Figure 5B).
Total amino-acid and collagen concentration as well as crystallinity indices appear to follow a common pattern and to be positively (although imprecisely) correlated with the quantitative preservation of DNA. Although one would expect that an increase in the CI would relate to an increase in the level of diagenesis of the bone (and thus to a decrease in collagen content of amino-acid concentration), our results apparently support the complete opposite (Figure 5). However, it should be noted that the range of variation observed for the CI of the mammoths is not significant and actually overlaps with that derived from modern bones.
Despite following a more comprehensive approach for describing DNA degradation by factoring in more than a single variable, i.e. sample age (17), the thermal age model (6) appears to be of limited usability in our study. For our six mammoth samples, increasing thermal ages are generally correlated with increasing quantities of DNA (more specifically mtDNA; Figure 5A) and with decreasing levels of DNA damage observed in mtDNA (Figure 5B), both results conflicting with predictions of the thermal age model. This unexpected outcome pinpoints the practical limitations of the thermal ages calculations. Indeed, of all the factors that contribute to the final determination of the effective depurination temperature (and thus the thermal age), only the sample age and its geographic location can be given with some degree of certainty. Other factors such as burial depth or relative humidity are difficult to assess, especially since little is known about the circumstances between the death of an animal and the retrieval of its remains. As these factors are generally unknown and thus the corresponding parameters impossible to acquire, default values have to be applied, resulting in thermal ages often restricted to the range of the 14C dates associated with the specimens. Although limited by the number of samples analyzed, our analyses demonstrate that there is no simple correlation between the absolute age of the mammoth samples and the preservation of their DNA (neither quantitatively nor qualitatively), not surprisingly as age is only one of many possible factors. The relevance of the thermal age as an analytical tool for predicting DNA preservation (41) is thus questionable for permafrost-preserved samples.
Concluding remarks
Our quantitative and qualitative analysis of DNA preservation in DNA extracts from permafrost-preserved mammoth bones has led to several new insights. (i) DNA recovered from bone is not only obtained during the enzymatic digestion step of the extraction but also to a significant extent released into the supernatant obtained during demineralization of the bone. (ii) Relative to the mtDNA, the nucDNA appears to be more concentrated in the SN fractions. Although there is evidence for different rates of preservation for both types of DNA, it appears to be likely that this is also the result of a size filtration effect. (iii) In spite of preservational similarities between the mammoth samples, the mt:nuc ratio appears to be extremely variable among samples, and the nuclear content thus poorly estimated by its mitochondrial counterpart. (iv) Apart from inevitable DNA damage, it appears to be also the richness in DNA in the living bone (or the piece of it that is eventually going to be extracted) that determines the amount of obtainable DNA; the latter factor seems to be as influential as the first one in our set of mammoth samples. (v) The preservation of permafrost preserved mammoths DNA is exceptional when compared with other documented qPCR data analyzed using Deagle et al.'s model. (vi) The commonly used methods to indirectly evaluate DNA preservation are fairly imprecise for fine scale determination of DNA preservation or even chiefly inappropriate for the permafrost preserved samples.
If applicable to a wider range of aDNA relevant specimens for which qPCR data are still pending, these findings could have general implications for the extraction of ancient DNA. We thus suggest that the DNA extraction method from bone be adjusted depending upon the scientific objectives. In cases where the recovery of any DNA is important, pooling the fractions (or extracting them together) will yield higher absolute amounts of DNA. If one is specifically interested in short nucDNA, targeting the SN fraction alone might be more relevant, whereas working on the PLT fraction alone will be more suited to any project aiming for the maximum amplifiable length.
This study once more demonstrates that the processes affecting the quality and quantity of DNA during diagenesis are still far from being understood. Thus additional efforts to meticulously characterize DNA damage and preservation will be necessary to gain a more comprehensive picture of the various aspects that contribute to the level as well as the state of DNA that we eventually can obtain from ancient specimens.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science and Engineering Research Council (grant #299103-2004); Social Sciences and Humanities Research Council grant (#410-2004-0579); the Canadian Research Chairs Program and McMaster University. Funding for open access charge: Natural Sciences and Engineering Research Council of Canada (NSERC).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Mike and Wendy Hackenberger from the Bowmanville Zoo for the Asian elephant bone sample as well as Bernard Buigues and Alexei Tikhonov for making the mammoth bone samples available to us. We thank David Harker and Matthew Collins for providing the thermal age data. We thank R. MacPhee, C. Flemming and D. Poinar for sampling.
REFERENCES
- 1.Debruyne R, Schwarz C, Poinar H. Comment on “Whole-Genome Shotgun Sequencing of Mitochondria from Ancient Hair Shafts”. Science. 2008;322:857. doi: 10.1126/science.1158565. [DOI] [PubMed] [Google Scholar]
- 2.Paabo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 3.Shemesh A. Crystallinity and diagenesis of sedimentary apatites. Geochim. Cosmochim. Acta. 1990;54:2433–2438. [Google Scholar]
- 4.Poinar HN, Hoss M, Bada JL, Paabo S. Amino acid racemization and the preservation of ancient DNA. Science. 1996;272:864–866. doi: 10.1126/science.272.5263.864. [DOI] [PubMed] [Google Scholar]
- 5.Poinar HN, Stankiewicz BA. Protein preservation and DNA retrieval from ancient tissues. Proc. Natl Acad. Sci. USA. 1999;96:8426–8431. doi: 10.1073/pnas.96.15.8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CI, Chamberlain AT, Riley MS, Stringer C, Collins MJ. The thermal history of human fossils and the likelihood of successful DNA amplification. J. Hum. Evol. 2003;45:203–217. doi: 10.1016/s0047-2484(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 7.Shapiro R. Damage to DNA Caused by Hydrolysis; from Chromosome Damage and Repair. New York: Plenum Publishing Corporation; 1981. [Google Scholar]
- 8.Ljungman M, Hanawalt PC. Efficient protection against oxidative DNA damage in chromatin. Mol. Carcinogen. 1992;5:264–269. doi: 10.1002/mc.2940050406. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T, Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 11.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 12.Jans MME, Nielsen-Marsh CM, Smith CI, Collins MJ, Kars H. Characterisation of microbial attack on archaeological bone. J. Archaeol. Sci. 2004;31:87–95. [Google Scholar]
- 13.Hofreiter M, Jaenicke V, Serre D, Haeseler Av, Paabo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert MT, Hansen AJ, Willerslev E, Rudbeck L, Barnes I, Lynnerup N, Cooper A. Characterization of genetic miscoding lesions caused by postmortem damage. Am. J. Hum. Genet. 2003;72:48–61. doi: 10.1086/345379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, Ovodov ND, et al. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:13578–13584. doi: 10.1073/pnas.0605327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Höss M, Jaruga P, Zastawny TH, Dizdaroglu M, Paabo S. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen AJ, Mitchell DL, Wiuf C, Paniker L, Brand TB, Binladen J, Gilichinsky DA, Ronn R, Willerslev E. Crosslinks rather than strand breaks determine access to ancient DNA sequences from frozen sediments. Genetics. 2006;173:1175–1179. doi: 10.1534/genetics.106.057349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handt O, Hoss M, Krings M, Paabo S. Ancient DNA: methodological challenges. Experientia. 1994;50:524–529. doi: 10.1007/BF01921720. [DOI] [PubMed] [Google Scholar]
- 19.Handt O, Krings M, Ward RH, Paabo S. The retrieval of ancient human DNA sequences. Am. J. Hum. Genet. 1996;59:368–376. [PMC free article] [PubMed] [Google Scholar]
- 20.Poinar HN, Schwarz C, Qi J, Shapiro B, Macphee RD, Buigues B, Tikhonov A, Huson DH, Tomsho LP, Auch A, et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 21.Malmstrom H, Svensson EM, Gilbert MTP, Willerslev E, Gotherstrom A, Holmlund G. More on contamination: The use of asymmetric molecular behaviour to identify authentic ancient human DNA. Mol. Biol. Evol. 2007;24:998–1004. doi: 10.1093/molbev/msm015. [DOI] [PubMed] [Google Scholar]
- 22.Deagle BE, Eveson JP, Jarman SN. Quantification of damage in DNA recovered from highly degraded samples – a case study on DNA in faeces. Front. Zool. 2006;3:11. doi: 10.1186/1742-9994-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zullo S, Kennedy JL, Gelernter J, Polymeropoulos MH, Tallini G, Pakstis AJ, Shapiro MB, Merril CR, Kidd KK. Eliminating mitochondrial DNA competition for nuclear DNA primers. Genome Res. 1993;3:39–45. doi: 10.1101/gr.3.1.39. [DOI] [PubMed] [Google Scholar]
- 24.Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 25.Kuch M, Gröcke DR, Knyf MC, Gilbert MTP, Younghusband B, Young T, Marshall I, Willerslev E, Stoneking M, Poinar H. A preliminary analysis of the DNA and diet of the extinct Beothuk: A systematic approach to ancient human DNA. Am. J. Phys. Anthropol. 2007;132:594–604. doi: 10.1002/ajpa.20536. [DOI] [PubMed] [Google Scholar]
- 26.Debruyne R, Chu G, King CE, Bos K, Kuch M, Schwarz C, Szpak P, Gröcke DR, Matheus P, Zazula G, et al. Out of America: ancient DNA evidence for a new world origin of late quaternary woolly mammoths. Curr. Biol. 2008;18:1320–1326. doi: 10.1016/j.cub.2008.07.061. [DOI] [PubMed] [Google Scholar]
- 27.Amelung W, Zhang X. Determination of amino acid enantiomers in soils. Soil Biol. Biochem. 2001;33:553–562. [Google Scholar]
- 28.Zhao M, Bada JL. Determination of [alpha]-dialkylamino acids and their enantiomers in geological samples by high-performance liquid chromatography after derivatization with a chiral adduct of o-phthaldialdehyde. J. Chromatogr. A. 1995;690:55–63. doi: 10.1016/0021-9673(94)00927-2. [DOI] [PubMed] [Google Scholar]
- 29.Weiner S, Bar-Yosef O. States of preservation of bones from prehistoric sites in the Near East: a survey. J. Archaeol. Sci. 1990;17:187–196. [Google Scholar]
- 30.Wright LE, Schwarcz HP. Infrared and isotopic evidence for diagenesis of bone apatite at Dos Pilas, Guatemala: palaeodietary implications. J. Archaeol. Sci. 1996;23:933–944. [Google Scholar]
- 31.Surovell TA, Stiner MC. Standardizing infra-red measures of bone mineral crystallinity: an experimental approach. J. Archaeol. Sci. 2001;28:633–642. [Google Scholar]
- 32.Collins MJ, Nielsen-Marsh CM, Hiller J, Smith CI, Roberts JP, Prigodich RV, Wess TJ, Csapo J, Millard AR, Turner-Walker G. The survival of organic matter in bone: a review. Archaeometry. 2002;44:383–394. [Google Scholar]
- 33.Noonan JP, Hofreiter M, Smith D, Priest JR, Rohland N, Rabeder G, Krause J, Detter JC, Paabo S, Rubin EM. Genomic sequencing of Pleistocene cave bears. Science. 2005;309:597–599. doi: 10.1126/science.1113485. [DOI] [PubMed] [Google Scholar]
- 34.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat. Protocols. 2007;2:1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 35.Binladen J, Wiuf C, Gilbert MTP, Bunce M, Barnett R, Larson G, Greenwood AD, Haile J, Ho SYW, Hansen AJ, et al. Assessing the fidelity of ancient DNA sequences amplified from nuclear genes. Genetics. 2006;172:733–741. doi: 10.1534/genetics.105.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green RE, Malaspinas A-S, Krause J, Briggs AW, Johnson PLF, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, et al. A complete neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robin ED, Ronald W. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell Physiol. 1988;136:507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- 38.Redi C, Garagna S, Zuccotti M, Capanna E. Genome Size: A Novel Genomic Signature in Support of Afrotheria. J. Mol. Evol. 2007;64:484–487. doi: 10.1007/s00239-006-0237-1. [DOI] [PubMed] [Google Scholar]
- 39.Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 40.Briggs AW, Stenzel U, Johnson PLF, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, Lachmann M, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert MT, Tomsho LP, Rendulic S, Packard M, Drautz DI, Sher A, Tikhonov A, Dalen L, Kuznetsova T, Kosintsev P, et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007;317:1927–1930. doi: 10.1126/science.1146971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.