Abstract
Bacterial growth requires equilibrated concentration of C, N and P sources. This work shows a phosphate control over the nitrogen metabolism in the model actinomycete Streptomyces coelicolor. Phosphate control of metabolism in Streptomyces is exerted by the two component system PhoR-PhoP. The response regulator PhoP binds to well-known PHO boxes composed of direct repeat units (DRus). PhoP binds to the glnR promoter, encoding the major nitrogen regulator as shown by EMSA studies, but not to the glnRII promoter under identical experimental conditions. PhoP also binds to the promoters of glnA and glnII encoding two glutamine synthetases, and to the promoter of the amtB-glnK-glnD operon, encoding an ammonium transporter and two putative nitrogen sensing/regulatory proteins. Footprinting analyses revealed that the PhoP-binding sequence overlaps the GlnR boxes in both glnA and glnII. ‘Information theory’ quantitative analyses of base conservation allowed us to establish the structure of the PhoP-binding regions in the glnR, glnA, glnII and amtB genes. Expression studies using luxAB as reporter showed that PhoP represses the above mentioned nitrogen metabolism genes. A mutant deleted in PhoP showed increased expression of the nitrogen metabolism genes. The possible conservation of phosphate control over nitrogen metabolism in other microorganisms is discussed.
INTRODUCTION
Bacterial growth requires an equilibrated concentration of carbon, nitrogen and phosphorus sources. Carbon, nitrogen and phosphate concentrations exert an important regulatory effect on primary and secondary metabolism in different bacteria including Streptomyces (1–5).
Phosphorus is an essential component of bacterial nutrition; expression of phosphate-regulated genes in Streptomyces species is modulated by the two-component system PhoR-PhoP (6). PhoR is the membrane sensor protein kinase which senses phosphate scarcity; PhoP is the response regulator which binds DNA and controls the transcription of genes belonging to the so-called pho regulon. PhoP was shown to control the expression of primary and secondary metabolism genes including actinorhodin and undecylprodigiosin biosynthesis genes (6,7). Binding of PhoP to the promoter regions of three different genes of the pho regulon pstS, phoU and phoRP was shown both in Streptomyces coelicolor (8) and S. natalensis (9). The PhoP-binding operator sequences of these genes as well as those present in the promoter regions of phoA and phoD of S. coelicolor (10) are composed of direct repeat units (DRu) of 11 nt. Operator sequences of other PhoP-regulated genes have been recently described in S. coelicolor and classified into three types of DRus organization with different degrees of complexity (11).
In bacteria a central role in nitrogen metabolism is played by glutamine synthetases that assimilate ammonium into the cellular organic nitrogen (NH3 + glutamate + ATP → glutamine + ADP + Pi) (2). The overall nitrogen metabolism is regulated in S. coelicolor by complex mechanisms that involve an apparent doubling of some structural and regulatory genes (12). There are two glutamine synthetase (GS) genes in S. coelicolor: glnA (13), that encodes the GS type I (GS-I), orthologous to that found in prokaryotes, and glnII (14), which encodes GS type II (GS-II), similar to eukaryotic GS. The glnII gene appears to be present in all Streptomyces species (15), but it is absent in the related actinomycetes Mycobacterium and Corynebacterium (12). The two types of GS enzymes differ in size, number of subunits, expression pattern and post-translational modification (2). In S. coelicolor, the adenylyltransferase GlnE down-regulates the GS-I enzyme by post-translational modification in response to excess of ammonium (16). The adenylated GS-I enzyme becomes inactive. GlnE can also deadenylate and restore the GS-I activity under ammonium limitation. This post-translational regulatory mechanism is widespread in bacteria. In Escherichia coli the GlnE activity depends on three proteins that are thought to be nitrogen sensing proteins. These are the GlnD protein, an uridylyltransferase/uridylyl-removing enzyme, and two proteins of the PII signal transduction superfamily, GlnB and GlnK (2,17). Both GlnB and GlnK are covalently modified—and thus regulated—by GlnD, and both PII proteins modulate the GlnE activity.
The GlnD and GlnK homologues of S. coelicolor were characterized by Hesketh et al. (18). In contrast to the enteric system, these proteins are not required for the GlnE-mediated regulation of the GS-I enzyme. The targets of the GlnK/GlnD system are not yet known (12). Genes encoding this system are clustered with the putative ammonium transporter gene amtB and form the operon amtB-glnK-glnD (19).
Two regulatory genes glnR and glnRII control expression of several nitrogen metabolism genes at the transcriptional level. The global nitrogen activator/repressor GlnR controls all the important routes for ammonium assimilation (19–22). GlnR in response to nitrogen limitation activates the transcription of glnA and glnII (encoding both GSs), as well as the transporter amtB and the putative nitrite reductase gene nirB. The binding motif of GlnR (GlnR box), has been characterized by footprinting assays of glnA, gdhA and nirB promoters, and by sequence analysis of 13 bound promoters (19,22).
The regulator gene glnRII is located 1 kb downstream of glnII, separated by two hypothetical coding sequences. The overall amino-acid sequence of GlnRII is similar to GlnR, and is nearly identical in the DNA recognition helixes. In fact, GlnRII also binds the promoter regions of glnA, glnII and amtB (19). Nevertheless, it is not strictly a functional homologue of GlnR and its role in nitrogen regulation is not yet clearly established (12).
Initial microarray studies (23) suggested that there might be a connexion between the phosphate control exerted by PhoP and the overall nitrogen regulation mediated by GlnR. It was, therefore, of great interest to study the relationships between these two pleiotropic regulators in the control of metabolism in S. coelicolor.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
The S. coelicolor strain M145 (24), which is the standard strain for the European Union STREAM (Streptomyces analysis of metabolism) project was used as the wild-type strain. The M145-derivative mutant strain S. coelicolor INB201 was obtained by replacement of the phoP coding sequence by the apramycin resistance cassette (F. Santos-Beneit et al., unpublished results). All strains were manipulated and conjugated according to standard procedures (24).
All the DNA materials are listed in Table 1. PCR using the indicated amplification primer pairs were done to clone promoter regions into adequate vectors. Fidelity of the PCR was checked by sequencing the entire insert. Vectors pGEM-T easy and pBS II KS + were used to clone the indicated amplicons (Table 1) for gel retarding and footprinting analyses. For luciferase reporter analysis, primers containing a NdeI-restriction site were designed in order to clone the promoters into the ATG codon of the luxA gene. Using this strategy, the cloned promoter sequences include the Shine-Dalgarno nucleotides and maintain the same distance to the start codon in the luciferase fusions than in the original gene.
Table 1.
List of primers and plasmids
| Primer | Sequencea | Promoter | Size of amplicon |
|---|---|---|---|
| PHO-37 | TCTAGAGGCTACGACGAGCGGGAAC | glnII | 316 |
| PHO-38 | GGATCCACGGGGCCACATCCTTCG | ||
| PHO-39 | TCTAGAGGAGAGCCACGATCCGATTG | glnA | 284 |
| PHO-40 | GGATCCCGGCGTTCTGGAACATCC | ||
| PHO-41 | TCTAGATCCCGAACTGCCCGACTC | amtB | 293 |
| PHO-42 | GGATCCATCGGCGTCTCCTCGTCG | ||
| glnR-1 | GCCGTACGGAGGAAGGTACG | glnR | 362 |
| CAR35 | TCAGGAGCAGCAGAGAACTCATC | ||
| CAR36 | GGCGGTCGGTTGCTCATG | glnR | 257 |
| glnRII-1 | GGATCCCCACGCACTGAGAGGAGTCTCCT | glnRII | 321 |
| glnRII-2 | TCTAGAATGAGACGTCAGCTCTTTCGCG | ||
| CAR57 | GTCAGGATCCGTCTCGGGATGCGGACGATTGG | glnR | 308 |
| CAR58 | ATGGTACCATATGCCCCACCTGCCTGGGACGGTTTG | ||
| CAR59 | GAACGGATCCGAGCCACGATCCGATTGC | glnA | 273 |
| CAR60 | ATGGTACCATATGGCTCCTCCTACTCCCGACCGT | ||
| CAR61 | TTCTGGATCCGTCCCACTTCGGACCGCTGATC | glnII | 307 |
| CAR62 | ATGGTACCATATGGCCACATCCTTCGGGGTGGGTCT | ||
| CAR63 | CCGTGGATCCGGCCGTACGCGATTTC | amtB | 420 |
| CAR64 | CTGGTACCATATGCGTCTCCTCGTCGTTG | ||
| 6FAM-F | CGACGTTGTAAAACGACGGCCAGT | Various | |
| Reverse | GGAAACAGCTATGACCATG | ||
| 6FAM-R | CAGGAAACAGCTATGAC | Various | |
| Forward | GTAAAACGACGGCCAGT | ||
| Plasmid | Featuresb | Reference | |
| pGEM-T-easy | Cloning vector, Ampr | Promega | |
| pBS II KS+ | Cloning vector, Ampr | Stratagene | |
| pBS II SK+ | Cloning vector, Ampr | Stratagene | |
| pLUXAR+neo | Conjugative-integrative promoter-probe vector, luxAB genes, Amr, Neor | Pérez-Redondo, unpublished | |
| pGEM-PglnII | PCR product from PHO-37 and PHO-38 cloned into pGEM-T-easy | This work | |
| pGEM-PglnA | PCR product from PHO-39 and PHO-40 cloned into pGEM-T-easy | This work | |
| pGEM-PamtB | PCR product from PHO-41 and PHO-42 cloned into pGEM-T-easy | This work | |
| pBS-PglnR | PCR product from glnR-1 and CAR35 cloned into pBS II KS+(EcoRV) | This work | |
| pBS-PglnR-b | PCR product from CAR35 and CAR36 cloned into pBS II KS+(EcoRV) | This work | |
| pBS-PglnRII | PCR product from glnRII-1 and glnRII-2 cloned into pBS II KS+(EcoRV) | This work | |
| pAR-N1 | PCR product from CAR57 and CAR58 cloned into pBS II SK+ | This work | |
| pAR-N2 | glnRp from pAR-N1 (−289 to +3) cloned into pLUXAR+neo (BamHI, NdeI) | This work | |
| pAR-N3 | PCR product from CAR59 and CAR60 cloned into pBS II SK+ | This work | |
| pAR-N4 | glnAp from pAR-N3 (−253 to +3) cloned into pLUXAR+neo (BamHI, NdeI) | This work | |
| pAR-N5 | PCR product from CAR61 and CAR62 cloned into pBS II SK+ | This work | |
| pAR-N6 | glnIIp from pAR-N5 (−287 to +3) cloned into pLUXAR+neo (BamHI, NdeI) | This work | |
| pAR-N7 | PCR product from CAR63 and CAR64 cloned into pBS II SK+ | This work | |
| pAR-N8 | amtBp from pAR-N7 (−401 to +3) cloned into pLUXAR+neo (BamHI, NdeI) | This work |
aRestriction sites introduced in amplicons are indicated by underline.
bPromoter coordinates are referred to the translation start site.
Streptomyces cultures were carried out in defined MG-3.2 medium containing 50 g l−1 starch, 60 mM glutamate, and 3.2 mM potassium phosphate. Total 106 spores ml−1 were used to inoculate 100 ml of MG medium in 500 ml baffled flasks and incubated at 30°C, 300 r.p.m. as indicated previously (25).
Electrophoretic mobility assays (EMSA)
DNA–PhoP interaction was tested in electrophoretic mobility shift assays (EMSA) as described previously (8). The promoters were excised from plasmids by restriction digestion and labelled at both ends with digoxigenin using the DIG Oligonucleotide 3′-end Labeling Kit, Second Generation (Roche). DNA fragments were incubated with different GST-PhoPDBD concentrations and DNA–protein complexes were resolved by PAGE.
DNase I footprinting assays
Pure GST-PhoPDBD protein was used for DNase I footprinting assays as previously described (8). DNA probes for glnRp, glnAp, glnIIp and amtBp were obtained by PCR using a 6-FAM-modified primer for labelling just one strand. Labelled and unlabelled primer pairs correspond to the forward and reverse M13 sequences listed in Table 1. The reaction mixtures included 9.3 nM of probe DNA and 2 μM of GST-PhoPDBD protein.
The labelled primers were used also for Sanger sequencing with the Thermo Sequenase Primer Cycle Sequencing Kit (GE Healthcare). Each reaction was loaded into an ABI PRISM 3130 sequencer together with the molecular standard Gene-Scan® 500 LIZ™ (Applied Biosystems). Electropherograms were analysed with PeakScanner v1.0 software (Applied Biosystems) to determine the protected sequence.
Luciferase assay and growth determination
The reporter luciferase activity was measured in a Luminoskan luminometer (Labsystems, Helsinki) as described previously (23,25). Riboflavine (10 μg ml−1) was added to the cell suspension in all samples to improve the sensitivity of luciferase assays, following the recommendation of Bachmann et al. (26). For dry weight determination, culture samples of 2 ml were washed twice with MilliQ water and dried at 80°C during 4 days.
‘Information theory’ analysis of binding sites
To calculate the information content (Ri value) of individual sequences (27), and to obtain logos and walkers for the analysis of binding sites we used the Delila package which include the makebk, encode, rseq, dalvec, makelogo, ri and lister programs (28,29). DNA sequences were scanned for binding sites using the RSA tools server (30) and Ri matrixes.
RESULTS
Transcriptional response of the nitrogen metabolism genes to phosphate limitation
We previously reported that phosphate limitation upregulated nitrogen metabolism genes in the ΔphoP mutant (23). Microarray results showed that glutamine synthetase gene glnII and genes of the operon amtB-glnK-glnD increased their transcription in response to phosphate downshift in the ΔphoP mutant, while no change was detected in the wild-type strain. The GlnA protein, detected in 2D gels, was overproduced in the ΔphoP mutant. These expression responses suggested that PhoP might be exerting a negative effect on expression of those nitrogen metabolism genes.
Inspection of the nucleotide sequences of the promoter regions indicated the presence of PHO-like sequences in the above mentioned nitrogen structural genes and in the glnR regulator gene. This suggested that the previously observed transcriptomic profiles were due to both direct and indirect regulation by PhoP. The direct control was checked by DNA-binding analysis of the glnA, glnII and amtB promoters using the purified DNA-binding domain of the PhoP response regulator (GST-PhoPDBD) (8). The promoter regions of the two regulatory genes, glnR and glnRII, were also analysed to explore a possible indirect control of the structural genes.
PhoP binds the glnR promoter but not the glnRII one
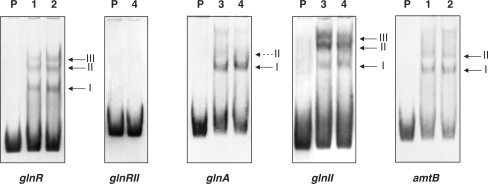
The glnR 5′-region was cloned by PCR as a fragment of 362 bp that also included 98 bp of the upstream coding sequence (CDS). Electrophoretic mobility shift assays (EMSA) of this fragment with the GST-PhoPDBD protein revealed the formation of three retarded bands (Figure 1). As reported previously (11), each retarded DNA–protein complex correspond to a number of protein monomers bound to the DNA fragment. The established model of the PhoP-binding site indicates that each PhoP monomer binds a direct repeat unit (DRu) of 11 nt. Two or three consecutive DRus form the core of the binding site. Once the core is occupied, further protein monomers can bind adjacent DRus, what account for the DNA–protein complexes of lower electrophoretic mobility (see the detailed analysis below).
Figure 1.
Analysis by EMSA of the promoters. Lane P, probe without protein; lanes 1–4, increasing concentrations of GST-PhoPDBD (0.125, 0.25, 0.5 and 1 µM, respectively). An excess (more than 1000-fold) of poly[d(I-C)] is included in every lane as internal control to avoid an unspecific binding of the protein to the DNA. Controls with competing excess of unlabelled probe are shown as Supplementary Data (Figure S1). The different shift bands are indicated by arrows. The assays were repeated three times.
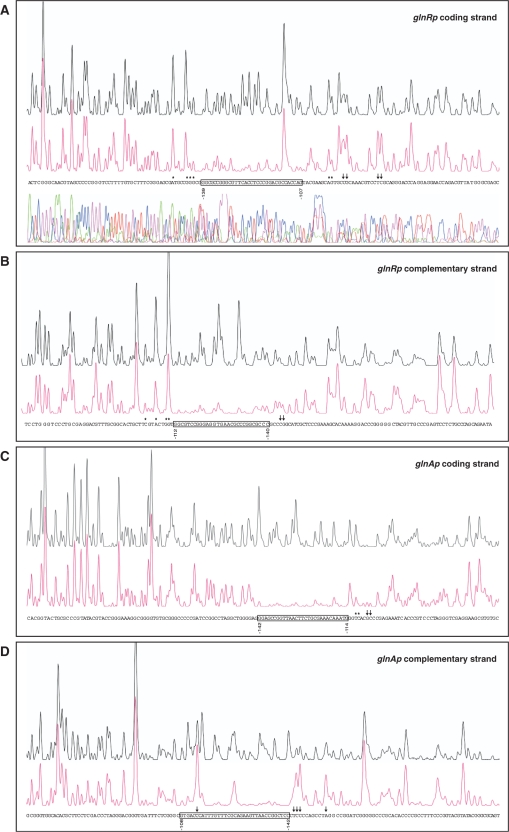
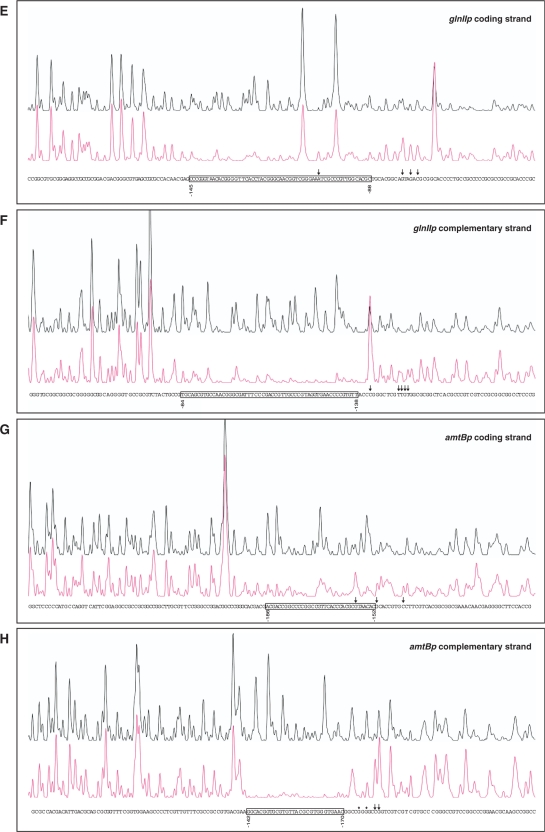
To locate the PhoP-binding site, we carried out DNase I footprinting experiments. Electrophoretic separation of digestion products was facilitated by using a smaller fragment of 257 bp that comprised only the glnR promoter sequence. The GST-PhoPDBD protein at a concentration of 2 μM protected from DNase I digestion a stretch of 33 nt located at positions −139 to −107 in the coding strand (all coordinates are referred to the translation start site; Figure 2A). This stretch comprised a DRu with sequence matching the first seven PHO box consensus bases (GTTCACC). In addition, some upstream and downstream nucleotides were protected to a lesser extent by protein binding. As previously observed in other PhoP footprintings (11), DNase I hypersensitive sites appeared next to the binding site at its 3′-end. The complementary strand showed protection from −112 to −140 nt, partial protections up to position −103, and hypersensitive sites at positions −144 and −145 (Figure 2B).
Figure 2.
DNase I footprints of the GST-PhoPDBD protein bound to the promoter regions of glnR (A, B), glnA (C, D), glnII (E, F) and amtB (G, H). In each panel, the upper electrophoregram (black line) is the control reaction. The protected nucleotide sequence is boxed; partially protected nucleotides (*), and hypersensitive sites (arrows) are also indicated. Sequencing reactions are not included except in panel A. Coordinates are from the translation start codon.
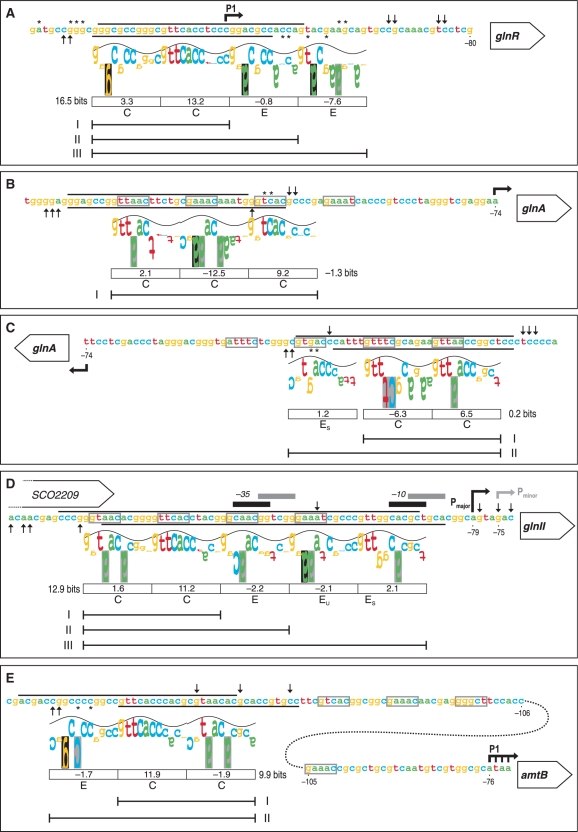
The promoter region of glnR contains three transcription start sites at positions −200, −170 to −168 and −119, which correspond to promoters P3, P2 and P1, respectively (21). Promoter P3 is active only during the exponential phase, whereas transcriptions originated from P2 and P1 promoters are detected at both exponential and stationary phases (31). Fink et al. (19) reported that only P2 and P3 are active in conditions of nitrogen limitation. The PhoP protection covers the distal P1 transcription start (Figure 3A). Thus, binding of PhoP probably blocks the glnR expression whatever promoter is active.
Figure 3.
‘Information content’ analysis of the new PhoP-binding sites identified in this work using the sequence walker method (29). The footprinting results of the promoter regions of glnR (A), glnA (B, C), glnII (D) and amtB (E) are summarized on the nucleotide sequences. The protected regions of the upper and bottom strands are indicated by the respective lines. Symbols for partially protected (asterisk) and hypersensitive sites (upward arrow, downward arrow) are as in Figure 2. For the PhoP-binding site in glnA, the two possible walkers are shown in the coding (B) and complementary strands (C). The walker limits are 2 bits, which is also the top of the sine wave, and −3 bits at the bottom. The height of each letter is the individual information value, determined from the Model I weight matrix (11). Negative values are represented by upside down letters. When a base does not occur at a given position in the set of sequences which forms the model, the letter background is black; a grey background indicates that the letter extends beyond the lower limit (−3 bits). The sine wave has a periodicity of 10.6 bases (the helical twist of B-form DNA) and serves to indicate major/minor groove contacts (28). The following features are bellow the walkers: boxes containing the Ri values for the above 11 nt direct repeat unit (DRu), the total Ri for the core site, letter codes for the DRu structure [C for core, E for extension, EU for extension unstable, ES for extension support (11)], and line segments comprising the DRus bound by PhoP monomers in the retarded complexes observed in Figure 1. The conserved 5-mer sequences of the GlnR-binding sites are boxed (grey lines). Bent arrows are the transcription start points. The −35 and −10 elements of major and minor glnII promoters are also shown.
Although computer searches of the promoter region of the second nitrogen regulatory gene glnRII did not reveal any conserved PHO DRu, we examined it for PhoP binding. Even at 1 μM of protein, the EMSA results did not show any binding under conditions identical to those used for glnR (Figure 1; see ‘Discussion’ section).
PhoP-binding sites overlap the GlnR boxes in both glutamine synthetase genes glnA and glnII
The 5′ sequences of the two glutamine synthetase genes of S. coelicolor were amplified by PCR. The cloned glnA fragment comprised the intergenic sequence (−220 to +16) and 36 bp of the upstream opposite CDS. The glnII fragment contained the full promoter region of 136 bp plus the last 105 bp of the upstream CDS. Both fragments were able to bind PhoP since they gave shifted bands in EMSA.
The glnAp gave a clear shifted band and a weak one of lower electrophoretic mobility (I and II, respectively; Figure 1). The footprinting assay of the glnA coding strand revealed a protected region extending from −142 to −114 and partial protection of nucleotides −111 and −110 (Figure 2C). The same region was protected in the complementary strand, from −142 to −108, although a hypersensitive site was evident at nucleotide −113 (Figure 2D). This protected region comprises the first and half of the second GlnR box that have been described in the glnA promoter (22). The GlnR boxes have been defined as composed of two conserved sequences of 5 nt separated by 6 variable nt (22) (see also Figure 3B–E). GlnR function as a transcriptional activator of glnA (19,22). Binding of PhoP at the glnA promoter region is likely to interfere with GlnR binding and possibly with RNA polymerase function since the glnA transcription start site is located at position −74 (20) (Figure 3B).
The gel mobility shift assays of the glnII promoter revealed three clear DNA–protein complexes. Complexes II and III showed stronger retardation (lower mobility) than complex I, and also showed a higher signal intensity (Figure 1).
As shown in Figure 2E and F, the GST-PhoPDBD protein protected a large nt stretch of the glnII promoter from DNase I digestion, extending from −145 to −88 in the coding strand, and from −138 to −84 in the complementary strand. In contrast to the glnA sequence, the coding strand of glnII exhibited a well-conserved DRu (Figure 3D). Also, the large protected nt stretch completely covers the two GlnR boxes described by Tiffert et al. (22). Two closely placed promoters have been described in the glnII gene. The main promoter should correspond to the major vegetative σ factor and its transcription start site is the adenine −79. The minor promoter shows putative elements of σ31 (14). This sigma factor, which is more active at stationary phase, recognizes promoters of actIII (an actinorhodin structural gene) and of hrdD (a homologue of Escherichia coli rpoD), and also the glnR P2 promoter (31). As shown in Figure 3D, the overlapping location of promoters and binding sites indicate that PhoP may repress glnII transcription interfering with both GlnR binding and promoter activity.
The promoter for the amtB-glnK-glnD operon contains separated PhoP and GlnR operators
The amtB gene encodes a putative ammonium transporter and forms an operon with glnK and glnD, which encode the PII protein and the PII nucleotidyl transferase respectively (18,19). The latter two proteins may be considered nitrogen metabolism sensors. Using RT–PCR experiments and a glnR mutant, Tiffert et al. (22) showed that GlnR activates the transcription of the amtB gene.
EMSA analyses of the DNA fragment containing the amtB 5′-region (−285 to +4) showed a clear shifted band and a second weak one with lower mobility (Figure 1). The DNase I protection of this fragment extended from −186 to −152 in the coding strand what contained a conserved DRu (Figure 2G). Results for the complementary strand showed a protected stretch from −142 to −170, and partial protection of the nucleotides −175 and −177 (Figure 2H).
In contrast to what we have found for the GS genes, the PhoP-binding site at the amtB promoter is placed upstream and separated from the GlnR boxes (Figure 3E). Three separated promoters (P3 to P1) have been identified in the amtB upstream region by low resolution S1 analysis (19). In principle, the location of the P1 transcription start site (−76 to −73, Figure 3E) suggests that PhoP does not control this promoter but the upstream ones P3 and P2. In this case PhoP might act as a ‘road-block’ preventing the expression from the upstream promoters.
Analysis of the complex operator structures
The PhoP-binding sites are formed by direct repeats units (DRu) of 11 nt, each one bound by a protein monomer. We have recently described the structures of DRus based on the ‘information theory’ analysis and on combined EMSA and footprinting results (11). The differentiated structures served to classify the PHO operators into three groups. Operators that produce a unique retarded band in EMSA are either class I, if formed by 2 DRus (i.e. one PHO box), or class II, if composed of three DRus. More frequently (as occurs in all cases reported here) PhoP binding produces two or more retarded bands in EMSA. Those sites belong to class III and comprise two or three conserved DRus that form the core of the site, and one or several DRus that extend the protein occupancy beyond the core.
Figure 3 includes the summary of the footprinting results. The protected regions were analysed by the sequence walker method (29) using the Model 1 matrix (11). In each walker, the letter height represents the conservation of the base and its contribution to the information content of the site (Ri value). Base conservation correlates with the number of contacts to the protein (32), and, when displayed as walkers, serves to identify the binding sequence.
The protected region in the glnR promoter comprised three DRus, and a partially protected fourth one (Figure 3A). The proposed DRu structure explains the formation of the three observed complexes in EMSA (Figure 1). A similar operator structure was observed in the phytase gene (SCO7697) (11).
The PhoP-binding site in the glnA promoter showed a less evident interpretation. In the coding strand a class II structure was suggested. This implies that the glnAp fragment should yield a unique retarded complex. Nevertheless, a second faint band appeared above the major retarded band in EMSA (Figure 1). Moreover, the highest conserved DRu is poorly protected, and the total Ri for this structure is negative (−1.3 bits, Figure 3B). When an accurate binding model is used, it has been demonstrated that the Ri value must be positive (29). In the complementary strand a more plausible structure was found because the core sequences show a low but positive Ri (0.2 bits), they were completely protected, and the two retarded bands can be explained (Figure 3C). The large protected sequence in the glnII promoter included five DRus (Figure 3D) that account for the formation of the three EMSA complexes.
The PhoP protected region in the amtB promoter covered a well conserved DRu that is flanked by two poorly conserved DRus. The two observed EMSA complexes indicate a structure composed of a 2 DRu core plus an extension repeat. The degree of protection and the walkers served us to decide which of the flanking DRus forms part of the core (Figure 3E).
Reporter studies: PhoP represses the transcription of nitrogen genes
In order to quantify the effect of PhoP on expression of the nitrogen-regulated genes, glnR, glnA, glnII and amtB promoter regions were cloned in pLUXAR+neo driving expression of the luxAB reporter gene. The resulting integrative plasmids pAR-N2 (glnRp), pAR-N4 (glnAp), pAR-N6 (glnIIp), pAR-N8 (amtBp) were introduced by conjugation into the wild-type M145 strain and into the INB201 (ΔphoP) mutant. Plasmids are stably maintained by integration into the Streptomyces chromosome ΦC31 attB site.
Cultures were grown in MG-3.2 medium (containing 3.2 mM phosphate), using three independent replicates. The cultures showed a diauxic growth with transition phase from 45 to 51 h that triggered the onset of antibiotic production; the ΔphoP mutant showed reduced growth as described in Santos-Beneit et al. (25).
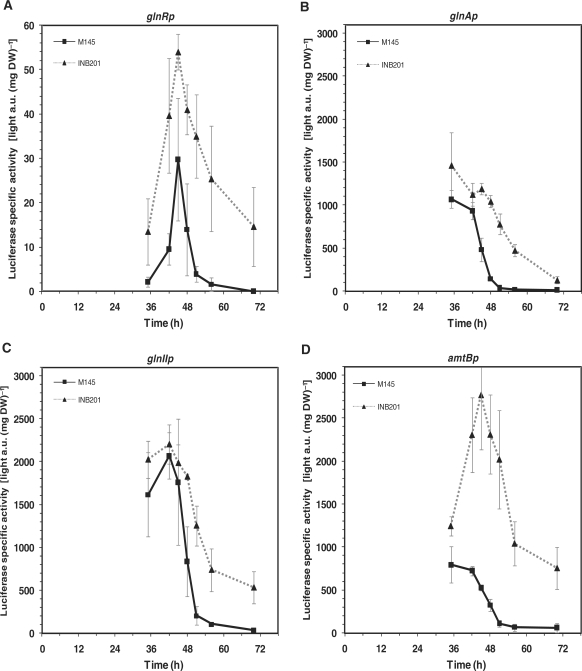
Using these reporter constructions it was clearly observed that the four promoter regions were repressed by PhoP since reporter enzyme higher activities were observed in the ΔphoP mutant with respect to parental strain at all sampling times (Figure 4A–D). The reporter expression patterns, varied among the promoters assayed. Thus, compared to the other assayed promoters (of phosphate transporter genes pstS, pitH1 and pitH2, and of glycerophosphodiesterase genes glpQ1 and glpQ2) [(23,25), Santos-Beneit, unpublished] the promoter region of the glnR gene produced usual or low activities across the time course of the culture. Indeed, in the wild-type strain the luminescence signals were below the instrument detection limit at 70 h (Figure 4A). In contrast, the structural glnA, glnII and amtB genes appeared to have very strong promoters. Among these, glnIIp showed the highest activities in the wild-type strain. Besides, maximum activities of the different promoters were reached at distinct growth phases. In both strains glnRp activities increased until the transition phase (45 h, Figure 4A), and decreased thereafter. For the glnA and amtB promoter activities, the highest values were at the exponential growth phase in the wild-type (35 h, Figure 4B and D). The glnIIp showed high activities during the first growth phase and a maximum activity at 42 h (at the initial transition phase, Figure 4C). In all cases, the wild-type activities dropped rapidly after the transition phase resulting in low values in the stationary phase. In contrast, deletion of the phoP gene caused slower promoter activity drops, as seen in the INB201 plots (Figure 4A–D).
Figure 4.
Promoter activity in S. coelicolor M145 (squares, solid lines) and ΔphoP mutant (triangles, dashed lines) of glnR (A), glnA (B), glnII (C) and amtB (D) using the luxAB genes as reporter. Error bars correspond to the standard error of triplicated cultures in MG-3.2.
In summary, we can conclude that PhoP represses directly the transcription of GS genes glnA, glnII, the putative ammonium transporter gene amtB, and the nitrogen regulatory genes glnD and glnK, which form part of the amtB operon. In addition, PhoP negatively controls these genes via repression of its activator gene glnR.
DISCUSSION
For many years it was known that optimal media for growth of Streptomyces species and secondary metabolite biosynthesis need to be equilibrated in their C/N/P/S ratios (1,33), but the molecular mechanism of the interactions between these major nutrients still remain obscure, although they are receiving increasing attention in some microorganisms (34–36).
Carbon and phosphate sources interact through several metabolic pathways, mainly in the glycolysis and different molecules serve as sensors of the C/P ratio in the cells. In enterobacteria carbon catabolite regulation is mediated by activation of the adenylate cyclase (forming cAMP) through interaction with the phosphorylated form of the IIA protein (IIAGlu) of the glucose translocation systems. In Bacillus subtilis, the interaction between C and P sources is mediated by fructose 1,6-bisphosphate (FBP) that interacts with the global regulator CcpA [reviewed by Sonenshein (35)]. Interestingly, CcpA causes repression of the phoPR promoter by binding to a novel transcription start site (37,38). This is a good example of interaction between two regulatory networks. In S. coelicolor the level of extracellular phosphate-binding protein PstS responds to both phosphate limitation (8,23) and some carbon sources like fructose or glucose (39).
There are no similar detailed reports of the interaction between the nitrogen and phosphate regulatory circuits. In the model actinomycete S. coelicolor the mechanism of phosphate control, mediated by the two-component PhoR-PhoP system, has been widely studied (4,6–8,11). Phosphate control of many pho regulon genes is mediated by binding of PhoP to PHO operators formed by direct repeats of 11-nucleotides (11). In initial transcriptomic studies we observed that several genes involved in nitrogen metabolism are regulated by phosphate (23).
The nitrogen-source regulon in S. coelicolor is controlled by two related regulatory proteins—GlnR and GlnRII (20,22).
GlnR is a master regulator in S. coelicolor that controls several genes involved in nitrogen metabolism in response to ammonium limitation. This response includes activation of glnA, glnII and amtB genes, and repression of the putative NADP-specific glutamate dehydrogenase gene gdhA (22). The GlnR regulon appears to be conserved in Mycobacterium and other actinomycetes (22,40).
As shown in this article, PhoP binds to operators located in the upstream regions of glnR, glnA, glnII and amtB. The complex structures of these operators can provide a tuning mechanism for the PhoP regulation. Expression from these promoter regions was drastically increased in the ΔphoP mutant, demonstrating that they are repressed by PhoP. The derepression was clearly observed using the luxAB reporter system throughout the time course of the culture and also in the transcriptomic analysis of the phosphate limitation response (23). These results revealed that there is a control of nitrogen metabolism by phosphate availability. This control is exerted indirectly by the binding of PhoP to the glnR promoter that in turn controls expression of the other nitrogen metabolism genes. However, PhoP also binds to the promoter regions of glnA, glnII and the amtB-glnD-glnK operon indicating that, in addition to the glnR-mediated regulation, PhoP also directly controls expression of the other five nitrogen metabolism genes. This double mechanism ensures a good degree of control of glnA, glnII and amtB-glnD-glnK by PhoP.
In S. coelicolor there is a second nitrogen regulator GlnRII that interacts with the glnA, glnII and amtB promoters (12,19). It is interesting that PhoP did not bind to the glnRII promoter and no PHO sequences were found in its promoter region. This result suggests that there is a nitrogen regulatory system that is independent of PhoP.
The PhoP negative control on nitrogen assimilation genes can be explained as a way to save the cell resources and to channel them to obtain phosphate from the medium when this nutrient is limiting. Indeed, the negative effect on growth of a high glutamine synthetase activity has been shown (41), and, as reported here, the S. coelicolor glnA and glnII promoters showed high activities (Figure 4). A response to phosphate limitation, mediated by an increased PhoP level, is to reduce expression of genes involved in N utilization.
An important question is how PhoP and GlnR interact with their respective operators in the nitrogen regulated promoters when the binding sequences are overlapping (e.g. glnA promoter) or non-overlapping (e.g. amtB promoter). We are further investigating if both operators in glnA are coincident or whether each of those two regulatory proteins recognize specific sequences in overlapping regions. Initial evidence suggests that in this case PhoP acts by preventing binding of GlnR to its specific operator in glnA (A. Sola-Landa et al., unpublished work). In the case of the amtB promoter PhoP appears to act as a ‘road-block’ for the transcription originating from upstream transcription start sites. This may explain the results observed in the ΔphoP mutant.
Streptomyces coelicolor is a habitant of the soil, an oligotrophic medium that is frequently limited in phosphate (3). An open question is if the phosphate government over nitrogen control is conserved in other microorganisms. A proteomic study on the global analysis of protein synthesis showed that the phosphate limitation decreases the expression of glnA in E. coli (42). Microarray analysis provided evidence that glnII and glnK are repressed after phosphate starvation in Shinorhizobium meliloti (43). These results are in agreement with the conservation of a phosphate control over N metabolism regulation, similar to that of S. coelicolor. We have also explored the existence of PHO boxes in nitrogen metabolism genes in E. coli. For this purpose, an information model was built based on the alignment of 25 PHO DRu compiled by Blanco et al. (44). Although no clear PHO boxes were detected in the glnA promoter region, there were candidate sequences in the promoters of the glnLG (ntrBC) two-component system that regulates glnA (2) and of the glutamine permease operon (glnHPQ) (45) (Figure 5).
Figure 5.

Putative PHO boxes in the E. coli genes glnH and glnL. (A) Alignment of the 25 direct repeat units (DRu) that form the known binding sites of PhoB (the orthologue regulator of Streptomyces PhoP), and the logo (28) that depicts the ‘information theory’ model. Information contents (Ri) of each DRu and site are shown. (B) Putative PHO boxes (composed of 2 DRu) found in the promoter regions of glnHPQ operon (glutamine permease) and glnLG (ntrBC) operon. Walkers and Ri values are determined by the model depicted above. Positions are referred to the translation start codon.
In conclusion, the nitrogen source and phosphate regulatory networks interact in different bacteria to provide an adaptation to changes in the available nitrogen and phosphate nutrients. S. coelicolor provides the first description of the control of a nitrogen regulon (GlnR) by the phosphate regulatory protein PhoP, what allows a fine coordination of the utilization of those two nutrient sources.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
‘Comisión Interministerial de Ciencia y Tecnología’ [BIO2003-01489, BIO2006-14853-C02-01]; the ‘Ministerio de Ciencia e Innovación’, Madrid [GEN2003-20245-C09-01]; the AECID (Agencia Española de Cooperación Internacional para el Desarrollo), ‘Ministerio de Asuntos Exteriores y de Cooperación’, Madrid [A/010257/07]; the ERA-NET SySMO Project [GEN2006-27745-E/SYS]; and the European Union (ACTINOGEN LSHM-CT-2004-005224). F.P.U. fellowship of the Ministerio de Ciencia e Innovación (Spain) (to K.A.); fellowship of the F.P.I. program (Ministerio de Ciencia e Innovación, Spain) (to F.S.B.). Funding for open access charge: Institute of Biotechnology of León.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the excellent technical help of B. Martín, J. Merino, A. Casenave and B. Aguado.
REFERENCES
- 1.Martín JF, Demain AL. Control of antibiotic biosynthesis. Microbiol. Rev. 1980;44:230–251. doi: 10.1128/mr.44.2.230-251.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiol. Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodgson DA. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 2000;42:47–238. doi: 10.1016/s0065-2911(00)42003-5. [DOI] [PubMed] [Google Scholar]
- 4.Martín JF. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J. Bacteriol. 2004;186:5197–5201. doi: 10.1128/JB.186.16.5197-5201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sola-Landa A, Moura RS, Martín JF. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl Acad. Sci. USA. 2003;100:6133–6138. doi: 10.1073/pnas.0931429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghorbel S, Kormanec J, Artus A, Virolle MJ. Transcriptional studies and regulatory interactions between the phoR-phoP operon and the phoU, mtpA, and ppk genes of Streptomyces lividans TK24. J. Bacteriol. 2006;188:677–686. doi: 10.1128/JB.188.2.677-686.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sola-Landa A, Rodríguez-García A, Franco-Domínguez E, Martín JF. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 2005;56:1373–1385. doi: 10.1111/j.1365-2958.2005.04631.x. [DOI] [PubMed] [Google Scholar]
- 9.Mendes MV, Tunca S, Antón N, Recio E, Sola-Landa A, Aparicio JF, Martín JF. The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metab. Eng. 2007;9:217–227. doi: 10.1016/j.ymben.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Apel AK, Sola-Landa A, Rodríguez-García A, Martín JF. Phosphate control of phoA, phoC and phoD gene expression in Streptomyces coelicolor reveals significant differences in binding of PhoP to their promoter regions. Microbiology. 2007;153:3527–3537. doi: 10.1099/mic.0.2007/007070-0. [DOI] [PubMed] [Google Scholar]
- 11.Sola-Landa A, Rodríguez-García A, Apel AK, Martín JF. Target genes and structure of the direct repeats in the DNA-binding sequences of the response regulator PhoP in Streptomyces coelicolor. Nucleic Acids Res. 2008;36:1358–1368. doi: 10.1093/nar/gkm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reuther J, Wohlleben W. Nitrogen metabolism in Streptomyces coelicolor: transcriptional and post-translational regulation. J. Mol. Microbiol. Biotechnol. 2007;12:139–146. doi: 10.1159/000096469. [DOI] [PubMed] [Google Scholar]
- 13.Wray LV, Fisher SH. Cloning and nucleotide sequence of the Streptomyces coelicolor gene encoding glutamine synthetase. Gene. 1988;71:247–256. doi: 10.1016/0378-1119(88)90041-8. [DOI] [PubMed] [Google Scholar]
- 14.Weißschuh N, Fink D, Vierling S, Bibb MJ, Wohlleben W, Engels A. Transcriptional analysis of the gene for glutamine synthetase II and two upstream genes in Streptomyces coelicolor A3(2) Mol. Gen. Genet. 2000;264:461–469. doi: 10.1007/s004380000315. [DOI] [PubMed] [Google Scholar]
- 15.Behrmann I, Hillemann D, Pühler A, Strauch E, Wohlleben W. Overexpression of a Streptomyces viridochromogenes gene (glnII) encoding a glutamine synthetase similar to those of eucaryotes confers resistance against the antibiotic phosphinothricyl-alanyl-alanine. J. Bacteriol. 1990;172:5326–5334. doi: 10.1128/jb.172.9.5326-5334.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fink D, Falke D, Wohlleben W, Engels A. Nitrogen metabolism in Streptomyces coelicolor A3(2): modification of glutamine synthetase I by an adenylyltransferase. Microbiology. 1999;145:2313–2322. doi: 10.1099/00221287-145-9-2313. [DOI] [PubMed] [Google Scholar]
- 17.Atkinson MR, Ninfa AJ. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 1998;29:431–447. doi: 10.1046/j.1365-2958.1998.00932.x. [DOI] [PubMed] [Google Scholar]
- 18.Hesketh A, Fink D, Gust B, Rexer H-U, Scheel B, Chater K, Wohlleben W, Engels A. The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 2002;46:319–330. doi: 10.1046/j.1365-2958.2002.03149.x. [DOI] [PubMed] [Google Scholar]
- 19.Fink D, Weißschuh N, Reuther J, Wohlleben W, Engels A. Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2) Mol. Microbiol. 2002;46:331–347. doi: 10.1046/j.1365-2958.2002.03150.x. [DOI] [PubMed] [Google Scholar]
- 20.Wray LV, Atkinson MR, Fisher SH. Identification and cloning of the glnR locus, which is required for transcription of the glnA gene in Streptomyces coelicolor A3(2) J. Bacteriol. 1991;173:7351–7360. doi: 10.1128/jb.173.22.7351-7360.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wray LV, Fisher SH. The Streptomyces coelicolor glnR gene encodes a protein similar to other bacterial response regulators. Gene. 1993;130:145–150. doi: 10.1016/0378-1119(93)90359-b. [DOI] [PubMed] [Google Scholar]
- 22.Tiffert Y, Supra P, Wurm R, Wohlleben W, Wagner R, Reuther J. The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 2008;67:861–880. doi: 10.1111/j.1365-2958.2007.06092.x. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez-García A, Barreiro C, Santos-Beneit F, Sola-Landa A, Martín JF. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics. 2007;7:2410–2429. doi: 10.1002/pmic.200600883. [DOI] [PubMed] [Google Scholar]
- 24.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 25.Santos-Beneit F, Rodríguez-García A, Franco-Domínguez E, Martín JF. Phosphate-dependent regulation of the low- and high-affinity transport systems in the model actinomycete Streptomyces coelicolor. Microbiology. 2008;154:2356–2370. doi: 10.1099/mic.0.2008/019539-0. [DOI] [PubMed] [Google Scholar]
- 26.Bachmann H, Santos F, Kleerebezem M, van Hylckama Vlieg JET. Luciferase detection during stationary phase in Lactococcus lactis. Appl. Environ. Microbiol. 2007;73:4704–4706. doi: 10.1128/AEM.02807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider TD. Information content of individual genetic sequences. J. Theor. Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 28.Schneider TD. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. [DOI] [PubMed] [Google Scholar]
- 29.Schneider TD. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res. 1997;25:4408–4415. doi: 10.1093/nar/25.21.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang JG, Hahn MY, Ishihama A, Roe JH. Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2) Nucleic Acids Res. 1997;25:2566–2573. doi: 10.1093/nar/25.13.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirny LA, Gelfand MS. Structural analysis of conserved base pairs in protein-DNA complexes. Nucleic Acids Res. 2002;30:1704–1711. doi: 10.1093/nar/30.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hobbs G, Frazer CM, Gardner DCJ, Flett F, Oliver SG. Pigmented antibiotic production by Streptomyces coelicolor A3(2): kinetics and the influence of nutrients. J. Gen. Microbiol. 1990;136:2291–2296. [Google Scholar]
- 34.Oh WS, Im YS, Yeon KY, Yoon YJ, Kim JW. Phosphate and carbon source regulation of alkaline phosphatase and phospholipase in Vibrio vulnificus. J. Microbiol. 2007;45:311–317. [PubMed] [Google Scholar]
- 35.Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 36.Commichau FM, Forchhammer K, Stülke J. Regulatory links between carbon and nitrogen metabolism. Curr. Opin. Microbiol. 2006;9:167–172. doi: 10.1016/j.mib.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Hueck CJ, Hillen W, Saier MH. Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res. Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 38.Puri-Taneja A, Paul S, Chen Y, Hulett FM. CcpA causes repression of the phoPR promoter through a novel transcription start site, PA6. J. Bacteriol. 2006;188:1266–1278. doi: 10.1128/JB.188.4.1266-1278.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Díaz M, Esteban A, Fernández-Abalos JM, Santamaría RI. The high-affinity phosphate-binding protein PstS is accumulated under high fructose concentrations and mutation of the corresponding gene affects differentiation in Streptomyces lividans. Microbiology. 2005;151:2583–2592. doi: 10.1099/mic.0.27983-0. [DOI] [PubMed] [Google Scholar]
- 40.Amon J, Titgemeyer F, Burkovski A. A genomic view on nitrogen metabolism and nitrogen control in mycobacteria. J. Mol. Microbiol. Biotechnol. 2008 doi: 10.1159/000159195. DOI: 10.1159/000159195. [DOI] [PubMed] [Google Scholar]
- 41.Zhang YP, Pohlmann EL, Conrad MC, Roberts GP. The poor growth of Rhodospirillum rubrum mutants lacking P-II proteins is due to an excess of glutamine synthetase activity. Mol. Microbiol. 2006;61:497–510. doi: 10.1111/j.1365-2958.2006.05251.x. [DOI] [PubMed] [Google Scholar]
- 42.VanBogelen RA, Olson ER, Wanner BL, Neidhardt FC. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J. Bacteriol. 1996;178:4344–4366. doi: 10.1128/jb.178.15.4344-4366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krol E, Becker A. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics. 2004;272:1–17. doi: 10.1007/s00438-004-1030-8. [DOI] [PubMed] [Google Scholar]
- 44.Blanco AG, Sola M, Gomis-Rüth FX, Coll M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure. 2002;10:701–713. doi: 10.1016/s0969-2126(02)00761-x. [DOI] [PubMed] [Google Scholar]
- 45.Nohno T, Saito T, Hong JS. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ) Mol. Gen. Genet. 1986;205:260–269. doi: 10.1007/BF00430437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.