Abstract
Here we report the analysis of dual G-quadruplexes formed in the four repeats of the consensus sequence from the insulin-linked polymorphic region (ACAGGGGTGTGGGG; ILPRn=4). Mobilities of ILPRn=4 in nondenaturing gel and circular dichroism (CD) studies confirmed the formation of two intramolecular G-quadruplexes in the sequence. Both CD and single molecule studies using optical tweezers showed that the two quadruplexes in the ILPRn=4 most likely adopt a hybrid G-quadruplex structure that was entirely different from the mixture of parallel and antiparallel conformers previously observed in the single G-quadruplex forming sequence (ILPRn=2). These results indicate that the structural knowledge of a single G-quadruplex cannot be automatically extrapolated to predict the conformation of multiple quadruplexes in tandem. Furthermore, mechanical pulling of the ILPRn=4 at the single molecule level suggests that the two quadruplexes are unfolded cooperatively, perhaps due to a quadruplex–quadruplex interaction (QQI) between them. Additional evidence for the QQI was provided by DMS footprinting on the ILPRn=4 that identified specific guanines only protected in the presence of a neighboring G-quadruplex. There have been very few experimental reports on multiple G-quadruplex-forming sequences and this report provides direct experimental evidence for the existence of a QQI between two contiguous G-quadruplexes in the ILPR.
INTRODUCTION
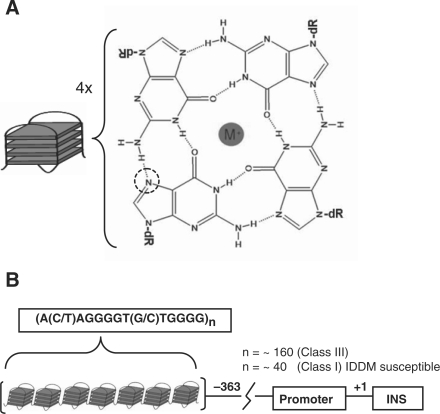
Guanine rich DNA can form quadruplex, tetraplex or G4 DNA by associating stacks of G-tetrads that are formed by Hoogsteen hydrogen bonding of four guanine nucleotides (Figure 1A). The most extensively characterized examples of quadruplex DNA are found in telomeres at the ends of eukaryotic chromosomes (1,2). Recent genome-wide analysis has identified a large number of potential quadruplex-forming sequences (3). Interestingly, these sequences have been found to occur with a higher abundance in the promoter regions (4), which suggests their potential involvement in regulatory processes. Several of these G-rich sequences have been correlated with gene function, particularly in proto-oncogenes where they are found in a higher abundance (5). Furthermore, other structural and functional studies have suggested a link between the presence of a G-quadruplex and the control of regulation of key genes (2,6–9) such as c-MYC (10,11), VEGF (12), bcl-2 (13), c-kit (14,15) and KRAS (16).
Figure 1.
(A) G-tetrad formed by Hoogsteen hydrogen bonding of guanine nucleotides (N7 position is marked with a dotted circle). Stacking of G-tetrads forms G-quadruplex or G4 DNA. (B) Schematic representation of the ILPR upstream of the INS gene. For simplicity it is depicted as a linear chain of identical quadruplexes.
Variable number of tandem repeats (VNTR) or minisatellite regions associated with many human diseases (17,18) often contain purine-rich segments, many of which have the potential to form noncanonical DNA structures. A VNTR region of particular interest is the insulin-linked polymorphic region (ILPR) that contains G-quadruplex and i-motif-forming sequences located at –363 bp upstream of the Insulin coding sequence (18–20) (Figure 1B). It contains tandem repeats of the most prevalent sequence ACAGGGGTGTGGGG (variant ‘a’) as well as other variants (‘b’–‘k’) (20,21). The number of these variant repeats has been correlated with the probability of developing insulin-dependent diabetes mellitus (IDDM or type I juvenile diabetes). For example, IDDM susceptible patients have significantly less number of repeats (∼40) than healthy individuals (∼160) (21–23).
Despite numerous studies on quadruplex-forming sequences in general, few studies have focused on two or more quadruplexes in the same molecule, which are most commonly found in telomeric DNA and other tandem repeat sequences such as the ILPR (24–30). While a sequence containing two ILPR repeats (ILPRn=2) has been shown to form an intramolecular G-quadruplex (19,20), no structural studies have focused on the ILPR with longer lengths (ILPRn ≥ 4) that are biologically more relevant. Based on simulation studies, Gupta and co-workers have predicted that longer ILPR sequences could potentially form a higher order quadruplex structure (31). Due to the lack of structural evidence on sequences that have the potential to form multiple quadruplexes, the current knowledge of quadruplex structures in these sequences is based mostly on simulation studies, and on the predictions from observations on single G-quadruplex forming units (27,31). However, the validity of these predictions has yet to be adequately tested.
Experimental studies conducted on multiple intramolecular quadruplex-forming sequences have been focused primarily on the telomere regions where different models have been postulated. Recently, Yu et al. (25) proposed that long human telomere sequences form a ‘bead on a string’ structure where each G-quadruplex exists as distinct noninteracting units. In contrast, based on crystal structure and simulation studies, a model of higher-order interactions between stacked telomeric G-quadruplexes has been proposed (27,32) and confirmed in a GGA triplet-repeat sequence (29,33). However, Petraccone et al. (34) in a recent biophysical and simulation study has suggested a different type of higher-order interaction involving loop–loop stacking between the two adjacent quadruplexes. In general, other higher-order quadruplex–quadruplex interactions (QQI) could also be envisaged, such as loop-mediated interactions via small molecules or intermolecular intercalation of quartet planes (28,35). However, there is still a paucity of experimental evidence for interactions between contiguous intramolecular quadruplexes (QQI).
Current understanding of multiple intramolecular quadruplex structure is often based upon knowledge gained from studies on single quadruplexes. However, formation of multiple intramolecular quadruplex structures, stacking or nonstacking, may be influenced by a neighboring quadruplex, although the extent of such effect is not well understood and is difficult to predict. Thus, it can be proposed that findings on single quadruplexes may not be necessarily extended to predict structures of multiple quadruplexes.
In this study, we have used native PAGE, dimethyl sulfate (DMS) footprinting, circular dichroism (CD) spectroscopy and mechanical unfolding at the single molecular level by optical tweezers to study a dual quadruplex forming ILPR sequence (ILPRn=4). Our results show that the conformation adopted by multiple intramolecular quadruplexes cannot be automatically predicted from structures of individual quadruplex units. Furthermore, our results suggest the existence of a QQI in the folded structure of the ILPRn=4. While higher-order structures have been predicted in the ILPR (31), this study provides experimental evidence in direct support of this prediction.
MATERIALS AND METHODS
Materials
DNA samples were purchased from Integrated DNA Technologies™ (www.idtdna.com) and purified by denaturing PAGE. Concentrations of the DNA were calculated using the UV absorbance value at 260 nm and the extinction coefficients for each single-stranded oligonucleotide were calculated by the nearest-neighbor approximation (36). The sequences of the oligonucleotides used in this study are of the following composition:
ILPRn: 5′-(ACAGGGGTGTGGGG)n, (n=1,2,3) ILPRn=4: 5′-(ACAGGGGTGTGGGG)2ACAGGGGTG TGGGGACAGGGGTGTGGGG 1Q56: 5′-(ACAGGGGTGTGGGG)2ATAATAATAAAT ATATAATAATAAATAT
CD spectroscopy
For the current study, 5 µM of the DNA samples were prepared in 10 mM Tris–HCl buffer pH 7.8 with 100 mM potassium chloride or 100 mM lithium chloride. Before measurement, the DNA samples were heated to 97°C for 10 min and cooled at a rate of 0.2°C/min to 25°C. CD spectra were recorded on a Jasco-810 spectropolarimeter (Easton, MD) using a quartz cuvette with a 1-mm optical path length at a temperature of 25°C. The spectra obtained were the average of three scans over a range of 200–320 nm at a 0.5-nm interval with a scan rate of 50 nm/min. The resulting spectra were then subtracted from a buffer only baseline and smoothed using a Savitzky–Golay function.
Thermal denaturation studies
For all melting experiments 5 µM of ILPR DNA in 10 mM Tris–HCl pH 7.8, 100 mM KCl was heated to 97°C for 10 min and annealed by cooling to 25°C at a rate of 0.2°C/min. The sample was then melted at an identical rate and CD values were recorded at 0.5°C intervals. Before melting, ∼100 µl of mineral oil was placed on top of the solution to eliminate evaporation. The CD melting experiments were performed on the previously mentioned spectropolarimeter with a 1-mm optical path length quartz cuvette and the temperature was controlled by a Jasco (model PFD-425S) peltier temperature controller (Easton, MD, USA).
Preparation of 5′ end radiolabeled DNA
DNA samples were radiolabeled at the 5′ end by incubating the DNA with T4 polynucleotide kinase (Promega™) and [γ-32P] ATP (Perkin Elmer™). The labeled products were then purified using denaturing PAGE.
Native PAGE
Radiolabeled oligonucleotides were prepared in 10 mM Tris–HCl pH 7.8 with 100 mM KCl. Before electrophoresis each sample was heated at 97°C for 10 min and cooled to 25°C at a rate of 0.2°C/min. The samples were analyzed on a 10% native acrylamide (19:1 acrylamide:bisacrylamide) gel containing 100 mM KCl and run in 1× Tris/Borate/EDTA buffer with 100 mM KCl at room temperature. The running buffer was periodically recirculated to maintain the salt concentration at the same level throughout the duration of the experiment.
DMS footprinting
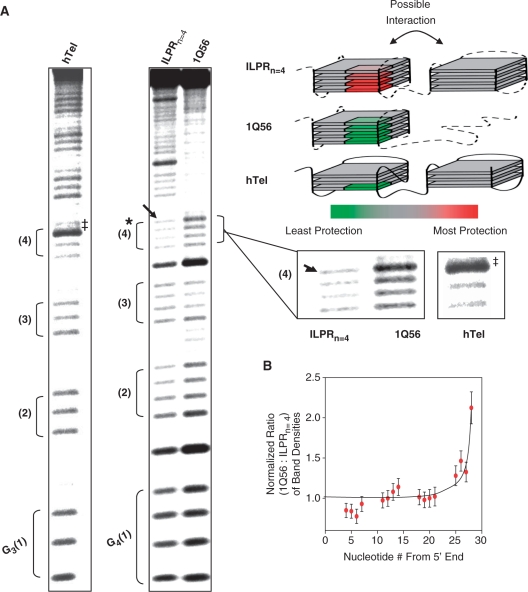
Oligonucleotide samples radiolabeled at the 5′-end were prepared in 10 mM Tris–HCl pH 7.8, 100 mM KCl with 1 µM of cold ILPR DNA. Mixed samples measuring 100 µl were then heated at 97°C for 10 min and cooled to 25°C at 0.2°C/min. The samples were then treated with DMS in a final concentration of 1% for 3 min. Samples in Supplementary Figure S3 were treated for 2 min to maintain single-hit kinetics of DMS in the absence of any salt. The reaction was stopped with 1.1 ml of stop buffer (2 M β-mercaptoethanol, 300 mM sodium-acetate, 250 µg/ml salmon sperm DNA) and immediately ethanol precipitated followed by washing with 70% ethanol. The DNA pellet was then dried and cleaved with piperidine followed by separation of the fragments on a 10% denaturing acrylamide (19:1 acrylamide:bisacrylamide) gel. DMS footprinting gel images were quantified using BioRad QuantityOne™ software. Band densities were normalized by three different ways: (i) normalizing to the first guanine from the 5′ end, (ii) normalizing to the guanines involved in the loops of the G-quadruplex and (iii) taking the sum of the density of all matching bands between the sequences ILPRn=4 and 1Q56. The ratio of the total density between the compared lanes was then used as a normalization factor. All methods yielded similar results and the method (iii) is used to generate the data depicted in Figure 7B.
Figure 7.
Dimethyl sulfate (DMS) footprinting of ILPRn = 4, 1Q56, and human telomere sequence (TTAGGG)8. (A) Phosphorimager scans of 10% denaturing PAGE. All of the samples were treated with DMS to a final concentration of 1% for 3 min in 10 mM Tris pH 7.8, 100 mM KCl. (*) Indicates guanines that are protected in the presence of a neighboring G-quadruplex in the sequence ILPRn = 4, compared to the single G-quadruplex forming 1Q56. (→) Indicates the guanine in ILPRn = 4 with the most protection and (‡) indicates guanine in hTel with enhanced cleavage. The first four G-stretches are marked sequentially from the 5′ end and expanded view of the fourth G-stretch in each of the sequences is shown on the side. A color scale is used to represent the level of protection with red being the most protected and green the least protected. Individual sequences are marked according to the color code to show protection or enhanced cleavage of the guanines. Loops in the quadruplexes are shown with dotted lines as their exact orientation is not known. (B) Quantitation of bands in the 1Q56 and ILPRn = 4 lanes (ratio of the corresponding band, (1Q56:ILPRn = 4 +/– SEM). All band densities were normalized to account for loading differences (see ‘Materials and Methods’ section for details about normalization).
Laser tweezers instrument
For the single molecule experiment, a diode pumped solid-state (DPSS) laser (1064 nm, 4 W, CW mode, BL-106C, Spectra-physics) was used as a laser source. Details on the laser tweezers instrument have been described previously (37). Briefly, a single laser source was used to create two laser traps consisting of P and S polarized light, respectively. The S polarized light was controlled by a steerable mirror (Nano-MTA, Mad City Labs) at a conjugate plane of the back focal plane of a focusing objective (Nikon CFI-Plan-Apochromat 60×, NA 1.2, water immersion, working distance ∼320 μm). The exiting P and S polarized beams were collected by an identical objective and detected by two position sensitive photodetectors (PSD, DL100, Pacific Silicon Sensor).
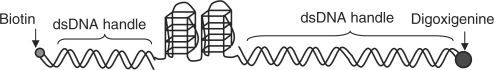
DNA construct for the single molecule studies
The DNA construct comprises of ILPR-59 (the underlined part in 5′ – CTA GAC GGT GTG AAA TAC CGC ACA GAT GCG (ACA GGG GTG TGG GG)4ACA GCC AGC AAG ACG TAG CCC AGC GCG TC) sandwiched between two dsDNA handles with unequal lengths. The longer and shorter dsDNA handles have digoxigenine and biotin attached to their free ends, respectively. The synthesis was performed according to previous report (37). Briefly, the ILPR-59 was annealed with XbaI oligo (5′-CGCATCTGTGCGGTATTTCACACCGT) and EagI oligo (5′GGCCGACGCGCTGGGCTACGTCTTGCTGGC) at the two ends in order to hybridize with the dsDNA handles. XbaI oligo was hybridized with the shorter dsDNA handle (615 bp) whereas the EagI oligo was hybridized with the longer dsDNA (2690 bp) (Scheme 1).
Scheme 1.
Illustration of DNA construct used for the mechanical unfolding experiments.
Single molecular experiments
For the single molecular experiment, the appropriate amount of DNA construct obtained above was incubated with streptavidin-coated polystyrene beads (diameter: 0.97 μm, Bangs Laboratory) for 1 h at room temperature and diluted to 700 μl working buffer (100 mM KCl, 2 mM EDTA, 10 mM Tris buffer, pH 8.0). The streptavidin coated beads linked with the DNA construct and the anti-digoxygenine coated beads (diameter: 2.17 μm, Spherotech) were injected separately into a reaction chamber via different channels.
These two types of beads were captured separately by the two laser traps. The two beads were rubbed against each other until the free end of a DNA molecule on the streptavidin-coated bead was tethered to the anti-digoxygenine coated bead via the digoxygenine and anti-digoxygenine complex. The two traps were then moved away from each other via the steerable mirror. A sudden drop in the tension was observed whenever there was an unfolding of a secondary structure. The force-extension curve for each tether was recorded in a Labview® program and the detail data treatment was performed using the programs in Matlab® and Igor® softwares. The single tether was confirmed by observing the single breakage of the tether during extension. Only these single tethered force-extension curves were considered for data analysis. To find the change in contour length during the rupture of a folded structure, individual force-extension curves were fitted with an extensive WLC formula (38),
where F is the force, T is absolute temperature, S is the stretch modulus, P is the persistence length, and x and L are end-to-end distance and contour length of the folded structure, respectively. The values of P and S at room temperature were taken from literature (38).
Prediction of probability of simultaneous unfolding
If the probability of unfolding a single domain at a particular force Fi is P(Fi), then the probability of unfolding both domains simultaneously (Psimultaneous) between the observed minimum (F1) and maximum (F2) unfolding force is given by
For continuous force range between F1 and F2,
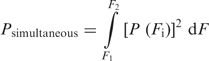 |
P(Fi) can be estimated from a Gaussian distribution of unfolding forces of a single domain between F1 and F2. To obtain Psimultaneous, numerical integration of the above function was performed with 1000 intervals from 5 to 55 pN.
RESULTS AND DISCUSSION
ILPR sequences indicate length-dependent structural features
To elucidate the effect of a G-quadruplex on the conformation adopted by its neighboring quadruplex, we decided to focus in depth on a multiple G-quadruplex-forming sequence containing four repeats of the predominant ILPR variant ‘a’ (ILPRn=4, see ‘Materials and Methods’ section for sequence details). Based upon the number of G-rich stretches, the ILPRn=4 can form two contiguous quadruplexes. We chose this sequence because the occurrence of homomeric repeats of any variants greater than four are very rare in the ILPR (data not shown).
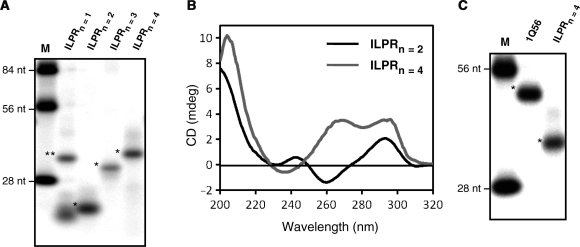
First, we used native gel electrophoresis to show that the ILPRn=2,4 form compact structures. Figure 2A shows the ILPR oligonucleotides of various lengths separated on a native gel with 100 mM KCl both in the gel and the running buffer. The ILPRn=2-4 (see ‘Materials and Methods’ section for sequences) showed an increased mobility compared to nonstructure-forming oligonucleotides of identical sizes (Figure 2A, lane M), indicating the formation of intramolecular structures. Upon quantitation of the bands in the gel, ILPRn=2–4 formed ∼99% intramolecular structures, with the exception of n=3 which formed ∼5% intermolecular structures. However, ILPRn=1 containing only two guanine stretches, incapable of forming an intramolecular G-quadruplex, showed mobilities consistent with either an intermolecular structure (see ** in Figure 2A) or the single stranded DNA form (see * in Figure 2A). Then, we used CD spectroscopy to confirm that the intramolecular structures observed in ILPRn=4 are G-quadruplexes. G-quadruplexes with a parallel folded topology have been shown in most cases to have a positive peak at ∼265 nm and a negative peak at ∼240 nm; while antiparallel folded topologies are generally associated with a positive ∼295-nm peak and a negative peak at ∼260 nm (39). In case of ILPRn=2 that is capable of forming one G-quadruplex, the CD spectrum in buffer containing 100 mM KCl is consistent with an antiparallel quadruplex structure (Figure 2B). However, with the addition of a second G-quadruplex forming unit to the ILPRn=2, which forms ILPRn=4, under identical conditions the CD spectrum implies the presence of parallel and antiparallel quadruplex forms (Figure 2B, see discussions below). This result demonstrates that the quadruplex conformation of ILPRn=4 may not be a simple addition of two ILPRn=2 quadruplex units. In addition, CD spectra of ILPRn=2,4 in 100 mM LiCl were absent of any quadruplex type features, demonstrating the dependence of quadruplex formation in the ILPR sequences on the monovalent cation (Supplementary Figure S1).
Figure 2.
Length-dependent quadruplex formation in ILPR sequences. (A) Phosphorimager scan of native PAGE (10%) of ILPR sequences of different lengths (n = 1–4). Lane M contains nonstructure forming oligonucleotides of 84, 56 and 28 nt in length. The position of bands in lanes ILPRn = 2–4 are consistent with the formation of intramolecular G-quadruplexes. Intermolecular and intramolecular structures are marked with (**) and (*) respectively. (B) Circular dichroism (CD) spectra of ILPR sequences (ILPRn = 2,4) in 10 mM Tris–HCl pH 7.8, 100 mM KCl. Peaks at ∼265 nm and ∼295 nm are indicative of a parallel and an antiparallel folded G-quadruplex topology respectively or a hybrid of the two (39). (C) Phosphorimager scan of native PAGE (10%) of the ILPR 56-nt single G-quadruplex forming mutant 1Q56 (see ‘Materials and Methods’ section for sequence) and wild-type ILPRn = 4. The 1Q56 contains a 5′ end that is the single G-quadruplex forming ILPRn = 2 sequence which is followed by a nonstructure forming 28-nt sequence.
The ILPRn=4 can potentially form two G-quadruplexes or a single G-quadruplex utilizing appropriate four G-stretches. To eliminate the latter possibility, we designed a 56-nt mutant of ILPRn=4 containing a quadruplex forming sequence 5′-(ACAGGGGTGTGGGG)2-3′ linked to a 28-mer nonstructure forming oligonucleotides at the 3′ end (1Q56, see ‘Materials and Methods’ for full sequence). The 1Q56 has the same length as the ILPRn=4, but can only form one G-quadruplex linked to an unstructured 3′ DNA tail. CD spectrum confirmed such structural conformation in 1Q56, which showed a ∼295 nm peak characteristic of an antiparallel quadruplex structure, and the spectral features at shorter wavelengths (40,41) implying the unstructured DNA tail (Supplementary Figure S2). This conformation of 1Q56 was further supported by DMS footprinting which revealed the protection of the four G-tracts at the 5′ end, most likely due to the formation of a G-quadruplex (10). Additionally, when equal concentrations of the 1Q56 and ILPRn=2 were analyzed, the magnitudes of the 295 nm peaks in both CD spectra were almost identical, suggesting the formation of equal amount of quadruplex structures in each of the sequences (Supplementary Figure S2). These data led us to conclude that the 1Q56 can form a single quadruplex and that the unstructured DNA tail in the 1Q56 has no effect on the adjacent quadruplex. As 1Q56 and ILPRn=4 are equal in length, but 1Q56 can only form a single quadruplex, the mobility of the 1Q56 in native gel can be used to indicate whether two G-quadruplexes are formed in ILPRn=4. The native gel in Figure 2C showed that both ILPRn=4 and 1Q56 had higher mobilities than the fully unstructured 56-nt marker; and that the mobility of the ILPRn=4 was greater than 1Q56. These results strongly suggested the formation of two quadruplexes in the ILPRn=4. Further evidence of the dual quadruplex formation came from DMS footprinting, which revealed that all eight G-tracts in ILPRn=4 were protected in the presence of potassium cations (Supplementary Figure S3), very likely due to the formation of two G-quadruplexes (10). Thus, results from DMS footprinting, CD, and native gel collectively established that the ILPRn=4 forms an intramolecular structure with two G-quadruplexes.
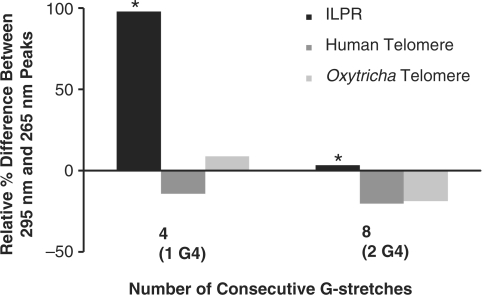
As mentioned before, the CD spectra showed a dramatic change between ILPRn=2 and ILPRn=4. This change is entirely different from that observed in the spectra between one and two G-quadruplex containing units in human and Oxytricha telomeres under nearly identical conditions (25,34) (Figure 3). In contrast to the ILPR sequences, as the length of Oxytricha (25) and Human telomere (25,34) sequences increased, the relative distribution of the CD peaks at 265 nm and 295 nm that suggested parallel and antiparallel strands respectively, remained almost identical (Figure 3). The length-dependent CD features in ILPR provides strong evidence to our hypothesis that conformation of two quadruplexes in ILPRn=4 cannot be extracted directly from individual quadruplex structures as suggested by CD signals in telomeres (25,34).
Figure 3.
Relative differences between 265 nm (parallel) and 295 nm (antiparallel) peaks for different lengths of ILPR, Oxytricha telomere, and Human telomere of one or two possible adjacent G-quadruplexes. Values were calculated by taking the difference between the 295 nm and 265 nm peaks. The resulting values were then normalized to the 295 nm peak for comparison. Values that are positive indicate a majority of antiparallel and the negative values a majority of parallel, while values near zero indicate equal intensities of both. Values from CD spectra corresponding to quadruplex structures for telomere DNA show a monotonous increase with length while the values of the ILPR drastically differ with change in repeat length (*). Values for long telomeric sequences were calculated from published results (25). Recent human telomeric CD from Petraccone et al. also showed a similar trend (34).
So far our results indicate that the ILPRn=4 forms an intramolecular structure with two contiguous quadruplexes. In addition, CD results from ILPRn=2–4 sequences show quadruplex structural features that suggest a quadruplex in cis may influence the conformation of its adjacent unit.
CD and single-molecule force measurements suggest the presence of hybrid parallel/antiparallel structures
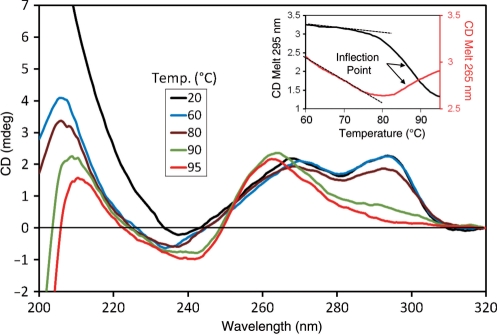
To investigate the effect of the neighboring quadruplexes on the conformation of the dual quadruplexes in the ILPRn=4, we performed CD melting of the sequence. At temperatures up to 80°C, the 295 nm and 265 nm peaks remained essentially unchanged. However, at temperatures above 80°C, there was a simultaneous change in both peaks with a concurrent increase in the 260 nm peak (Figure 4). The 295 nm and 265 nm melting showed clear transitions with identical inflection points (T1/2 = 87°C, inset Figure 4) calculated from the first derivative. It should be noted that, the deconvolution of the 265 nm melting is more complicated as several factors (change in parallel/antiparallel) quadruplex features (positive and negative peaks at ∼265 nm, respectively) and ssDNA formation (positive peak at ∼260 nm) with increasing temperature) contribute to its magnitude. The appearance of the 260 nm peak with concurrent loss of the 295 nm and 265 nm peaks has been observed in the formation of unstructured ssDNA during simultaneous melting of quadruplex strands (41,42). Thus, it appears that the antiparallel and parallel strands in ILPRn=4 quadruplexes melt simultaneously. Similar simultaneous melting observed previously on a human telomeric sequence has been attributed to a hybrid-type intramolecular G-quadruplex (42), therefore, quadruplexes in the ILPRn=4 may adopt a hybrid conformation.
Figure 4.
Temperature-dependent change in CD spectra of ILPRn = 4. Inset: CD melting curves at 265 nm and 295 nm. Dotted lines represent sloping baselines of 265 nm and 295 nm melting curves.
Previously, similar CD melting of a single ILPRn=2 quadruplex by our group revealed two different transitions, which were assigned to the melting of distinct parallel and antiparallel G-quadruplexes, respectively (37). It should be noted that the ILPRn=2 CD spectrum is dependent on the rate of annealing with slower rates favoring the antiparallel structure and faster rates favoring the parallel structure (data not shown). In the current study, quadruplexes formed in the ILPRn=4 were annealed slowly to match the conditions used for the multiple quadruplex forming telomere sequences (25). From our results so far, it is possible that both quadruplexes in the ILPRn=4 adopt a hybrid conformation, which will then be entirely different from the coexisting mixture of parallel and antiparallel G-quadruplex structures observed in ILPRn=2 (37).
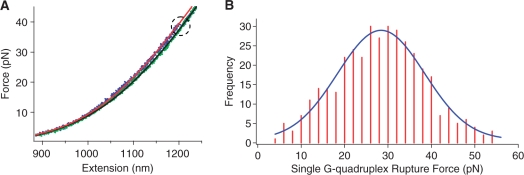
The formation of hybrid G-quadruplexes in ILPRn=4 is fully supported by the single molecular studies using laser tweezers. Previously, we have applied laser tweezers to mechanically unfold G-quadruplex structures in the ILPRn=2 (37). The presence and property of specific quadruplex species can be revealed from the histograms of unfolding forces of these species. Using the same approach (see ‘Materials and Methods’ section), we have observed a total of three types of events that corresponded to the unfolding of one or two G-quadruplexes in a DNA construct containing the ILPRn=4 sequence (see ‘Materials and Methods’ for the details of the DNA construct). The first type showed a single rupture event with a change in contour length (ΔL) <15 nm in the force-extension curve (Figure 5A). The value of ΔL is consistent with that of a 25-mer ILPR G-quadruplex (37), suggesting only one G-quadruplex is present in the DNA construct. The histogram of the rupture force for this event showed a single peak centered at 28.4 ± 0.4 pN (Figure 5B). Previously, we observed that the ILPRn=2 containing a single G-quadruplex unit could be mechanically unfolded either at 22.6 pN or 36.9 pN (37), two forces corresponded to unfolding of parallel and antiparallel G-quadruplexes, respectively. The single unfolding force observed in ILPRn=4 was located between these two values, strongly suggesting that the G-quadruplex formed in the ILPRn=4 was neither purely parallel nor purely antiparallel. Instead, it most likely assumed a single hybrid G-quadruplex conformation with both parallel and antiparallel strand orientations.
Figure 5.
(A) Force-extension curves of the 59-mer ILPR sequence (see ‘Materials and Methods’ section for detailed sequence) showing a single rupture event. The rupture event is indicated inside a dotted circle. The red and green lines represent WLC model (see text) fittings for extension and relaxing curves, respectively. (B) Histogram of the rupture forces from (A). The data are fitted with a Gaussian curve shown in blue.
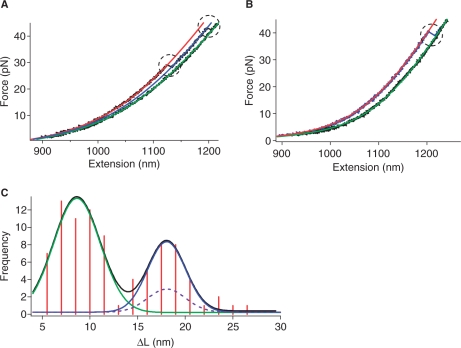
The second type of mechanical unfolding revealed two sequential unfolding events (Figure 6A). Each event had the ΔL < 15 nm, indicating two G-quadruplexes were unfolded successively. Force distribution histogram for either of the quadruplexes during this sequential unfolding was similar to that of the first unfolding type, suggesting the hybrid conformation for quadruplexes. Therefore, our mechanical unfolding experiments provided compelling evidence to support the finding from CD studies that G-quadruplexes with hybrid conformation may exist in ILPRn=4. This reinforces our proposal that the G-quadruplex conformation in ILPRn=4 cannot be simply derived from the structure of quadruplex in ILPRn=2.
Figure 6.
Force–extension curves of the 59-mer ILPR depicting (A) sequential unfolding events and (B) two G-quadruplexes unfolded together. The rupture events are indicated by dotted circles. The red and green lines represent WLC model fittings for extension and relaxing curves, respectively. The blue line in (A) is the WLC model fitting for the extension curve after unfolding of the first G-quadruplex. (C) Histogram of the change in contour length (ΔL) for simultaneous and sequential rupture events. The black line represents a two-peak Gaussian fitting. The green line is the Gaussian fitting for the ΔL of the single G-quadruplexes unfolded during the sequential rupture; while the blue line is the Gaussian fitting for the ΔL of two G-quadruplexes unfolded together. The dashed curve is the predicted probability for simultaneous rupture of two unrelated G-quadruplexes.
The force-extension curve shown in Figure 6B represented the third type of rupture events. It had only a single rupture whose ΔL was between 15 and 30 nm. Since this value was twice the value obtained from the rupture of a single G-quadruplex, it could be inferred that the rupture represents simultaneous unfolding of two G-quadruplexes. These two G-quadruplexes may exist independently or via QQI.
In contrast to the biochemical finding that majority of the ILPRn=4 forms two G-quadruplexes, mechanical unfolding experiments revealed that one G-quadruplex was more favored in the same ILPR sequence, perhaps due to the constraints imposed on the ILPRn=4 by the flanking dsDNA handles (see Scheme 1 and ‘Materials and Methods’ section). Compared to almost all of the DNA constructs used in current systems to study quadruplexes (43), the constraints presented in our system provided a unique resemblance to the in vivo situation where G-rich regions are almost always flanked by dsDNA regions.
Analysis of the simultaneous unfolding events suggests the existence of QQI in ILPRn=4
To clarify whether QQI exists between the two G-quadruplexes during the simultaneous unfolding of quadruplexes in ILPRn=4, we plotted the histogram of change in contour length (ΔL) for both sequential and simultaneous ruptures. The histogram clearly showed two populations that can be well fitted by a two-peak Gaussian equation (Figure 6C, notice the two ΔL peaks at 8.6 ± 0.3 nm and 18.1 ± 0.5 nm). Within experimental error, the ΔL of the simultaneous unfolding (18.1 ± 0.5 nm) was twice that of individual ruptures in the sequential unfolding events (8.6 ± 0.3 nm). This result confirmed that the simultaneous rupture of two G-quadruplexes occurred in ILPRn=4. Our calculations (see ‘Materials and Methods’ section) further showed that the predicted probability for the two independent G-quadruplexes to be ruptured simultaneously was 10.96% (see dotted line in Figure 6C for the expected population). However, the probability of the simultaneous unfolding observed here was 33.77% (notice the population fitted with the blue line Figure 6C), a value significantly higher than that predicted from the concurrent unfolding of the two independent G-quadruplexes (10.96%). This discrepancy can be well explained if QQI exists between the two G-quadruplexes in the ILPRn=4. The increased tension during the stretching of the ILPRn=4 might destroy this interaction, leading to cooperative unfolding of both G-quadruplexes. Similar interpretation has been used to explain the hairpin–hairpin interaction between two RNA hairpins in a mechanical unfolding investigation (44).
DMS footprinting reveals nucleotides possibly involved in the QQI
To further investigate the potential QQI predicted by the single molecule studies, we used DMS footprinting to probe the N7 of guanines (45) involved in the G-tetrads (Figure 1A).
In order to explore directly the possible structural effect due to the presence of an adjacent quadruplex, we compared the DMS footprints of the ILPRn=4 to the 1Q56 that forms only one G-quadruplex at the 5′ end. Compared to DMS treatment in the absence of K+ in which G-quadruplex formation is minimal, when 100 mM K+ was added, both sequences showed significant protection of nearly all Gs except the ones in the TGT loops and in the 5′ terminal G-tract [G4(1), Supplementary Figure S3]. It is not unusual to see enhanced cleavage at the 5′ and 3′ ends of G-quadruplexes as the terminal nucleotides are more accessible to DMS (11) possibly due to ‘breathing’ of the structure. The more pronounced cleavage of the two terminal G-tracts [G4(1) and G4(4) in Figure 7A] in 1Q56 conforms to this pattern, suggesting that the nonstructured segment at the 3′ tail does not contribute any protection to the terminal Gs of the G-quadruplex. However, ILPRn=4 only shows enhanced cleavage at the 5′ end [G4(1)], but not the 3′ end [G4(4)] of the 5′ terminal G-quadruplex, most likely due to the presence of the downstream G-quadruplex. The effect of the neighboring G-quadruplex is more obvious after normalizing the bands of ILPRn=4 against those of 1Q56, which clearly demonstrates a maximal protection of the last G in the G4(4) of the ILPRn=4, but not 1Q56 (Figure 7B, see ‘Materials and Methods’ section for details on quantitation; see ‘→’ denoting the last G in Figure 7A). These observed protections in the 5′ end G-quadruplex can be ascribed either to the presence of the neighboring quadruplex itself, or to the QQI between the two tandem quadruplexes.
To determine the probable cause of the observed protection, we intended to compare ILPRn=4 to the most widely studied example of the human telomere (TTAGGG)8, or hTel. Under DMS footprinting conditions identical to the ILPRn=4 treatment, the first four G-stretches from the 5′ end of the hTel [G3(1)-G3(4)] showed footprinting patterns very similar to the 1Q56 (Figure 7A). Remarkably, the last guanine of the fourth G-stretch [see ‘‡’ in the G3(4)] showed strikingly enhanced methylation (or less protection) akin to the 1Q56, which is markedly different from that observed in the ILPRn=4. In the most recent model of the dual quadruplex in hTel (34), the QQI is through the loop nucleotides without the involvement of the terminal guanine of the 5′ G-quadruplex. This may explain the enhanced methylation of the terminal guanine possibly due to ‘breathing’ akin to that of a DNA duplex (46,47). Such ‘breathing’ could also be contributed by the bending or flexibility in the linker region (ACA) between the two quadruplexes in ILPRn=4. Compared to the hTel, the better protection of the terminal guanines in the ILPRn=4 strongly suggests that the two quadruplexes in ILPRn=4 are more closely associated to effectuate a better suppression of the structural ‘breathing’, the lack of which leads to the easier methylation of the guanines in current hTel models (25,32,34). The presence of different linkers in hTel and ILPRn=4 suggests the important contribution of linker composition toward the suppression of ‘breathing’. Interestingly, there is a similar increase in methylation at the 5′ end of the first G-quadruplexes of both hTel and ILPRn=4, which not only supports the ‘breathing’ at that end, but also implies a relatively constrained fourth G-stretch [G4(4)] in the ILPRn=4, a possible result from the QQI in the ILPR. Therefore, combining with the 1Q56 DMS footprinting results and the single molecular data, we suggest that there is a QQI, which may be different from current models in hTel (25,32,34).
Structural features of multiple ILPR G-quadruplexes
In RNAs and proteins, higher-order structures are commonplace, however, in the context of DNA, there is significantly less structural diversity. Several higher-order intermolecular G-quadruplex structures, such as interlocking or ‘slipped’ G-quadruplexes (48), G-octaplex (35), G-wire formation (49) or Frayed Wires (50), have been reported. Although, many of these structures are very stable, the higher-order intermolecular interaction seems improbable in vivo where intramolecular interactions are likely to prevail. However, only very few studies, including those on the ILPR (24), have focused on ‘intramolecular’ multi G-quadruplex forming sequences. Although, lacking in specific details of the interactions, simulation studies on multimeric ILPR sequence have predicted intramolecular higher-order structures (QQI) (31).
The exact nature of the proposed QQI is unclear at this point. It is possible that the protection is due to a direct contact involving the fourth G-stretch. Alternatively, the protection may be attributed to a distal interaction that increases the rigidity of the 5′ quadruplex structure to suppress the ‘breathing’ of its terminal G-stretch (see discussion above).
Based upon our data, we propose that the ILPRn=4 sequence has the following characteristics: (i) quadruplexes in the ILPRn=4 adopt a hybrid structure containing both parallel and antiparallel strands based on CD and single molecular studies, which elucidates the fact that the conformation of quadruplexes in ILPRn=4 is not a simple addition of two ILPRn=2 units, and (ii) presence of a QQI that may lead to a more rigid G-stretch between the two quadruplexes in the ILPRn=4.
Potential biological significance
Higher-order quadruplex structures in the ILPR could be postulated to have diverse biological consequences. The ILPR has been shown to have extreme polymorphism with a ∼90% heterozygosity rate (18). To explain this phenomenon it has been hypothesized that replication errors in the ILPR and other tandem repeat regions are resulted from the inhibition of replication by G-quadruplex or other noncanonical DNA structures (17,18). Experimental evidence for this hypothesis has been provided by Gupta and co-workers who demonstrated in vitro that inhibition of replication can be ascribed to the structure formation in the ILPR in a manner that is dependent on the number of variant ‘a’ repeats (23,31). Since, they also proposed that higher-order quadruplex structure may form in the ILPR with larger number of repeats, it is tantalizing to speculate that the higher-order structures may contribute to the observed increase in replication errors. Additionally, the transcription factor Pur-1 and the Insulin protein have been shown to bind specifically to ILPR G-quadruplexes (24,51). The formation of higher-order quadruplex structures via QQI may therefore, affect the binding of various proteins, which potentially can affect gene regulatory processes.
Conclusion
We have shown that the ILPRn=4 capable of forming dual G-quadruplexes adopts a folded structure through a possible QQI. DMS footprinting suggests that the QQI in the ILPR is different from that observed in the telomere sequence. In addition, it is clear from our study that knowledge gained from single quadruplex structures may not always be automatically extended to describe the conformation in multiple quadruplexes, whose structures must merit individual attention. This work will begin to elucidate the structural basis of the polymorphism in the ILPR as well as other mini-satellite regions, and will also provide insights into the basic mechanisms of the contribution of these regions to diseases such as IDDM. However, the contribution of sequence and length polymorphism to the quadruplex structure and more importantly, to the biological function of the ILPR, is still not well understood, and will become important subjects for future investigations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Kent State University (KSU) and an Ohio Board of Regent grant (to S.B. and H.M.); a New Faculty Award Program at Camille and Henry Dreyfus Foundation (to H.M.); and NIH 810008116 (to H.M. and S.B.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Scott Strobel, Dr Fred Walz, and Mark Morris for their critical reading of this manuscript.
REFERENCES
- 1.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Cambridge: RSC Publishing; 2006. [Google Scholar]
- 3.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 9.Huppert JL. Hunting G-quadruplexes. Biochimie. 2008;90:1140–1148. doi: 10.1016/j.biochi.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-Quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 18.Krontiris TG. Minisatellites and human disease. Science. 1995;269:1682–1683. doi: 10.1126/science.7569893. [DOI] [PubMed] [Google Scholar]
- 19.Hammond-Kosack MCU, Kilpatrick MW, Docherty K. The human insulin gene-linked polymorphic region adopts a G-quartet structure in chromatin assembled in vitro. J. Mol. Endocrinol. 1993;10:121–126. doi: 10.1677/jme.0.0100121. [DOI] [PubMed] [Google Scholar]
- 20.Hammond-Kosack MCU, Dobrinski B, Lurz R, Docherty K, Kilpatrick MW. The human insulin gene linked polymorphic region exhibits an altered DNA structure. Nucleic Acids Res. 1992;20:231–236. doi: 10.1093/nar/20.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond-Kosack MCU, Docherty K. A consensus repeat sequence from the human insulin gene linked polymorphic region adopts multiple quadriplex DNA structures in vitro. FEBS Lett. 1992;301:79–82. doi: 10.1016/0014-5793(92)80214-2. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy GC, German MS, Rutter WJ. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat. Genet. 1995;9:293–298. doi: 10.1038/ng0395-293. [DOI] [PubMed] [Google Scholar]
- 23.Catasti P, Chen X, Moyzis RK, Bradbury EM, Gupta G. Structure-function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 24.Lew A, Rutter WJ, Kennedy GC. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu HQ, Miyoshi D, Sugimoto N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006;128:15461–15468. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- 26.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haider S, Parkinson GN, Neidle S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008;95:296–311. doi: 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Yamazaki S, Osuga H, Sugiyama H. The recognition of higher-order G-quadruplex by chiral cyclic-helicene molecules. Nucleic Acids Symp. Ser. 2006;50:183–184. doi: 10.1093/nass/nrl091. [DOI] [PubMed] [Google Scholar]
- 29.Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 30.Pedroso I, Duarte L, Yanez G, Burkewitz K, Fletcher T. Sequence specificity of inter- and intramolecular G-quadruplex formation by human telomeric DNA. Biopolymers. 2007;87:74–84. doi: 10.1002/bip.20790. [DOI] [PubMed] [Google Scholar]
- 31.Catasti P, Chen X, Mariappan S, Bradbury E, Gupta G. DNA repeats in the human genome. Genetica. 1999;106:15–36. doi: 10.1023/a:1003716509180. [DOI] [PubMed] [Google Scholar]
- 32.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 33.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petraccone L, Trent JO, Chaires JB. The tail of the telomere. J. Am. Chem. Soc. 2008;130:16530–16532. doi: 10.1021/ja8075567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo J, Adachi W, Umeda S, Sunami T, Takenaka A. Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats. Nucleic Acids Res. 2004;32:2541–2549. doi: 10.1093/nar/gkh575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantor CR, Myron M, Warshaw HS. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleolides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 37.Yu Z, Schonhoft J, Dhakal S, Bajracharya R, Hegde R, Basu S, Mao H. ILPR G-quadruplexes formed in seconds demonstrate high mechanical stabilities. J. Am. Chem. Soc. 2009;131:1876–1882. doi: 10.1021/ja806782s. [DOI] [PubMed] [Google Scholar]
- 38.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Clark CL, Cecil PK, Singh D, Gray DM. CD, absorption and thermodynamic analysis of repeating dinucleotide DNA, RNA and hybrid duplexes [d/r(AC)]12.[d/r(GT/U)]12 and the influence of phosphorothioate substitution. Nucleic Acids Res. 1997;25:4098–4105. doi: 10.1093/nar/25.20.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kypr J, Vorlickova M. Circular dichroism spectroscopy reveals invariant conformation of guanine runs in DNA. Biopolymers. 2002;67:275–277. doi: 10.1002/bip.10112. [DOI] [PubMed] [Google Scholar]
- 42.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl Acad. Sci. USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wartell RM, Benight AS. Fluctuational base-pair opening in DNA at temperatures below the helix-coil transition region. Biopolymers. 1982;21:2069–2081. doi: 10.1002/bip.360211011. [DOI] [PubMed] [Google Scholar]
- 47.Putnam BF, Van Zandt LL, Prohofsky EW, Mei WN. Resonant and localized breathing modes in terminal regions of the DNA double helix. Biophys. J. 1981;35:271–287. doi: 10.1016/S0006-3495(81)84789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishnan-Ghosh Y, Liu D, Balasubramanian S. Formation of an interlocked quadruplex dimer by d(GGGT) J. Am. Chem. Soc. 2004;126:11009–11016. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- 49.Marsh TC, Vesenka J, Henderson E. A new DNA nanostructure, the G-wire, imaged by scanning probe microscopy. Nucleic Acids Res. 1995;23:696–700. doi: 10.1093/nar/23.4.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Batalia MA, Protozanova E, Macgregor RB, Erie DA. Self-assembly of frayed wires and frayed-wire networks: nanoconstruction with multistranded DNA. Nano Lett. 2002;2:269–274. [Google Scholar]
- 51.Connor AC, Frederick KA, Morgan EJ, McGown LB. Insulin capture by an insulin-linked polymorphic region G-quadruplex DNA oligonucleotide. J. Am. Chem. Soc. 2006;128:4986–4991. doi: 10.1021/ja056097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.