Abstract
Large differences in plant genome sizes are mainly due to numerous events of insertions or deletions (indels). The balance between these events determines the evolutionary direction of genome changes. To address the question of what phenomena trigger these alterations, we compared the genomic sequences of two Arabidopsis thaliana lines, Columbia (Col) and Landsberg erecta (Ler). Based on the resulting alignments large indels (>100 bp) within these two genomes were analysed. There are ∼8500 large indels accounting for the differences between the two genomes. The genetic basis of their origin was distinguished as three main categories: unequal recombination (Urec)-derived, illegitimate recombination (Illrec)-derived and transposable elements (TE)-derived. A detailed study of their distribution and size variation along chromosomes, together with a correlation analyses, allowed us to demonstrate the impact of particular recombination-based mechanisms on the plant genome evolution. The results show that unequal recombination is not efficient in the removal of TEs within the pericentromeric regions. Moreover, we discovered an unexpectedly high influence of large indels on gene evolution pointing out significant differences between the various gene families. For the first time, we present convincing evidence that somatic events do play an important role in plant genome evolution.
INTRODUCTION
Both the existence and viability of living organisms depend on their ability to survive under continuously changing environmental conditions. Because of that, their genomes have to continuously evolve in order to fulfil adaptation constraints and enable a reproductory success. In the case of land plants, their inability to move greatly increases the impact of genome plasticity required to meet this challenge. This situation is reflected in the rates of occurrence for structural genome modifications and polyploidization events observed in plant species (in contrast to animals).
The recent bioinformatic studies of genome structure, based mainly on the whole-genome sequence data of Arabidopsis and rice, revealed a major influence of large scale duplications on plant genome evolution (1). While it is known that polyploidizations cause the simultaneous multiplication of all genes in the genome—thus building the base for their subsequent functional divergence—these events remain relatively rare. In this context, the discrete yet much more frequent insertions and deletions (indel events) seem to have greater impact on genome size, structure and functionality. Unfortunately, our present knowledge of genome changes on the level of accidental insertion and deletion events remains relatively rudimentary. The above-mentioned events can be attributed to several different evolutionary mechanisms, including, in particular, the activity of transposable elements (TEs), unequal homologous recombination (Urec) and illegitimate recombination (Illrec). Although substantial amounts of data have been collected on TEs, since Barbara McClintock described first TEs (2), the global studies depicting chromosomal distributions of various TE classes are based generally on sequence data from a single line/variety of a species and are usually not analysed in the context of different mechanisms of genome evolution (3,4). In the case of Urec—a mechanism postulated as a major force for both expansion and reduction of tandemly arrayed genes, the published reports are mainly based on the analyses of particular gene families (4–7). Thus, the overall action of the Urec process in a broader whole-genome context remains largely unknown. Apart from the above, there is also insufficient data on its role in the removal of TEs from the genome (8). Finally, almost nothing is known about the role of ILLrec (also termed nonhomologous end joining—NHEJ) events in the whole genome background (9). Although the process was found to be of major importance for the inactivation and deletion of repeated and noncoding sequences, it was not investigated apart from the case of conserved motifs in TEs (8), plant resistance genes (7) and pseudogenes (10). In light of the above, it seems intriguing to compare various indel-generating mechanisms in a genome-scale manner, in order to study their relative impact and significance on short time-scale evolutionary changes.
As it is, although many recently published articles consider genome variation at the SNP level in Arabidopsis (11–15), relatively few concentrate on large indels. This is most often due to difficulties in gathering numerous sequences of a length sufficient for large indel identification. Recently, Clark et al. (15) used hybridization to high-density oligonucleotide arrays to resolve the sequence polymorphism among 20 diverse A. thaliana lines. This analysis revealed a large number of polymorphic regions, however, the strategy applied did not allow for an accurate discrimination of large indels (and mechanisms of their origin) from highly diverged regions (15). Moreover, because of sequence length, predominant portion of large indels (starting from several hundred base pairs in length) cannot be efficiently detected with most recent high-throughput sequencing strategies (16). On the other hand, the importance of indel-based polymorphism for genetic and practical approaches is growing, even as new analytical technologies are being constructed within this area (17–19). For the very same reasons, the knowledge about the mechanisms and overall genetic basis and evolutionary trends of large indels occurrence is of major importance.
Though larger indels are much less abundant than small ones in the plant genome, nevertheless, they greatly influence the genome size, as well as its structural and functional evolution. In the case of Arabidopsis such an analysis could be performed by comparing genomic data from two A. thaliana lines, Col, which was subjected to BAC-based sequencing afforded by the Arabidopsis Genome Initiative (21), and Ler, a line partially shotgun-sequenced by Cereon Genomics (currently part of the Monsanto Co.) (22) Thanks to such an approach there is a unique opportunity to catch the corresponding mechanisms of genome restructurization in the act and investigate the ways in which they operate in a chromosome. In our previous study we investigated large indels (≥100 bp in length) for four chromosomal segments using these data collections (23). In this work we performed a more comprehensive whole genome analysis based on a novel heuristic approach to reconstructing genome intervals affected by indel events. In addition to the identification of large indels and assignment of corresponding evolutionary mechanisms, we studied their occurrence in coding sequences, as well as analysing the indels’ impact on gene evolution and genome size (by correlating indel distributions with a number of genome features).
MATERIALS AND METHODS
Assignment of Ler contigs to Col chromosomal coordinates and identification of large indels differentiating Col and Ler lines: estimation of the level of synonymous substitutions
Both Col and Ler sequences were downloaded from the TAIR website. Initially, the contigs were filtered to remove those sequences dominated by repeat sequences and transposable elements (TEs). The rejected cases constituted these contigs, where over 70% of the sequence was covered by one or more known repeats or TEs [as determined by RepeatMasker (A.F.A. Smit, R. Hubley and P. Green, RepeatMasker Open-3.0. 1996 to 2004; http://www.repeatmasker.org); search run in ‘sensitive’ mode versus Repbase database, ver. 8.12 (24)]. Subsequently, for each of the accepted Ler contigs a BLASTZ (25) comparison versus all Col chromosomes was carried out. Afterwards, the computations were carried out for each chromosome strand (i.e. two strands per chromosome) separately using the following heuristic approach (in an attempt to map the contig using its best preserved areas first).
First, a contig was divided into intervals according to the representation of its areas (as mapped by the parts of individual ungapped BLASTZ alignments) on the chromosome strand (see Figure S1). Second, the trusted intervals (seeds) were chosen from those intervals which did not overlap with any repeats/TEs, were over 60 bp in length, over 90% in sequence identity and were covered only by a single aligned part of the chromosome (i.e. were the one and only ‘good quality image’ of the contig part on the chromosome).
Proceeding from the trusted set, further intervals were filled proceeding from the 5′ to 3′ end of the contig, minimizing the introduced gaps (as long as a fragment could be added within 99 bp of an already existing fragment). For filling the intervals with corresponding chromosome strand stretches, only ungapped alignments of greater than 50% sequence identity and of more than 100 bp length were considered. This last step was repeated, until no further intervals satisfying the condition could be added.
The end result was a single, gapped alignment representing the mapping of contig to the chromosome. For each one of the constructed alignments (two for each strand of each chromosome) a coverage parameter was calculated as the sum of products constituted of aligned intervals’ lengths and the respective alignment sequence identities.
The intervals dominated by repeats/TEs (over 90% in repeats/TEs) were excluded from the coverage calculation. The contig mapping with highest coverage (if any) was chosen as the assignment of Ler contig to Col chromosomal coordinates. A collection of custom Perl scripts was used to implement and execute the algorithm. We checked for inversions by filtering indels versus the opposite chromosome strands (labelling the cases where indel was missing on one strand but present on the opposite strand, as inversions). Finally, large indels were inferred directly from gaps in the sequence of length between 100 and 20 000 bp.
Eighty-two pairs of coding sequences from both lines were aligned using ClustalX (29). Numbers of synonymous substitutions per synonymous site (Ks) were estimated using Nei–Gojobori (p-distance) method implemented in MEGA4 (20).
Identification of mechanisms responsible for indels
For all non-terminal indels (identified as insertions in the Col line) several additional properties were analysed in order to identify the corresponding mechanisms of their origin.
First, insertion coordinates were compared with the map of repeated sequences constructed for all five chromosomes RepeatMasker. If some repeats were detected, several parameters were resolved including their number, size, borders, orientation, relative position to insertion, level of interruption/truncation, repeats name and class.
Second, the flanking regions of the indel were tested whether they contain highly similar (and likely homologous) sequences of >50 bp and >95% sequence identity.
Based on this information, a heuristic procedure resolving molecular mechanism of indel origin (mainly on the basis of TE presence, terminal repeats and presence of diverged fragments stemming from unequal crossing over) was constructed using spreadsheet functions in the Excel program (Microsoft Co.)
In summary, indels were divided into 15 categories. These include: ‘intact TE’, which corresponds to indels entirely occupied by an intact transposable element; ‘novel/modified TE’, which corresponds to indels that have transposon-related both ends (from the same TE class), and some additional internal parts of transposon origin, thus could be either a rare unclassified so far TE, or a TE which was modified by succeeding mutation; ‘LTR-derived’, which corresponds to clear LTR element insertions; ‘TE with additional modifications’ and ‘truncated TE(s)’, which both comprise of indels caused by TE insertion/excision with some smaller mutation (internal deletions or cutting off, respectively); ‘a few TEs’ caused by the insertion of more than one transposable element; ‘deletion of TE(s)-containing fragment by Urec’, ‘deletion of a TE fragment by Urec’ and ‘deletion of TE(s) by Urec’, which correspond to the removal of transposons by Urec; ‘Urec within TE-free sequences’; ‘Col solo-LTR’ and ‘Ler solo-LTR’, which correspond to the insertions of the LTR element following the removal of its internal part by Urec between LTRs; ‘deletion of a TE fragment by ILLrec’; ‘ILLrec’, caused by illegitimate recombination (mainly deletions), and ‘unknown’, when we were not able to clearly identify the corresponding mechanism(s) of indel origin. Additionally, in the case of ILLrec-based indels we manually checked indels larger than 1 kb to exclude any potential phenomenon that could escape detection (e.g. unknown TEs detected by the BlastX screening of transposases and reverse transcriptases against the Plant UniProt database). Finally, all 15 categories were further combined in four general categories (Unk, TE, Urec, ILLrec).
Verification of contig assignment, indels detection and mechanism identification
The final parameters for contig assessment and indels detection were established after preliminary testing. The results obtained were verified by two means. First, 50 randomly selected contigs that were determined to have non-terminal Col indels (insertions in the Col or deletions in the Ler line) were manually checked if they were appropriately assigned to the chromosome and chromosomal region, and whether they were correctly resolved as indel-possessing regions. A molecular mechanism responsible for individual indel creation was further verified by extracting the corresponding Col chromosomal sequence covering the indel region with 1 kb up- and downstream borders [EMBOSS package (26)]. Furthermore, the sequences were screened for repeated sequences (RepeatMasker), the regions of homology suggesting a recombination-based mechanism [Blast 2 sequences (27)] and annotated genes (TAIR; www.arabidopsis.org). The second procedure was based on experimental indel verification. For this purpose 19 non-terminal Col indels were randomly selected. However, because of standard PCR amplification limits, indels shorter than 2 kb were used for this analysis. The sequences of the Col region covering an indel were used to design PCR primers using the Primer3 program (28). To enable the recognition of sites, the primers were required to be at least 30 bp apart from the indel breakpoints and the corresponding Col and Ler sequences of the primer sites had to be identical (no mismatches were allowed). Moreover, the primers were checked if they had a unique hybridization site in the Col genome (by BLASTN against AGI whole genome). The seeds of Col-0 (CS1093) and Ler-0 (CS20) lines were supplied by ABRC.
The Col and Ler genomic regions corresponding to each indel were amplified by a PCR using standard reaction conditions and 0.5 U of Taq DNA polymerase (Fermentas, Vilnius, Lithuania). PCR products were verified by agarose gel electrophoresis and cloned to pGEM-T Easy vector (Promega GmbH, Mannheim, Germany) following sequencing using standard T7 and SP6 primers. In a few cases, the PCR products were directly sequenced using the designed primers. The final sequences were compared with corresponding data of Col pseudochromosome and Ler contigs by ClustalX (29).
Data normalization procedure
The number of indels detected depended in part on the Ler contig coverage for corresponding Col chromosomal location. To eliminate this effect we applied the data normalization procedure that estimated a number of indels by multiplying the indels number detected by contig coverage for a particular chromosomal section. After preliminary testing, the optimized resolution for the data analysis was set up at 1.5 kb (smaller resolutions gave an inconsistent result because of the too infrequent number of mutations per section). Thus, contig coverage was calculated by dividing the section size (1.5 Mb) by the total length (in bp) of non-overlapping contigs assigned for the section.
Averaging the records for chromosomal arms
Eight chromosomal arms (the NOR-carrying arms of chromosomes 2 and 4 were excluded from the investigation) were divided into sections of 1 Mb starting from the centromere. Centromere positions were established from TAIR data. For the sections, the contig coverage was counted individually in order to enable a data normalization procedure (see ‘Data normalization procedure’ section). In the final step of this stage, the histogram analysis of indels distribution for particular indel-generating mechanisms was performed. Its results were normalized and the average number of mutations estimated for each section from the summarized data of chromosomal arms. Similar analyses were conducted to evaluate average indel length (median) for particular categories, and to estimate the total length of mutated regions for Urec-, ILLrec- and TE-derived indels (in this case the normalization procedure was applied as well). It should be noted that the first nine sections spanning 5 Mb from the centromere were counted based on data from all eight arms, but further sections were counted from the records of a decreased number of chromosomal arms, because of differences in their length.
Screening for indels within genes and corresponding coding sequences (CDSs)
In order to identify these genes, which were affected by an indel event, gene annotation were downloaded from the TAIR website (TAIR7 Genome Release) and compared with indel data. The gene list obtained was filtered against transposon-related sequences by two means. First, all Arabidopsis genes were filtered out if their annotations contained terms as follows: ‘reverse transcriptase’, ‘transpos’, ‘retroelement’, ‘pseudogene’, ‘ribonuclease H’ or ‘virus’. Second, the remaining coding sequences were screened against repeats by RepeatMasker. If the stretch of a sequence in gene CDS that was detected to be homologous to TE was longer than 100 bp, the gene was excluded as potentially TE-related (although by this mean we could theoretically exclude some genes that evolved by transposon exonization, there is no data reporting this process in Arabidopsis, presumably due to the relatively low abundance of repeats, and very compact gene organization). Following this filtering the curated list of genes affected by indels was used to study the mechanisms responsible for indels generation. This approach resulted in a generation of four indel-harbouring gene lists, namely Unk, TE-affected, Urec-affected and ILLrec-affected. The detailed analysis of transcriptional activity was performed by a comparison of these lists with data from Yamada et al. (30). The indel-mutated gene list (in this case undivided into indel-generated mechanism categories) was further used to study to what extent the evolution of various gene families was affected by indel-generating processes. For this, we used gene family representations given in the TAIR data. In cases of receptor-like kinases and disease resistance genes (genes with NBS domain) we employed the corresponding data from Shiu et al. (31), and from the NIBLRRS Project data (32). Lastly, the F-box genes were obtained from HMMpfam records.
Data used for correlation studies
The position of genetic markers and their physical locations in the Arabidopsis genome were obtained from Singer et al. (33) RI map. Recombination rates were calculated as the genetic distance (in cM/50 kb) between pairs of neighbouring informative SFP markers and plotted versus the average physical distance between the same markers. Then, the mean recombination frequency was calculated for chromosomal sections used in the study (with 1.5 Mb windows). GC level was established for particular chromosomal sections using the geecee program supplied in the EMBOSS package (26). For an analysis of TAGs we used data from Rizzon et al. (34), but filtered against the transposable element-related sequences, as described above. The number of TAGs for each 1.5-Mb chromosomal section was counted and divided by the total number of non-TAG genes for the section. Methylation levels were kindly provided by Xiaoyu Zhang and Steve Jacobsen (35). These were recalculated by adding values for particular chromosomal sections. Two groups of data corresponding to two methods of methylation site identification were applied: methylcytosine immunoprecipitation [mCIP; (35)]. In all the correlations calculated in the present work we used data obtained for particular chromosomes instead of averaged records for a chromosomal arm. Statistics were performed using the WINKS SDA Software (Texasoft, Cedar Hill, TX).
Analysis of recently active TE
From indels identified as ‘intact TE’ (detailed category), ‘LTR-derived’, ‘Col solo-LTR’ and ‘Ler solo-LTR’, we selected those cases where indels were covered in ≥95% by a repeat. TE corresponding to an indel could not be truncated (TE with one or two ends shorter by more then 20 bp was considered as truncated), and no additional insertion of other TEs was permitted, however, we accepted small deletions within TEs (their accumulative length had to be <100 bp).
RESULTS
Assignment of Ler contigs to Col pseudochromosomes and identification of indels
The sequence data of 81 306 Ler contigs were downloaded from the Monsanto Co. database and assigned to Col chromosomes by using a Perl script developed especially for this purpose. In total, we were able to assign 55 151 contigs (67.8%) with the total length of 69.6 Mb amounting to 58.4% coverage of the Columbia genome. The rejected contigs either consisted of repeated elements which resulted in ambiguous assignments to a number of chromosome locations, or did not fit into any location, presumably being placed in the unsequenced parts of the Columbia genome. Though the number of unassigned contigs seems to be large, in fact it is not, especially when we take into consideration that 23.3% of the Columbia genome is still not sequenced (36), and the number of repeated elements was estimated to be ∼10% (21). Not all chromosomal locations were covered by contigs at the same level, especially the pericentromeric regions and centromeres themselves having a relatively low number of contigs assigned (Figure S2). The list of Ler contigs assigned to Col chromosomes, with their location and orientation, is presented in Table S1.
Based on BlastZ-generated alignments of the Ler-Col sequence, the discontinuities in the alignments of 100–20 000 bp were identified. This analysis resulted in 6636 insertions or deletions distinguishing the two Arabidopsis accessions (lines). Of them, 2871 were described as non-terminal indels, as both 5′ and 3′ borders and were identified in the corresponding sequence alignments. The majority of these (2201) were insertions in Col (or deletions in Ler). The discrepancy in the number of non-terminal insertions in Col and Ler is due to the relatively short length of Ler contigs and singletons; on the average the contigs were only 1261 bp long. For this reason, a majority of insertions in Ler could be detected as terminal indels only. As terminal indels do not give the opportunity to analyse their length, borders and mechanisms of origin, only non-terminal insertions in the Col line were selected for further analysis (Table S2).
The error rate of the Ler contigs assignment to the Col chromosomes and indels identification was 5% as assessed from a manual verification on the sample of 100 randomly selected non-terminal indels (insertion in Col/deletion in Ler). In addition, 19 indel sites were randomly selected for detailed, experimental verification. Out of these, in three cases we obtained unspecific PCR products for one of the two accessions. The remaining 16 indel sites were successfully amplified for both lines and the resulting products were sequenced, and aligned using the ClustalX program. In all cases, the results confirmed the predicted indel polymorphism. These 16 indels verified were submitted to GeneBank under accessions EU737117–EU737148.
To estimate the divergence time between Col and Ler, we selected 82 genes that were completely covered by Ler contigs, and used them to calculate levels of synonymous substitutions (Ks) between the two accessions. Because of the very high level of sequence similarity between the two lines, 40 gene pairs were not informative, as they showed no synonymous subsitutions. However, there is a conspicuous secondary peak in the age distribution centred around Ks = 0.006, which corresponds to Col-Ler split (Table S3). Using an estimated rate of Ks of 1.5 per silent site per billion years (38), the lines diverged ∼200 000 years ago.
Characterization and mechanisms of indels origin within Arabidopsis Col and Ler accessions
As already stated, we identified 2201 non-terminal insertions in Col accession. Taking into consideration chromosome coverage by the contigs (see data normalization procedure in ‘Materials and Methods’ section), and the error rate of 5%, the hypothetical total number of insertions (we will use term ‘insertion’ operationally as it could be a deletion in the other genome) in the Col genome it was estimated to be ∼4300 (4514 × 95%). Although we do not have complete information about a number of non-terminal insertions in Ler, we can quite accurately calculate the number of smaller non-terminal insertions in this line. For insertions of the 100–200 bp sequence, the probability that their number would be significantly reduced because of Ler contig length is relatively low. For 942 cases of 100–200 bp-long insertions, 425 (45%) cases were insertions in Ler. Thus we can make an assumption, that the frequency of insertions in Ler accession is similar, and conclude that there are ∼8500 indels of 100–20 000 bp that differentiate the two Arabidopsis lines. Taking total genome size into account 120 Mb, a single large indel occurs every 14.2 kb.
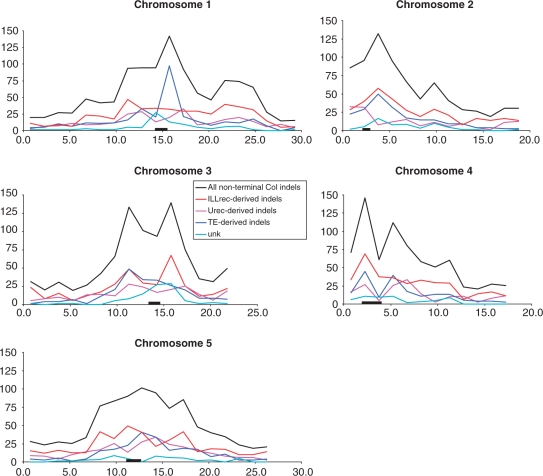
For non-terminal indels we were able to determine their length, and elucidate the probable evolutionary forces responsible for their appearance. All of the three main mechanisms (TE, Urec, and ILLrec) have some characteristics, which enable discrimination between them. However, in many cases an individual indel analysis delivered evidence of two or even more mechanisms being involved. Hence, we developed a heuristic approach, that takes into account a number of parameters and performs some additional analyses, such as the location of repeated elements within an insertion and searching for homologous sequences within genomic regions under consideration. Here, for simplicity, we focus on general mechanisms, and refer to the detailed categories in particular cases only, where it can be of a special interest. The presumable general mechanisms responsible for indels were divided into three classes: ILLrec, which includes sequence homology-independent recombination pathways, TE, which correspond to new insertions or the excisions of transposable elements (both perfectly conserved and slightly modified mainly by illegitimate recombination) and Urec, which originates from homologous recombination events involving two identical or highly similar sequences of >50 bp length. Unclear cases were classified to be in the unknown origin (Unk) category (this fourth class consists of mutations likely generated by mechanisms that can not be assigned to any of the above for some reason; it was not used in the analysis of mechanisms generating indel mutations). The distribution of various indel categories along particular chromosome is shown in Figure 1. Full information describing non-terminal indels detected is presented in Table S2.
Figure 1.
Indel distribution along Arabidopsis chromosomes. The x-axis represents the physical distance (Mb) along the chromosome. The y-axis represents the normalized numbers of indels counted for each 1.5 Mb chromosomal section. The bar represents the centromeric region.
In the presented whole-genome analysis, it has been estimated that the ratios of particular indel-generating mechanisms were different: the highest number of indels was derived from ILLrec events (42.9%), while the numbers of Urec and TE-generated indels were similar (24.4% and 26.4%, respectively) (Table 1). Considering the mean sizes of indels, the largest indels were generated by Urec events (median = 2924), smaller by TE-insertion/excision events (median = 1314) and much smaller by ILLrec (median = 215) (Table 1).
Table 1.
Indels number and size
| Mechanism (general) |
Total | ||||
|---|---|---|---|---|---|
| Unk | Urec- derived indels | ILLrec- derived indels | TE-derived indels | ||
| Number detected | 151 | 569 | 980a | 526a | 2201 |
| Percentage detected | 6.9 | 25.9 | 44.5a | 23.9a | |
| Number estimated | 285 | 1101 | 1936 | 1192 | 4514 |
| Percentage estimated | 6.3 | 24.4 | 42.9 | 26.4 | |
| Median size | 1531 | 2924 | 215 | 1314 | 682 |
Results and estimations for insertions in Col accession
aIn 15 cases indels were due to LTR insertion followed by their removal by Urec, thus they are double counted in columns describing the involvement of the mechanisms studied.
A detailed, histogram-based analysis of indel size distributions shows exponential decay characteristics for events generated by illegitimate recombination. In the case of unequal recombination-derived indels the same plot reaches the highest value at ∼500 bp, but the frequency of mutations for this category is still relatively high up to ∼6 kb (Figure S3). This result suggests a relatively broad range of size distribution for the Urec category of indels. TE-derived indels vary widely in size range with a few peaks within a 150–2600 bp range, a strong peak at ∼5 kb and a number of hits at ∼8 and ∼10 kb. The above-mentioned areas correspond to non-autonomous DNA transposons, copia-like retroelements and autonomous DNA transposons/gypsy retroelements, respectively (Figure S3).
In this work we only analysed indels of >100 bp in length. For the same reason, the number of indels resulted from ILLrec events is likely to be underestimated (the fact reflected in its distribution on the corresponding histogram). The distributions mean and median were 394 bp and 215 bp, respectively. If the deletions smaller than 100 bp had been included, these two values would have been shifted to a much lower size, as evidenced by Bennetzen et al. (37) [compare also with Ref. (10)]. Fortunately, two other types of indels (TE- and Urec-derived) give much larger sizes, and thus the inaccuracy resulted from size distribution is presumably not significant.
Indel distributions along the chromosome varies with respect to indel category
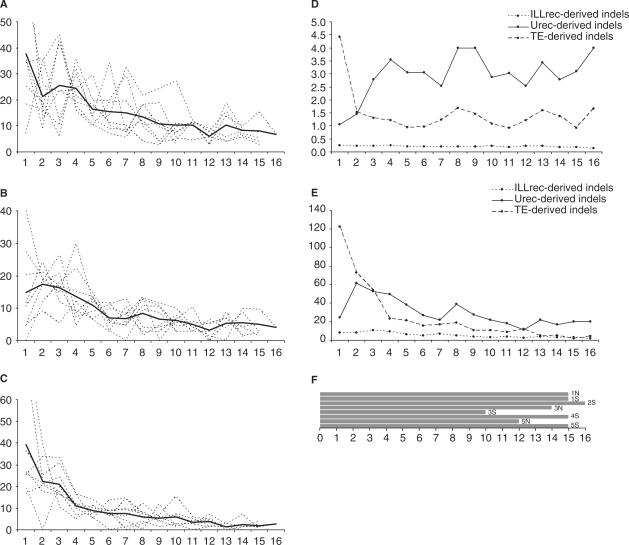
The indel distributions along a chromosomal arm are often disrupted by additional peaks, some of which stem from ancient rearrangements events (see below). In order to analyse these distributions more accurately, we decided to eliminate the chromosome-specific bias by averaging the distributions for individual chromosomal arms (see ‘Materials and Methods’ section). This analysis revealed some general trends of indel accumulation (Figure 2A–C). The Kruskal–Wallis test confirmed that particular indels categories are not identical with respect to location (H = 9.61, P = 0.009) and the Tukey multiple comparison procedure identified the differences between the distribution of ILLrec- and TE-derived indels, and ILLrec- and Urec-derived indels, but not for Urec- and TE-related indels (at the 0.05 significance level). The number of TE-derived indels increases slowly from a telomere and approaches exponential growth some 2 Mb ahead of a centromere with the precise maximum at the first section from the centromere. Conversely, distributions of Urec-derived indels have distinct courses along the chromosome: accumulation of these indels increases gradually along the chromosomal arm, starting from telomere and reaching the maximum about 1.5 Mb from the centromere, then subsequently decreasing. Still different, the ILLrec-derived indels demonstrate a bimodal distribution of intermediate form between TE- and Urec-related distribution curves. The number of ILLrec-derived indels increases towards the centromere reaching two maxima, the first of which lies about 2.5 Mb ahead of the centromere and the second forming a pericentromeric peak.
Figure 2.
Distributions of indel numbers (A–C), sizes (D) and their combined length (E). The x-axis represent the physical distance (Mb) along an averaged chromosomal arm calculated in 1 Mb sections, starting from (peri)centromeric region towards telomeres. (A) The y-axis represents the normalized numbers of ILLrec-derived indels. The averaged data shown as a solid black line, while component data for eight chromosomal arms depicted in dashed lines. (B) The y-axis represents the normalized numbers of Urec-derived indels. The averaged data shown as a solid black line, while component data for eight chromosomal arms are depicted in dashed lines. (C) The y-axis represents the normalized numbers of TE-derived indels. The averaged data shown as a solid black line, while component data for eight chromosomal arms depicted in dashed lines. (D) Medians of indel sizes in kb plotted on y-axis with respect to particular indel-generating mechanisms. (E) Sum of indel length in kb plotted on y-axis with respect to particular indel-generating mechanisms. Data were normalized by contig coverage. (F) Length of eight chromosomal arms taken into account in A, B, C, D, and E. The chromosomal arms notation according to the chromosome number and position of an arm (N corresponds to North, S corresponds to South).
In order to identify the underlying causes of emerging chromosomal distributions we analysed a number of pairwise correlations between particular indel types and some chromosome characteristics (Table 2). For ‘all non-terminal indels’ category a negative correlation was found with gene density and GC-level, whereas a positive correlation was indicated with TE frequency. This fact is indicative of the expected selection pressure on functional, gene-rich regions. As expected, all three individual indel categories (ILLrec-, Urec- and TE-derived indels) are also negatively correlated with gene density and GC-level, and positively correlated with TE frequency. It is however noteworthy that the strength of these relationships varies (see Table 2). Apart from the above-mentioned correlations, in particular the Urec-derived indels were found to be strongly correlated with tandemly arrayed genes (TAG) frequency, and with recombination rate.
Table 2.
Spearman's rank correlation coefficients between different indels categories and gene density, TE frequency, recombination frequency, GC level, TAGs and DNA methylation level (mCIP)
| All non- terminal Col indels | ILLrec- derived indels | Urec- derived indels | TE-derived indels | Indels within genes | |
|---|---|---|---|---|---|
| Gene density | –0.877*** | –0.793*** | –0.639*** | –0.873*** | –0.202 |
| TE frequency | 0.912*** | 0.791*** | 0.685*** | 0.897*** | 0.264* |
| Recombination frequency | 0.060 | 0.039 | 0.234* | 0.043 | 0.374*** |
| GC-level | –0.291** | –0.255* | –0.420*** | –0.335** | –0.586*** |
| TAGs | 0.237* | 0.159 | 0.499*** | 0.268* | 0.610*** |
| mCIP | 0.882*** | 0.782*** | 0.627*** | 0.870*** | 0.200 |
*P < 0.05; **P < 0.01; ***P < 0.001.
To address the question as to what impact the particular mechanisms of indels generation have on genome shape, we analysed distributions of indel sizes and their combined length along an averaged chromosomal arm (Figure 2D and E, respectively). In the case of ILLrec-derived indels it appears their sizes do not change along the arm. On the contrary, Urec-derived indels tend to be much shorter close to the centromere. This is also reflected in the sum of indel lengths in centromere proximity (Figure 2E). Mirroring but reverse to this trend, TE-derived indels become the largest close to centromere. Medians and the combined length of Urec-related mutations are about twice as large as TE-related mutations along an arm, but compensate ∼2 Mb from the centromere around the centromere. It should be noted that indel size fluctuations within centromeric and pericentromeric regions are not accidental, as they are derived from a relatively large number of events (from 13 to 126 indel events for each first twelve sections from a centromere for each mechanism).
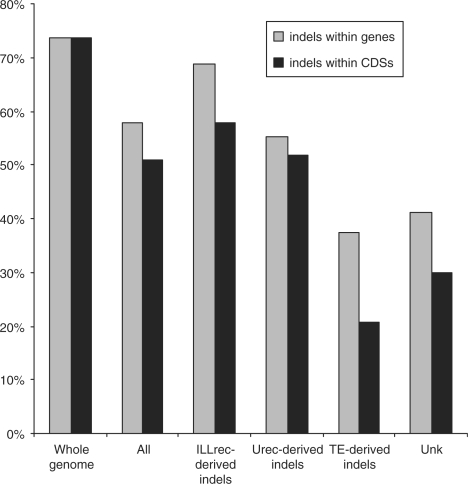
Indel events within genes and CDSs
We also analysed indels that occurred specifically within protein-encoding genes. For this purpose the gene annotation data were scanned to exclude genes related to known TEs (see ‘Materials and Methods’ section). Altogether, we were able to identify 1185 different genes that were affected by 894 indels (some mutations covered more than one gene). The majority of these genes possess indels within coding regions (908 cases, 76.6%). Both types of recombination (ILLrec, Urec) have the highest influence on gene content (Table 3). A similar analysis for coding regions revealed an even greater role of unequal homologous recombination (Table 3), with the length of the overlapping gene-indel region reaching its maximum value for unequal recombination (Table 3).
Table 3.
Indels within genes and coding regions
| ILLrec- derived indels | Urec- derived indels | TE-derived indels | Unk | Total | |
|---|---|---|---|---|---|
| Within genes | |||||
| No. | 400 | 392 | 63 | 41 | 894b |
| Percentage | 44.7 | 43.8 | 7.0 | 4.6 | |
| Meana | 364 | 1266 | 1078 | 1019 | 950 |
| Mediana | 178 | 925 | 586 | 747 | 572 |
| Within CDSs | |||||
| No. | 194 | 375 | 32 | 32 | 631b |
| Percentage | 30.7 | 59.4 | 5.1 | 5.1 |
aThe value determines overlapping sequences of indels and genes.
bIn two cases the indel was due to LTR insertion followed by its removal by Urec, thus it is double counted.
When analysing the distribution of indels within genes, we found they positively correlate with both TAG frequency and recombination frequency, and negatively correlate with the GC level (Table 2).
We also checked the impact of large indels on gene transcriptional activity. For this purpose the genes and CDSs affected by indels were divided on the basis of whether they are or are not transcribed [based on data from ref. (30)]. Altogether, out of 1184 indel-affected genes, 1148 have the transcriptional activity tested by Yamada et al. (30). For these genes 57.9% (665 cases) were transcribed. Results for a similar analysis for particular mechanisms are shown in Figure 3, and more detailed data are shown in Table S4. The number of genes and CDSs harbouring indels, which are/are not transcriptionally active varies significantly among four indel-generated mechanism classes (χ2 = 37.10, df = 3, P < 0.001, and χ2 = 23.55, df = 3, P < 0.001, respectively). For all genes/CDSs harbouring indels, the frequency of active elements is significantly lower than for the whole genome data (Table S5). However, this analysis was performed based on insertion in the Col line (deletion in Ler) and transcriptional data for the Col line, as well. This means, that we cannot reliably expect the result to show the real level of gene inactivation due to indel events, although the overall trends are unlikely to be affected. These tendencies suggest that TE insertions, in particular, have a much stronger effect on gene activity than other mechanisms of the indel generation.
Figure 3.
Genes (defined as sequences from 5′ to 3′ untranslated regions (UTRs); including exons, introns and UTRs) and CDSs (defined as translated part of the gene from start codon to stop codon and excluding introns) affected by indels at transcriptional level. Percentage of transcriptionally active genes (y-axis) for particular indel categories (x-axis) given in relation to total number of silenced genes. ‘Whole genome’ represents percent of transcriptionally active genes/CDSs within the genome, as tested by Yamada et al. (30), and ‘All’ illustrates transcriptionally active genes in all genes/CDSs harbouring indels. Data for particular indel categories are also shown.
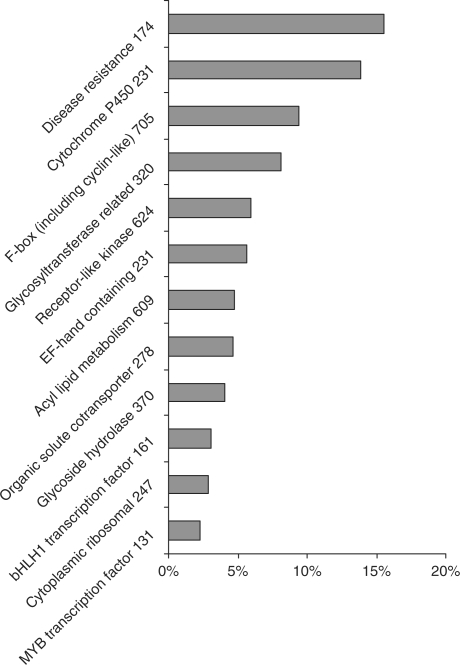
Moreover, we attempted to analyse how often the indels are detected in different gene families. The number of indel-harbouring genes varies between different gene families (χ2 = 74.3, df = 11, P < 0.0001—precise results are shown in Figure 4). The most frequently mutated gene families were disease resistance genes and cytochrome P450, whereas the rarest mutations were found within functionally constrained genes encoding transcription factors and cytoplasmic ribosomal proteins.
Figure 4.
Gene families affected by indels. Percentages of indel-harbouring genes (y-axis) given in relation to total group representation in the genome (x-axis). ‘Disease resistance’ corresponds to all the genes with NBS domain.
Recently active transposable elements in the Arabidopsis genome
The study gave us the unique opportunity to find out these TEs, which were actively mobile at least in one line sometime during the past ∼200 000 years after Col-Ler split. We will refer to them as ‘recently active’. Investigation of the TE category let us disclose correct, non-truncated transposons. As we focused on insertions in the Col line (=deletions in Ler), all retroelements and RC/Helitron class transposons detected were active in the Columbia accession, because of their transposition cycle. Other types of DNA transposons detected could be recently active either in Col (a case of an insertion), or in Ler (a case of an excision) line. Altogether, after using a conservative criterion to minimize the introduction of false positives, we detected 166 transposable elements, out of which the retroelements constitute 40.4%, and the DNA transposons account for the remaining 59.6% (see Table 4, for details). Not surprisingly, detected TE appeared to be relatively unaltered as compared to their corresponding consensus sequence in RepBase: median values for percent of substitution, percent of deletion and percent of insertion were 3.05, 0.45 and 0.00, respectively.
Table 4.
Recently active transposable elements detected by a comparison of Col/Ler accessions
| Number of elements detected | Percent of elements detected | |
|---|---|---|
| Retroelements | 67 | 40.4 |
| SINEs | 3 | 1.8 |
| LINEs/L1 | 4 | 2.4 |
| Copia | 40a | 24.1 |
| Gypsy | 20a | 12.1 |
| DNA transposons | 99 | 59.6 |
| Pogo | 4 | 2.4 |
| En-Spm | 1 | 0.6 |
| MULE (MuDR) | 33 | 19.9 |
| Harbinger | 2 | 1.2 |
| RC/Helitron | 44 | 26.5 |
| ATTIRX1 | 4 | 2.4 |
| hAT | 4 | 2.4 |
| Mariner | 4 | 2.4 |
| Unclassified | 3 | 1.8 |
| Total | 166 |
Those correspond to intact elements, which were inserted/excised from one line in comparison with the other.
aThe value includes solo-LTRs.
Within the retroelements subset, the Copia-like transposons are twice as common as the Gypsy group, while LINE and SINE elements seem to occur only rarely. A relatively small number of Gypsy elements that were detected in comparison with Copia elements could be attributed mainly to the very strong preference of these retrotransposons for incorporation within pericentromeric regions—for which the number of assigned Ler contigs is relatively low. It is also in accordance with previous results (4).
Within DNA transposons two main classes, namely Helitrons and MULEs (Mutator-like transposable elements), were determined to be recently active. Both varieties have been proven to acquire and fuse fragments of plant genes (37,39,40), which may indicate this process is of importance in the Arabidopsis genome, presumably enhancing its plasticity.
We detected 25 cases of indels containing solo-LTRs. Out of these 13 cases were caused by an insertion in the Columbia line followed by its subsequent removal by Urec. In the same dataset there are 60 clear cases of LTR insertion in Col line (both copia and gypsy class). From our results it can be estimated that 21.7% (13 × 60) of LTRs were removed from the Columbia genome by unequal recombination between their terminal repeats within the last ∼200 000 years. However, this estimate should be treated with caution as the number of solo-LTRs detected is relatively low and no normalization procedure that takes into consideration chromosomal position could be applied. Keeping this in mind, our general conclusion is that Urec is not efficient enough to fully counteract genome expansion by LTR insertions, especially for chromosomal regions in close proximity to the centromere, where Urec's activity is relatively low.
DISCUSSION
One of the most apparent results of our studies concerns the indel chromosomal distribution. The striking difference in their occurrence within pericentromeric regions in comparison to chromosome arms was found. Taking into consideration the origin of indels, this phenomenon is due to either TE insertions or recombination-based mechanisms (Figure 2A–C). The regions with a highly elevated level of indels comprise about 4 Mb counting from the centromere towards the telomere, and correspond directly to chromosomal regions of elevated numbers of transposable elements. Wright et al. (41) already provided convincing evidence that the accumulation of TEs in pericentromeric regions is the result of strong selection against the TE disruption of gene function. Thus, it seems reasonable to argue that both ILLrec- and Urec-derived indels, due to their frequency, counteract TE-based genome expansion (42,43); thus (peri)centromeric regions appear to be an evolutionary ‘battlefield’ between transposons and recombination-based events. This conclusion is also supported by the inferred strong positive correlations between TE frequency and ILLrec/Urec-originated indels distribution (0.79 and 0.69, respectively). These two types of mechanisms seem to counteract genome expansion in different fashions and are pointed out below.
Unequal recombination is insufficient in the removal of transposable elements within centromeres
A closer look at the indel distribution along chromosome arms (Figure 2A and B) demonstrates a difference between ILLrec- and Urec-derived events. The frequency of Urec-related indels, although high within pericentromeric regions, decreases close to the centromere (Figure 2B). Moreover, the distribution of indels generated by Urec is significantly correlated with recombination frequency (0.234, P < 0.05). It is well documented that centromeric regions have a strong reduction in rates of recombination (44,45), a phenomenon likely connected with the high level of DNA methylation (35). Interestingly, the average length of mutations generated by the Urec mechanism is about 3-fold shorter within the first 2 Mb from the centromere, than along the chromosome arm (Figure 2D). The low numbers and sizes of Urec-related indels within these regions are reflected in the total length of modified DNA (Figure 2E).
Another piece of evidence emerged from our investigation of retrotransposon insertions and their elimination by Urec through solo-elements: the number of solo-elements appears to be relatively low in comparison with intact ones (∼1:5). This contradicts the previous report of Devos et al. (42), who found the ratio of solo-LTRs to intact elements in Arabidopsis is ∼1:1. This discrepancy is presumably due to the distinct data sets applied. Devos et al. (42) analysed all the intact elements within the genome, as a consequence finding many more solo-LTRs within the chromosomal arms, where Urec is relatively efficient; in contrast we detected only these events that took place in the Col genome after Col-Ler separation. Based on these observations we suggest unequal recombination is inefficient in the removal of TE in the proximity of centromere.
Somatic events play an important role in plant genome evolution
Contrary to Urec-, ILLrec-derived indels are not correlated with recombination frequency and accumulate within centromeric regions (Figure 2A). Their average sizes do not depend on chromosomal position (Figure 2D). Our data suggest that DNA methylation levels do not inhibit this mechanism of indel occurrence. It should be emphasized that ILLrec-related indels being shorter than 100 bp are much more common, than larger ones [(37); see also Figure S3]. Moreover, at least 34% of indels in the TE-related category have some traces of ILLrec action, as they are truncated and fragmented. As a consequence, the impact of ILLrec indels on genome size is much larger than could be deduced from Figure 2F. Thus, our final suggestion is that ILLrec takes over the function of Urec in genome reduction within centromeres. It is tempting to speculate its main role is not the removal of TEs, but their inactivation by disturbing the sequence of their coding regions. In plants, unlike in animals, there are no germ lines, hence most of somatic cells can potentially develop towards generative cells. As it is widely accepted that ILLrec is active mainly in somatic cells, where the availability of homologous sequences is limited (9), it is noteworthy that somatic events are much more essential for plant genome evolution that previously supposed. It is probable that the much higher plasticity of plant genomes in comparison with animal ones is, in part at least, a consequence of the broader influence of somatic events on plant genome evolution.
Large indels have an unexpectable high effect on gene evolution
Out of 2201 indels detected in our study, there were 894 (40.6%) and 631 (28.6%) affected genes and their coding regions, respectively. Contig coverage for chromosomal arms, which are rich in coding sequences, is much higher than for (peri)centromeric ones (Figure S2), thus the contribution of indels affecting genes within all indels is in fact several times lower. Even with this reservation, however, the result reveals a surprisingly significant influence of indels on gene evolution, and indicates their role in these processes as comparable to their function in the deletion of repeated sequences. Both Urec- and ILLrec-derived indels appear to shape gene structure much more frequently than TE-based indels (Table 3). This finding is in a line with previous data on gene evolution in plants (5–7). On the other hand, the coding regions are mainly affected by Urec-originated events (59.4% of all the cases; Table 3), and the average length of indel-affected gene sequences is a few times larger for Urec-, than for ILLrec-based indels (Table 3). Again, it should be noted that the polymorphisms described in this study were derived and based on insertions in the Col line (or deletion in Ler) and existing Col gene annotations; thus presumably some genes that have been disrupted by indels were not identified, as they do not exist in present annotations. In human pseudogenes, ILLrec generates deletions three times more often than insertions (10), and it is suggested in regard to larger events (such as those caused by Urec) that the trend could in fact be opposite to the one observed here (46). Hence, we must conclude that the effect of ILLrec on coding sequences could have been underestimated.
The occurrence of indels within genes correlates strongly with the distribution of tandemly arrayed genes (TAGs), a finding which is not surprising, as these mutations are the main force of TAGs birth-and-death life cycle (5). On the other hand, we found a strong negative correlation with GC-level and positive with recombination frequency. These two values are related to each other (47) and indicate that the occurrence of indel mutations within genes is governed by recombination-dependent events, and takes place usually during meiotic crossing-over. This finding is in accordance with previous data (9).
By current annotation, genes lacking expression support are overrepresented within genes harbouring insertions in the Col line relative to Ler (Figure 3). The difference is statistically significant (Table S5), however, it cannot be concluded where the transcriptional silencing is an effect, and where a cause of indels. Conversely, there are significant discrepancies between particular indel categories. Genes interfered by TE insertion/excision events exhibit transcriptional inactivity almost twice as often as ILLrec-affected ones (Table S4, Figure 3). This phenomenon is presumably connected to indel size (Table 1), although other reasons, such as DNA methylation around TEs have been suggested (35).
Moreover, we investigated the frequency of indel appearance in particular gene families. In order to compare indel polymorphism with more global descriptions of polymorphism (SNPs and polymorphic regions) described by Clark et al. (15), we selected the same gene families for this study. The overall trend between these two data collections is similar. In both cases, the disease resistance gene family emerges as the most polymorphic group of genes, while the cytoplasmic ribosomal proteins and transcription factors exhibit the lowest levels of polymorphism. Contrary to Clark et al. (15), the mutated genes with the F-box domain were found to be less prevalent. This is probably due to the very stringent criteria we applied to the exclusion of TE-related genes, which eliminated a large fraction of genes within the category.
Indel distributions along the chromosome may reflect breakpoints of ancient rearrangements
The distribution of indels along all the chromosomes is strongly influenced by their centromeric location, where the main indel peaks occur. However, there are some additional peaks within chromosomal arms. The most apparent of these is an additional indel peak in the middle of the south arm of chromosome 1 (normalized indel number counts to 75.54; Figure 1). The region under investigation exhibits normal gene density, GC-content, and there seem to be no remnants after ancient local rearrangements (initially deduced from a comparison with homeologous chromosomal regions). Still, there is a significant increase in TEs number, and TAGs within this region (data not shown). Together with the majority of ILLrec indels within the region, this observation points to an ancient chromosomal fusion event, which took place during the reduction of A. thaliana chromosome number. To assess this conclusion, we compared the chromosomal location of the indel peak with results from comparative genetic (48–53) and physical (54,55) mapping, which both support the hypothesis. It should be emphasized that this particular chromosomal fusion event has been suggested to be the most recent one in the Arabidopsis genome's history [fusion/breakage 3; (51)].
Considering the number of indels, the next noncentromeric peaks are located on chromosomes 5, 2 and 4 (normalized indel number of 85.51, 64.96 and 60.52, respectively; Figure 1). In all these cases, the peaks are mainly the result of ILLrec-derived indels, and correspond to regions with elevated numbers of TEs. For chromosomes 5 and 2, a comparison with a comparative genetic map revealed that the peaks correspond to other regions of ancient chromosomal fusion breakpoints [fusion/breakage 1 and 2, respectively, according to Ref. (51)]. In contrast, no obvious correlations with any known genomic features for the region on chromosome 4 were detected.
Furthermore, we double-checked the chromosomal fusion breakpoints in order to infer whether any remnants of these events could be detected on the level of indel distribution. Besides chromosomes 1, 2 and 5, other breakpoints are located within chromosomal regions embedded in pericentromeric indel peaks, making it difficult to identify. It is worth noting that we could expect any remnants of the fusions only in the case of chromosomes 1, 2 and 5, where the ancient pericentromeric regions had been taken apart: in fusions that resulted in the formation of present chromosomes 3 and 4 only telomeric sequences could be expected. We also found that it is impossible to detect remnants of more ancient chromosomal rearrangements at the level of indel distribution, though they can be easily detected by bioinformatic (23,56,57) and hybridization approaches (58,59), where coding sequences are mainly involved.
Recently, Lysak et al. (54) proposed a mechanism by which chromosome number reduction might proceed and suggested that the ancient pericentromeric region had been lost during chromosomal fusions. However, our study suggests that traces related to ancient pericentromeric regions can be detected at least in some cases (e.g. increased levels of TEs and accumulation of ILLrec-derived indels within breakpoint regions). Therefore, it seems reasonable to conclude that some parts of the pericentromeric heterochromatin domains were temporarily retained after the fusion event, and the succeeding process of their removal was relatively slow. Hence, the underlying mechanism of ancient centromere loss merits further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
State Committee for Scientific Research [PBZ-MNiSW-2/3/2006/19 and PBZ-MNil-2/1/2005 to J.S.]. Funding for open access charge: Department of Biotechnology, Faculty of Biology, Adam Mickiewicz University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank W. Makalowski for the discussion and comments on the manuscript. Our students, M. Kot, Z. Andrzejewska and H. Galganska are acknowledged for their technical assistance in the experimental verification of indel polymorphism. The Ler sequence dataset was kindly provided by the Monsanto Co. The main computational analyses were performed on a computing cluster kindly made available by J. Chełkowski, as part of the ‘Transgenesis and genomics of crop plants’ Science Network activities. We are grateful to X. Zhang and S.E. Jacobsen for providing DNA methylation data for the Arabidopsis genome and to S.H. Shiu for the receptor-like kinase gene family data. The list of Arabidopsis disease resistance genes was obtained from the NIBLRRS Project website.
REFERENCES
- 1.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr. Opin. Plant. Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl Acad. Sci. USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le QH, Wright S, Yu Z, Bureau T. Transposon diversity in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2000;97:7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira V. Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol. 2004;5:R79. doi: 10.1186/gb-2004-5-10-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyers BC, Kaushik S, Nandety RS. Evolving disease resistance genes. Curr. Opin. Plant Biol. 2005;8:129–134. doi: 10.1016/j.pbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Yandeau-Nelson MD, Xia Y, Li J, Neuffer MG, Schnable PS. Unequal sister chromatid and homolog recombination at a tandem duplication of the A1 locus in maize. Genetics. 2006;173:2211–2226. doi: 10.1534/genetics.105.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wicker T, Yahiaoui N, Keller B. Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J. 2007;51:631–641. doi: 10.1111/j.1365-313X.2007.03164.x. [DOI] [PubMed] [Google Scholar]
- 8.Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 2005;56:1–14. doi: 10.1093/jxb/eri025. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Gerstein M. Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 2003;31:5338–5348. doi: 10.1093/nar/gkg745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 2003;13:1250–1257. doi: 10.1101/gr.728603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid KJ, Torjek O, Meyer R, Schmuths H, Hoffmann MH, Altmann T. Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor. Appl. Genet. 2006;112:1104–1114. doi: 10.1007/s00122-006-0212-7. [DOI] [PubMed] [Google Scholar]
- 13.Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:1289–1299. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borevitz JO, Hazen SP, Michael TP, Morris GP, Baxter IR, Hu TT, Chen H, Werner JD, Nordborg M, Salt DE, et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2007;104:12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 16.Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;12:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borevitz J. Genotyping and mapping with high-density oligonucleotide arrays. Methods Mol. Biol. 2006;323:137–145. doi: 10.1385/1-59745-003-0:137. [DOI] [PubMed] [Google Scholar]
- 18.West MA, van Leeuwen H, Kozik A, Kliebenstein DJ, Doerge RW, St Clair DA, Michelmore RW. High-density haplotyping with microarray-based expression and single feature polymorphism markers in Arabidopsis. Genome Res. 2006;16:787–795. doi: 10.1101/gr.5011206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salathia N, Lee HN, Sangster TA, Morneau K, Landry CR, Schellenberg K, Behere AS, Gunderson KL, Cavalieri D, Jander G, et al. Indel arrays: an affordable alternative for genotyping. Plant J. 2007;51:727–737. doi: 10.1111/j.1365-313X.2007.03194.x. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 21.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 22.Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziolkowski PA, Blanc G, Sadowski J. Structural divergence of chromosomal segments that arose from successive duplication events in the Arabidopsis genome. Nucleic Acids Res. 2003;31:1339–1350. doi: 10.1093/nar/gkg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 27.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 28.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 31.Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KFX, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer T, Fan Y, Chang HS, Zhu T, Hazen SP, Briggs SP. A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet. 2006;2:e144. doi: 10.1371/journal.pgen.0020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput. Biol. 2006;2:e115. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Hosouchi T, Kumekawa N, Tsuruoka H, Kotani H. Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res. 2002;9:117–121. doi: 10.1093/dnares/9.4.117. [DOI] [PubMed] [Google Scholar]
- 37.Bennetzen JL. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 2005;15:621–627. doi: 10.1016/j.gde.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol. Biol. Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 39.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 40.Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 41.Wright SI, Agrawal N, Bureau TE. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 2003;13:1897–1903. doi: 10.1101/gr.1281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devos KM, Brown JK, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vitte C, Bennetzen JL. Eukaryotic transposable elements and genome evolution special feature: Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl Acad. Sci. USA. 2006;103:17638–17643. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 45.Haupt W, Fischer TC, Winderl S, Fransz P, Torres-Ruiz RA. The centromere1 (CEN1) region of Arabidopsis thaliana: architecture and functional impact of chromatin. Plant J. 2001;27:285–296. doi: 10.1046/j.1365-313x.2001.01087.x. [DOI] [PubMed] [Google Scholar]
- 46.Gregory TR. Insertion-deletion biases and the evolution of genome size. Gene. 2004;324:15–34. doi: 10.1016/j.gene.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 47.Drouaud J, Camilleri C, Bourguignon PY, Canaguier A, Bérard A, Vezon D, Giancola S, Brunel D, Colot V, Prum B, et al. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 2006;16:106–114. doi: 10.1101/gr.4319006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boivin K, Acarkan A, Mbulu RS, Clarenz O, Schmidt R. The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 2004;135:735–744. doi: 10.1104/pp.104.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuittinen H, de Haan AA, Vogl C, Oikarinen S, Leppala J, Koch M, Mitchell-Olds T, Langley CH, Savolainen O. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics. 2004;168:1575–1584. doi: 10.1534/genetics.103.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yogeeswaran K, Frary A, York TL, Amenta A, Lesser AH, Nasrallah JB, Tanksley SD, Nasrallah ME. Comparative genome analyzes of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 2005;15:505–515. doi: 10.1101/gr.3436305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koch MA, Kiefer M. Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species – Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana. Am. J. Bot. 2005;92:761–767. doi: 10.3732/ajb.92.4.761. [DOI] [PubMed] [Google Scholar]
- 52.Henry Y, Bedhomme M, Blanc G. History, protohistory and prehistory of the Arabidopsis thaliana chromosome complement. Trends Plant Sci. 2006;11:267–273. doi: 10.1016/j.tplants.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 53.Kawabe A, Hansson B, Hagenblad J, Forrest A, Charlesworth D. Centromere locations and associated chromosome rearrangements in Arabidopsis lyrata and A. thaliana. Genetics. 2006;173:1613–1619. doi: 10.1534/genetics.106.057182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl Acad. Sci. USA. 2006;103:5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lysak MA, Pecinka A, Schubert I. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res. 2003;11:195–204. doi: 10.1023/a:1022879608152. [DOI] [PubMed] [Google Scholar]
- 56.Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell. 2000;12:1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Babula D, Kaczmarek M, Barakat A, Delseny M, Quiros CF, Sadowski J. Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol. Genet. Genomics. 2003;268:656–665. doi: 10.1007/s00438-002-0782-2. [DOI] [PubMed] [Google Scholar]
- 59.Ziolkowski PA, Kaczmarek M, Babula D, Sadowski J. Genome evolution in Arabidopsis/Brassica: conservation and divergence of ancient rearranged segments and their breakpoints. Plant J. 2006;47:63–74. doi: 10.1111/j.1365-313X.2006.02762.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.