Abstract
In addition to the better known guanine-quadruplex, four-stranded nucleic acid structures can be formed by tetrads resulting from the association of Watson–Crick base pairs. When such association occurs through the minor groove side of the base pairs, the resulting structure presents distinctive features, clearly different from quadruplex structures containing planar G-tetrads. Although we have found this unusual DNA motif in a number of cyclic oligonucleotides, this is the first time that this DNA motif is found in linear oligonucleotides in solution, demonstrating that cyclization is not required to stabilize minor groove tetrads in solution. In this article, we have determined the solution structure of two linear octamers of sequence d(TGCTTCGT) and d(TCGTTGCT), and their cyclic analogue d<pCGCTCCGT>, utilizing 2D NMR spectroscopy and restrained molecular dynamics. These three molecules self-associate forming symmetric dimers stabilized by a novel kind of minor groove C:G:G:C tetrad, in which the pattern of hydrogen bonds differs from previously reported ones. We hypothesize that these quadruplex structures can be formed by many different DNA sequences, but its observation in linear oligonucleotides is usually hampered by competing Watson–Crick duplexes.
INTRODUCTION
Quadruplex DNA structures are attracting extensive interest from many research areas, ranging from molecular and structural biology to supramolecular chemistry and nanotechnology (1). These structures may play a role in a number of biological processes [for reviews see (1–3)], and are attractive targets for drug design, especially in cancer chemotherapy (4). Evidence of quadruplex formation in vivo has been found recently by electron microscopy (5), and by generation of quadruplex-specific antibodies (6,7). In addition, recent bioinformatics studies have shown the high prevalence of quadruplex forming structures in promoter regions of many prokaryotic genes (8).
The most studied four-stranded motif in DNA is the G-quadruplex, formed by two or more planar G-tetrads, where four guanines are paired through both their Watson–Crick and Hoogsteen sides. These quadruplexes may contain other kind of tetrads, such as those consisting only of adenine (9), thymine (10,11) and cytosine (12), or tetrads containing Watson–Crick base pairs (13–18). When tetrads formed by two Watson–Crick base pairs are found in the context of a G-quadruplex, the interaction between the two base pairs is always through their major groove side. On the contrary, tetrads formed by minor groove association of Watson–Crick base pairs have always been found in structures where no G-tetrads are present, and give rise to a totally different kind of quadruplex. Whereas major groove tetrads are planar, in the minor groove association the two base pairs have a mutual inclination of around 30–40°. Most probably the inclination between the two base pairs makes these tetrads incompatible with pure guanine tetrads.
Implication in biological processes of quadruplex structures stabilized by minor groove tetrads has not been proven so far. However, it has been proposed that these tetrads may be responsible for the four-stranded structures observed in poly(CA)·poly(TG) fragments (19), and, in general, in processes where homologous DNA recognition is required, such as genetic recombination (20). The association of DNA duplexes through minor groove contacts was proposed many years ago as an initial step in recombination prior to strand exchange (21). Other processes in which this unusual quadruplex motif can be involved are the expansion of sequence repeats. It has been proposed that quadruplex formation in sequences containing repeats may be responsible of disturbing the DNA metabolism and the subsequent genetic instabilities associated with the expansion of the repeats (22). However, many repeats do not contain guanine-rich sequences and cannot form classical G-quadruplexes.
Quadruplex structures stabilized by minor groove tetrads were first found in the crystallographic dimeric structure of the linear heptamer d(GCATGCT) (23). However, self-associating cyclic oligonucleotides have been the best models to study this DNA motif so far. In the crystallographic and solution structures of d<pCATTCATT>, two cyclic octamers self-associate by forming two minor groove A:T:A:T tetrads (24,25). In the solution structures of d<pTGCTCGCT> and d<pCGCTCATT>, dimerization occurs by two minor groove G:C:G:C (25) and G:C:A:T (26) tetrad, respectively. A different type of G:C:G:C minor groove tetrad has been also observed in the dimeric structure of d<pCCGTCCGT> (27), and in hybrid complexes between linear and cyclic oligonucleotides (28). Similar minor groove tetrads have been found between symmetry-related molecules in the crystallographic structures of several DNA and DNA–RNA hybrid duplexes (29,30). In some cases, these tetrads are found involving terminal G and C residues that do not form base pairs with their own duplex but with symmetry-related ones, forming junction-like quadruplexes. Such structures have been found in pure DNA crystals and in several bisintercalative complexes involving acridine derivatives (31–35). In these complexes, the acridine derivatives do not intercalate in the usual way, but interact with the terminal nucleotides of four DNA duplexes forming a large intercalation platform between two minor groove G:C:G:C tetrads.
Minor groove tetrads may also be present in ordered nanostructures based on GC or AT pairing. High resolution scanning tunnelling microscopy studies in adlayers formed by coadsorption of guanine and cytosine on a graphite surface have revealed the formation of well-ordered periodic structures in the solid/liquid interface (36). Such structures have been attributed to alternate arrangements of GC base pairs through their major and minor groove sides. Very similar arrangements have been observed with AT base pairs (37).
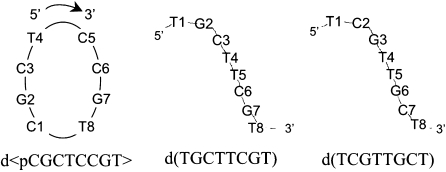
Within our efforts to explore other possible base pair arrangements susceptible of forming minor groove tetrads, we found that the cyclic octamer d<pCGCTCCGT> adopts a dimeric structure stabilized by these tetrads. This non-repetitive sequence is especially interesting because their linear analogues cannot form duplex-like structures. We explored several linear oligonucleotides of sequences derived from the cyclic one, and found that d(TGCTTCGT) and d(TCGTTGCT) adopt similar dimeric quadruplex structures, stabilized by the same type of minor groove tetrad. In this article, we report on the 3D structure and stability of the cyclic oligonucleotides d<pCGCTCCGT>, and its two linear analogues d(TGCTTCGT) and d(TCGTTGCT) (see Scheme 1 for numbering). In the three cases, the stabilization occurs through a C:G:G:C tetrad not observed before. The two intermolecular GC Watson–Crick base pairs interact through their minor groove sides, but in this case the additional hydrogen bonds that stabilize the tetrad are different than those observed in other minor groove tetrads, expanding the structural polymorphism of this kind of tetrads.
Scheme 1.
Sequences of the oligonucleotides studied in this work
MATERIALS AND METHODS
Experimental details
Linear oligonucleotides were synthesized by standard phosphoroamidite chemistry, and the cyclic one was made as reported by Alazzouzi et al. (38). Samples for NMR experiments were dissolved (in Na+ salt form) in either D2O or 9:1 H2O/D2O (25 mM sodium phosphate buffer, pH 7) in absence or presence of salts (100 mM NaCl, 10 mM MgCl2). All NMR spectra were acquired in Bruker spectrometers operating at 600 and 800 MHz, equipped with cryoprobes and processed with the TOPSPIN software. In the experiments in D2O, presaturation was used to suppress the residual H2O signal. A jump-and-return pulse sequence (39) was employed to observe the rapidly exchanging protons in 1D H2O experiments. NOESY (40) spectra in D2O were acquired with mixing times of 100, 200 and 300 ms. TOCSY (41) spectra were recorded with the standard MLEV-17 spin-lock sequence and a mixing time of 80 ms. In most of the experiments in H2O, water suppression was achieved by including a WATERGATE (42) module in the pulse sequence prior to acquisition. The spectral analysis program SPARKY (43) was used for semiautomatic assignment of the NOESY cross-peaks, and quantitative evaluation of the NOE intensities.
Circular dichroism spectra at different temperatures were recorded on a Jasco J-810 spectropolarimeter fitted with a thermostated cell holder. UV spectra were collected on a Jasco V-550 spectrometer fitted with a thermostated cell holder. For TDS (thermal differential spectra) experiments, UV spectra were recorded at low (5°C) and high (75°C) temperature. CD and UV spectra were recorded in 25 mM sodium phosphate buffer, pH 7, with 100 mM NaCl and 10 mM MgCl2. For melting experiments, the samples were initially heated at 90°C for 5 min, and slowly cooled to room temperature and stored at 4°C until use.
NMR constraints
The intrinsic ambiguity between inter- and intramolecular distances in dimeric structures could be overcome by using similar procedures as those employed in previously calculated structures of the same family (25,26), see Supplementary Data. Most of the cross-peaks could be directly assigned on the basis of their similarities with the related structures of d<pTGCTCGCT> (25), and d<pCCGTCCGT> (27). The very few cross-peaks (<5%) for which assignment remained ambiguous were not included in the structure calculations.
Initial calculations were performed with qualitative distance constraints (classified as 3, 4 or 5 Å), and the resulting structures were then refined by employing more accurate distance constraints obtained from a complete relaxation matrix analysis with the program MARDIGRAS (44). Error bounds in the interprotonic distances were estimated by carrying out several MARDIGRAS calculations with different initial models, mixing times and correlation times, as described in previous works. In addition to these experimentally derived constraints, Watson–Crick hydrogen bond restraints were used. Target values for distances and angles related to hydrogen bonds were set as described from crystallographic data.
Torsion angle constraints for the sugar moieties were derived form the analysis of J-coupling data obtained from DQF-COSY experiments. Since only the sums of coupling constants were estimated, rather loose values were set for the dihedral angle of the deoxyriboses (δ angle between 110° and 170°, ν1 between 5° and 65°, and ν2 between –65° and –50°).
Structure determination
Structures were calculated with the program DYANA 1.4 (45) and further refined with the SANDER module of the molecular dynamics package AMBER 7.0 (46). Initial DYANA calculations were carried out on the basis of qualitative distance constraints. The resulting structures were used as initial models in the complete relaxation matrix calculations to obtain accurate distance constraints, as described in the previous paragraph. These structures were taken as starting points for the AMBER refinement, consisting of an annealing protocol in vacuo, followed by long trajectories where explicit solvent molecules are included and using the Particle Mesh Ewald method to evaluate long-range electrostatic interactions. The specific protocols for these calculations have been described elsewhere (25,47). The AMBER-98 force-field (48) was used to describe the DNA, and the TIP3P model was used to simulate water molecules (49). Analysis of the representative structures as well as the MD trajectories was carried out with the programs Curves V5.1 (50), and MOLMOL (51).
RESULTS
d<pCGCTCCGT>, d(TGCTTCGT) and d(TCGTTGCT) form dimeric structures
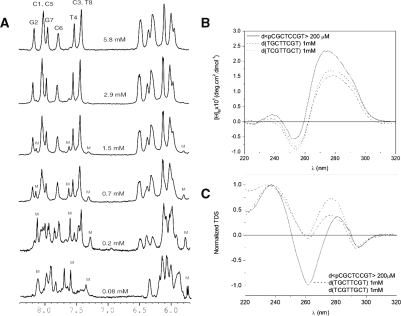
CD and TDS spectra are shown in Figure 1. The NMR spectra of the three oligonucleotides exhibit a strong dependence on temperature and oligonucleotide concentration, indicating that the structures are not monomeric (Figures 1 and 2). The sharp imino signals observed between 13.0–14.0 p.p.m. suggest the formation of GC Watson–Crick base pairs. In the case of the cyclic oligonucleotide, the conformational constraint induced by the cyclization impedes the formation of higher order structures with Watson–Crick base pairs. Although, in principle, the two linear oligonucleotides might form such high-order structures, the close similarity of their NMR spectra with that of d<pCGCTCCGT> and other oligonucleotides of the same family studied in previous works, led us to conclude that the structures adopted by the three oligonucleotides are dimeric. This result was confirmed by non-denaturing polyacrylamide gel electrophoresis (see Supplementary Figure S1).
Figure 1.
The 1D NMR spectra of d<pCGCTCCGT> in D2O at 5°C and different oligonucleotide concentrations (25 mM sodium phosphate buffer, pH 7, M stands for monomeric form) (A). CD (B) and TDS (C) spectra of d<pCGCTCCGT>, d(TGCTTCGT) and d(TCGTTGCT) (25 mM sodium phosphate buffer, pH 7, 100 mM NaCl, 10 mM MgCl2).
Figure 2.
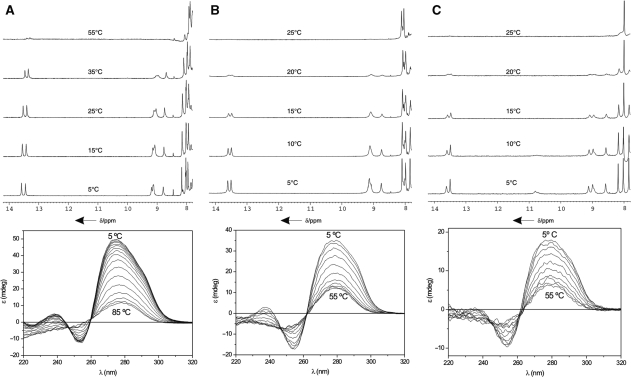
Top: 1D NMR spectra of d<pCGCTCCGT> (A), d(TGCTTCGT) (B) and d(TCGTTGCT) (C) in H2O at different temperatures (25 mM sodium phosphate buffer, pH 7, 100 mM NaCl, 10 mM MgCl2). Bottom: CD spectra at different temperatures, 25 mM sodium phosphate buffer, pH 7, 100 mM NaCl, 10 mM MgCl2.
Thermal denaturation
Unfolding transitions were followed by NMR and CD spectroscopy. The melting behavior of d<pCGCTCCGT> is similar to that of related cyclic oligonucleotides (52). Exchangeable protons are observed even at temperatures close to the denaturation midpoint, suggesting slow dissociation kinetics (Figure 2, Top). Interestingly, similar behaviour is observed in the melting transitions of the linear oligonucleotides, although they melt at lower temperatures than d<pCGCTCCGT>. This fact suggests that the relatively slow dissociation is not due to the cyclization of the sugar-phosphate backbone, but may be a general feature of quadruplex stabilized by minor groove tetrads.
Denaturation was also followed by circular dichroism (Figure 2, Bottom). In the three cases, the CD spectra at low temperature exhibit a large positive band around 275 nm and a negative band around 255 nm. Both bands disappear progressively as the temperature is increased, with the maximum of the positive band shifting to larger wavelengths.
Thermodynamic parameters for these denaturation transitions were estimated from the variation of the unfolding midpoint with the oligonucleotide concentration, from CD experiments (53). At 100 mM NaCl and 10 mM MgCl2 concentration, the estimated ΔG0298 values are −39 kJ/mol for the cyclic oligonucleotide and −15 kJ/mol for the linear ones (Table 3).
Table 3.
Thermodynamic parameters characterizing the folding of different cyclic and linear oligonucleotides that form minor groove quadruplex structures
| Sequence | Tetrad | ΔH0 (kJ/mol) | ΔS0 (J/mol K) | ΔG0 (kJ/mol) | Tm (K)a (5 mM oligo) |
|---|---|---|---|---|---|
| d(TGCTTCGT)b | C:G:G:C | −208 | −649 | −15 | 300 |
| d(TCGTTGCT)b | C:G:G:C | −217 | −677 | −15 | 301 |
| d<pCGCTCCGT>b | C:G:G:C | −184 | −487 | −39 | 346 |
| d<pTGCATGCA>c | G:C:G:C(direct) | −224 | −624 | −38 | 335 |
| d<pAGCTAGCT>c | G:C:G:C(direct) | −226 | −673 | −25 | 315 |
| d<pTGCTCGCT>d | G:C:G:C(direct) | −187 | −520 | −32 | 331 |
| d<pCCGTCCGT>e | G:C:G:C(slipped) | −221 | −625 | −35 | 330 |
| d<pCGCTCATT>f | G:C:A:T | −104 | −320 | −9 | 287 |
| d<pCATTCATT>d | A:T:A:T | −183 | −570 | −13 | 298 |
aTm values, normalized to 5 mM oligonucleotide concentration, have been calculated from the adjusted thermodynamic parameters.
b100 mM NaCl and 10 mM MgCl2, 25 mM phosphate buffer pH 7.
cData from Nuria Escaja, PhD Thesis. University of Barcelona, 1999; 25 mM phosphate buffer pH 7.
dData from ref. (25); 25 mM phosphate buffer pH 7.
eData from ref. (27); 25 mM phosphate buffer pH 7.
fData from ref. (26); 25 mM phosphate buffer pH 7.
NMR assignment
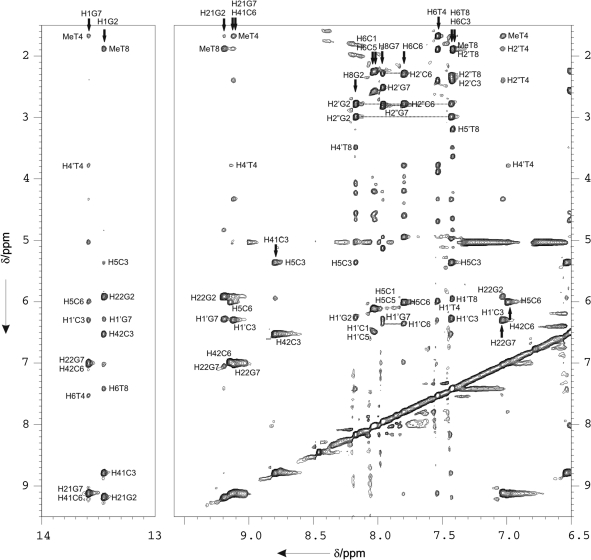
Sequential assignments of exchangeable and non-exchangeable protons were conducted following standard methods. Some regions of the 2D NOESY spectrum of d<pCGCTCCGT> in H2O are shown in Figure 3. Many spectral features are common to other structures of this family studied by NMR. In the three cases, the number of signals in the spectra corresponds to 8 nt. This indicates that the dimer is symmetric, since all the corresponding protons in each subunit of the dimer are magnetically equivalent and, consequently, have the same chemical environment. Since residues 1–8 are equivalent to residues 9–16, in this section, we will use the numbering 1–8 for the discussion of the NMR assignment, and the numbering 1–16 for describing intermolecular NOEs. All intra-nucleotide H1′-base NOEs are medium or weak, indicating that the glycosidic angle in all the nucleotides is in an ‘anti’-conformation. Strong sugar-base sequential connections were observed between residues 2 → 3, and 6 → 7. Sequential NOEs between residues 3 → 4 and 7 → 8 are medium in the H1′-base region, and very weak in the H2′/H2″-base region (see Figure 3 and Scheme 1). Almost all resonances were identified, including some H5′/H5″ protons. The complete assignment lists are shown in Tables S1–S3 in the Supplementary Data, and have been deposited at the Biological Magnetic Resonance Bank (BMRB).
Figure 3.
Two regions of the NOESY spectrum (Tm = 150 ms) of d<pCGCTCCGT> in H2O (4 mM oligonucleotide concentration, 100 mM NaCl, T = 5°C, pH 7). Watson–Crick base pairing can be established from the H1G-H42C, H1G-H41C and H1G2-H5C cross-peaks for each of the GC base pairs. Sequential assignment pathways for non-exchangeable protons are shown. Cross-peaks are labelled according to the spin systems numbering shown in scheme 1.
Interestingly, different chemical shifts are observed for H4′/H5′/H5″ protons of the nucleotides in the first position of the loops, which are stacked over the base pairs. For these three sequences, one of the thymines stacks on a purine residue and H4′/H5′/H5″ protons resonate at higher field than usual, while the other thymine stacks on a pyrimidine residue showing expected chemical shifts.
The exchangeable proton spectra are particularly informative. Amino protons of all cytosines were assigned from their NOE cross-peaks with the H5 protons. For d<pCGCTCCGT>, as shown in Figure 3, strong cross-peaks are observed between the two amino protons of C3 and C6 with the two imino signals between 13.0 and 14.0 p.p.m., corresponding to G2 and G7, respectively. The amino resonances of the guanines are also labeled. This NOE pattern is characteristic of GC Watson–Crick base pairs, which are all intermolecular: C3-G10 and C11-G2, for the first set of cross-peaks, and C6-G15 and C14-G7, for the second one (see numbering scheme for the dimers in Figure 4). Strong NOE signals between the amino protons of G2 and H1′ proton of the G7, and between the amino protons of G7 and the H1′ proton of C3 are observed. These cross-peaks are particularly informative since NOE signals between amino protons of G7 and the H1′ proton of C3 can be only explained as intermolecular contacts (G7-C11 and C3-G15). Thymine imino protons are observed around 10.0–11.0 p.p.m. region, indicating that in all cases the thymines are not base-paired. Although all these iminos exchange rapidly with the bulk water, the signals of thymine iminos in the first position of the loops are relatively sharp and can be identified in experiments recorded with a jump-and-return pulse sequence. On the contrary, signals of iminos of thymines in the second position of the loops of d(TGCTTCGT) and d(TCGTTGCT) are very broad or undetectable (see Supplementary Figure S2).
Figure 4.
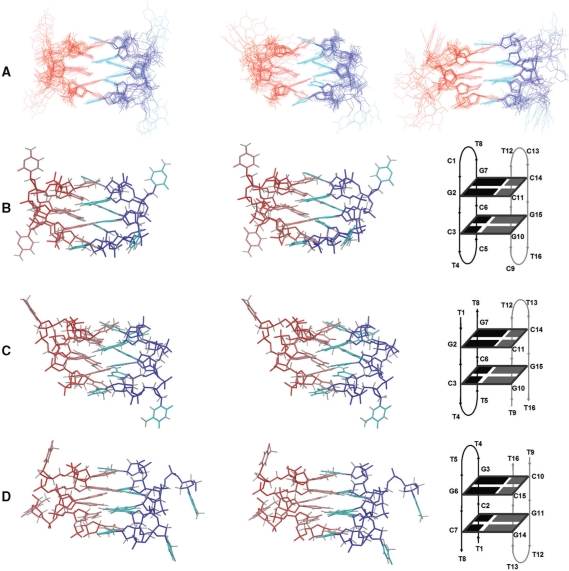
Superposition of the 10 refined structures of d<pCGCTCCGT>, d(TGCTTCGT) and d(TCGTTGCT) (A), and stereoscopic views of the average structure of d<pCGCTCCGT> (B), d(TGCTTCGT) (C) and d(TCGTTGCT) (D). Red and blue indicate different molecules. The sugar-phosphate backbone is indicated in darker colors. Note that the perspective in (C) and (D) is not the same, but has been selected to show the two C:G:G:C tetrads in the same orientation.
NOESY spectra of d(TGCTTCGT) and d(TCGTTGCT) are shown in the Supplementary Figures S4 and S5. The spectra of both oligonucleotides are very similar to their cyclic analogue, with akin patterns of NOE cross-peaks.
Experimental constraints and structure calculations
As mentioned in the Materials and Methods section, the intrinsic ambiguity between inter- and intramolecular cross-peaks was resolved as in previous studies of structures from this family. A summary of the final distance constraints obtained by a complete relaxation matrix calculation is shown in Table 1 and in Supplementary Figures S6–S8. In these figures and in the following discussion we will use the numbering scheme of Figure 4, where residues are numbered from 1 to 8 for the first subunit, and from 9 to 16 for the second subunit. Inspection of DQF-COSY cross-peaks suggests that the major conformers of all deoxyriboses are in the general S-domain. However, DQF-COSY spectra had to be recorded at 5°C, and the large line width of the proton signals at low temperature prevented quantitative estimation of the J-coupling values. Consequently, no dihedral constraints were included in the structure calculation.
Table 1.
Experimental constraints and calculation statistics
| d(TGCTTCGT) | d(TCGTTGCT) | d<pCGCTCCGT> | |
|---|---|---|---|
| Experimental distance constraints | |||
| Total number | 318 | 336 | 214 |
| Intra-subunit | 240 | 274 | 162 |
| Intra-residue | 152 | 182 | 94 |
| Sequential | 52 | 54 | 38 |
| Range > 1 | 36 | 38 | 30 |
| Inter-subunit | 78 | 62 | 52 |
| RMSD | |||
| All bases well-defineda | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.4 ± 0.1 |
| All heavy atoms well-defineda | 0.8 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 |
| All heavy atoms | 2.3 ± 0.5 | 2.9 ± 0.6 | 2.5 ± 0.5 |
| Sum of violations (Å) (average and range) | 7.0 (6.6–7.6) | 8.0 (7.6–8.3) | 2.5 2.2–2.9 |
| Max. violation (Å) (average and range) | 0.38 (0.35–0.49) | 0.49 (0.43–0.58) | 0.25 0.22–0.27 |
| Av. NOE energy (kcal/mol) | 27 | 37 | 8 |
| Range of NOE energies (kcal/mol) | 25–30 | 38–43 | 7–10 |
aAll except residues 1, 5, 9 and 13.
The structures were calculated by using restrained molecular dynamics methods with experimental constraints. Initial structures were calculated with the program DYANA and then refined with the AMBER package following the protocols described in the Materials and methods section. Except for the residues in positions 1 and 5, and the corresponding ones in the symmetry-related subunit, all residues in the final structures are well defined, with an RMSD of around 1 Å. These values are even lower when only atoms in the bases are considered (Table 1). The structures of the two linear oligonucleotides are slightly better defined than the cyclic one. This is due to the lower number of distances constraints of the latter. The final AMBER energies and NOE terms are reasonably low in all the structures, with no distance constraint violation >0.5 Å. Atomic coordinates have been deposited in the Protein Data Bank (accession numbers PDB: 2k8z, 2k90 and 2k97).
Description of the structures
In all three cases, the resulting structures are symmetric dimers, as expected from the complete degeneration of the signals in their NMR spectra. This symmetry is reflected in the average geometrical parameters shown in the Supplementary Tables S4–S6. As can be seen in Figure 4, the arrangement of the two subunits in the dimer is antiparallel, and can be interconverted by a binary symmetry axis perpendicular to the plane of the figure. The structure is stabilized by four intermolecular GC Watson–Crick base pairs, which associate to each other by their minor grooves sides, forming two C:G:G:C tetrads (Figure 5C). All glycosidic angles are ‘anti’, with values ranging from −110° to −152°. In addition to the six Watson–Crick hydrogen bonds, each tetrad is stabilized by two additional hydrogen bonds. In the case of d<pCGCTCCGT> and d(TGCTTCGT), the two tetrads are C11:G2:G7:C14 and C3:G10:G15:C6, according to the numbering scheme shown in Figure 4B and C. In the first tetrad, an additional hydrogen bond is formed between an amino proton of G2 and the N3 of the neighbouring G7. The other hydrogen bond occurs between an amino proton of G7 and the O2 of C11 in the adjacent base pair. The same hydrogen bonds are formed between the equivalent residues in the second tetrad: amino proton(G10)-N3(G15) and amino proton(G15)-O2(C3). An identical pattern of hydrogen bonds is formed in the structure of d(TCGTTGCT), but in this case the two tetrads are C7:G14:G11:C2 and C15:G6:G3:C10, according to the numbering in Figure 4D. Note that the perspective in Figure 4D has been selected to show the tetrads in the same orientation in the three molecules. The additional hydrogen bonds are now: amino proton(G14)-N3(G11) and amino proton(G11)-O2(C7) for the first tetrad, and amino proton(G6)-N3(G3) and amino proton(G3)-O2(C15) for the second one.
Figure 5.
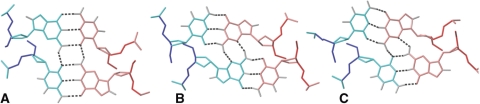
The three known minor groove tetrads involving GC base pairs. (A) Direct minor groove G:C:G:C tetrad observed in the dimeric solution structures of d<pTGCTCGCT>, (B) slipped minor groove tetrad found in the solution structure of d<pCCGTCCGT>, (C) C:G:G:C tetrad observed in d<pCGCTCCGT>, d(TGCTTCGT) and d(TCGTTGCT).
All guanine amino protons are buried in the core of the structure and lie very close to sugar moieties. For example, the amino protons of G7 are close to the opposite C11 residue, and in G2, the amino protons are near the adjacent G7 residue, giving rise to the particular NOE contacts aforementioned. As in other minor groove tetrads, the two base pairs are not in the same plane (Figure 4). In the case of d<pCGCTCCGT> and d(TGCTTCGT), the mutual inclination between adjacent base pairs is around 40°, but in the case of d(TCGTTGCT), this value is near 20°. In general, the central steps in all the structures exhibit rise values of approximately 3.2–3.4 Å, with the exception of the GC steps of d(TGCTTCGT), which have rise values around 4.2 Å. The twist values of these central steps range between 25° and 38° in all the structures. Other local helical parameters are given in Table S7 in the Supplementary Data.
Other structural features are very similar to previously studied structures of this family. Thymines in the first position of the loops form two caps at both ends of the stacks, and interact with each other through hydrophobic contacts between their methyl groups. Interestingly, the thymines in position 8 in the linear oligonucleotides adopt the same conformation as in the cyclic oligonucleotide, in spite of their position at the 3′-termini. The 5′-terminal thymines are disordered, though. A list of backbone angles is provided in the Supplementary Tables S4–S6. Backbone angles are not as well defined as in other structures of the same family studied previously in our group, due to the lower number of distance constraints and the lack of stereospecific assignment of H5′/H5″ protons. The average values of the well-defined ones (order parameter >0.9) are within the usual values found in right-handed double-stranded DNA. The very tight turn between residues in the loops causes several phosphate groups to be very close to each other. This close proximity is a general characteristic of this family of structures, and gives rise to two extremely narrow grooves with a width of around 1 Å (considering P–P distance 7 Å and a sum of van der Waals radii of 5.8 Å). The width of the other two grooves is similar to that of the major groove in a B-DNA duplex.
DISCUSSION
Comparison with other structures of the family
The strong similarity between the structures of d(TGCTTCGT), d(TCGTTGCT) and d<pCGCTCCGT> is apparent from the RMSDs between their C1′ atoms shown in Table 2 and, whose average values are 2.3 Å for all C1′, and 0.8 Å for the C1′ atoms of the well-defined nucleotides in the central core of the dimers. The comparison with other members of this family of structures is also shown in Table 2, and indicates that all are globally very similar, especially when the C1′ atoms of the central core of the molecule are compared. This confirms our previous observation that minor groove tetrads impose important restrictions on the 3D structures of this class of quadruplexes.
Table 2.
Atomic RMSDs (Å) between the structures of several oligonucleotides stabilized by minor groove tetrads
| d(TGCTTCGT) | d(TCGTTGCT) | d<pCGCTCCGT> | d<pTGCTCGCT> | d<pCATTCATT> | d<pCGCTCATT> | d<pCCGTCCGT> | |
|---|---|---|---|---|---|---|---|
| d(TGCTTCGT) | 2.8 | 2.3 | 2.3 | 2.6 | 2.3 | 2.6 | |
| d(TCGTTGCT) | 1.1 | 1.8 | 1.4 | 1.4 | 2.9 | 1.9 | |
| d<pCGCTCCGT> | 0.6 | 0.8 | 1.2 | 1.9 | 1.9 | 0.9 | |
| d<pTGCTCGCT> | 0.6 | 0.8 | 0.8 | 1.2 | 2.4 | 1.2 | |
| d<pCATTCATT> | 0.8 | 1.1 | 0.7 | 0.8 | 2.6 | 1.8 | |
| d<pCGCTCATT> | 0.9 | 1.3 | 0.9 | 1.1 | 0.6 | 1.7 | |
| d<pCCGTCCGT> | 0.6 | 0.9 | 0.7 | 0.8 | 0.9 | 1.0 | |
| d(GCATGCT) | 0.7 | 0.9 | 0.8 | 0.5 | 0.8 | 1.0 | 0.9 |
Upper: RMSD between all C1′. Lower: RMSD between the C1′ of the nucleotides involved in the tetrads.
In spite of the high global similarity between all the structures, the three oligonucleotides studied in this work present some important differences. Since in this case the sequences are not repetitive, the structures are less symmetric than in other oligonucleotides studied earlier. Although the two subunits forming the dimer are completely equivalent to each other, the two stacks are different: one is formed by GC steps and the other by CG steps. Twist values in the GC and CG steps of the three oligonucleotides are between 25° and 37°. These values are intermediate between the average twist of 26° for d<pTGCTCGCT> (25), where all the steps are GC, and the 41° value for d<pCCGTCCGT> (27), where all the steps are CG. Rise values are generally around 3.3 Å, slightly higher than in the previously studied structures of d<pTGCTCGCT> and d<pCCGTCCGT>.
Characteristic downfield chemical shifts are observed for some of the H4′ and H5′/5″ protons of residues in the first position of the loops. These unusual downfield shifts are due to the proximity of the purine bases in the 3D structure, which are two positions away in the sequence (in the 5′→3′ direction). This occurs in loops with the sequence 5′-NTNG-3′, and not with those with 5′-NTNC-3′. The same effect has been observed before in the NMR spectra of the related oligonucleotides, and seems to be a general characteristic of these tightly turned loops.
The C:G:G:C minor groove tetrad
As mentioned above, the oligonucleotide structures stabilized by minor groove tetrads are very similar. However, the nature of the minor groove tetrad and the type of interactions that stabilize them are quite different in each case. In A:T:A:T and G:C:A:T tetrads, the stabilization arises from a cation in the centre of the molecule, which probably coordinates the O2 atoms of the surrounding thymines (24,26). In G:C:G:C tetrads, this stabilization is due to additional hydrogen bonds between the adjacent GC base pairs. Two different patterns of inter base pair hydrogen bonds have been found so far: the direct alignment, where the interaction between the base pairs occurs through two bifurcated amino(G)-O2(C) hydrogen bonds (Figure 5A), and the slipped alignment, where the interaction takes place through two amino(G)-N3(G) hydrogen bonds (Figure 5B). The former alignment has been observed in the structures of the cyclic octamer d<pTGCTCGCT> and the linear heptamer d(GCATGCT), the latter is present in the structure of d<pCCGTCCGT> and in the heterodimer d<pCAGTCCCT> + d<pCCTTCGGT>. In all these structures the two GC base pairs forming the tetrad run opposite to each other. On the contrary, in the 3D structures of the three oligonucleotides studied in this work, the tetrads are formed by two GC base pairs with a parallel orientation, giving rise to a C:G:G:C tetrad. As shown in Figure 5C, the stabilization arises from extra hydrogen bonds between guanine amino protons not involved in Watson–Crick hydrogen bonds and the cytosine O2 and the guanine N3 of the adjacent base pair. Contrary to previously studied G:C:G:C minor groove tetrads (Figures 5A and B), the C:G:G:C tetrad is not symmetric (Figure 5C). In other words, the two base pairs involved in the C:G:G:C tetrad are not equivalent to each other.
Thermal stability
Values for the free energy of conformational stability, ΔG, have been experimentally determined for the dimeric structures of several oligonucleotides stabilized by different types of minor groove tetrads. Such values are compiled in Table 3. Although not all the data have been obtained under identical buffer conditions, comparison of ΔG values of dimeric structures formed by cyclic oligonucleotides indicates that the direct and the slipped G:C:G:C tetrads and the C:G:G:C tetrad provide an stabilization effect of a similar magnitude. In these tetrads, with direct hydrogen bonds between base pairs, the stabilization is clearly higher than in the case of A:T:A:T and G:C:A:T tetrads that require a cation coordination.
Although in previous studies, minor groove G:C:G:C tetrads have been observed in cyclic oligonucleotides at very low ionic strength (25 mM sodium phosphate) (27,52), a higher ionic strength tends to stabilize these structures because the repulsion between phosphate charges is partially alleviated. In the case of the linear octamers d(TGCTTCGT) and d(TCGTTGCT), their stability is quite lower than that of their cyclic analogue, and a buffer of moderate salt concentration is required for their study by NMR. Although the dimeric structure of d<pCGCTCCGT> can be observed in 25 mM sodium phosphate buffer (data not shown), it has been studied under the same buffer conditions as its linear analogues for a fairer comparison between the three structures.
Interesting conclusions can also be drawn from the comparison between ΔG values of cyclic and linear oligonucleotides stabilized by the same type of minor groove tetrad. The difference in thermal stability between d<pCGCTCCGT> and d(TGCTTCGT) and d(TCGTTGCT) allows for an estimation of the stabilization effect produced by the cyclization of the sugar-phosphate chain, which is around 24 kJ/mol. In the light of these data, and assuming that the stabilization produced by the cyclization is sequence independent, the linear analogues of the cyclic oligonucleotides that form G:C:G:C tetrads (direct or slipped) may form similar quadruplex structures. In contrast, in the absence of other stabilization agents, G:C:A:T and A:T:A:T tetrads do not confer enough stability for quadruplex formation in linear oligonucleotides of this size.
It is interesting to notice that neither d(TGCTTCGT) nor d(TCGTTGCT) form monomeric hairpins. However, they do adopt stable hairpin-like structures through dimerization. The fact that sequences that do not form monomeric hairpins can adopt stable hairpin-like structures through dimerization may have implications in biological processes where hairpin–hairpin interaction is being invoked, such as DNA repeat expansions (22,54). It has been proposed that DNA hairpins and slipped structures that occur in sequences containing repeats lead to errors in replication and cause expansions and deletions of the repeating track (55), since the relative stability of single DNA hairpins is insufficient to explain the expansion tendency of the different repeats (54).
Minor groove tetrads in linear oligonucleotides
Although minor groove tetrads were first observed in the crystallographic structures of the linear heptamer d(GCATGCT) (23), and, more recently, in hybrid complexes between linear and cyclic oligonucleotides (28), this is the first time that they have been observed in solution in a dimeric structure formed by two linear oligonucleotides. Because of the high physiological relevance of linear DNAs, we have been searching for minor grooves tetrads in linear oligonucleotides during the last few years. We focused in sequences able to form G:C:G:C tetrads, such as the heptamer d(GCATGCT) and linear analogues of the cyclic oligonucleotides d<pCCGTCCGT> and d<pTGCTCGCT>, where this motif had been previously found. In most cases, we found that the NMR spectra were not consistent with single well-defined structures.
The observation of this DNA motif in solution with linear oligonucleotides is particularly problematical because, contrary to other quadruplexes stabilized by G-tetrads or intercalated CC+ base pairs, most sequences that can form a minor groove quadruplex are also able to form antiparallel Watson–Crick duplexes (sometimes with bulges or mismatches) or small hairpins. The thermodynamic and structural data obtained in the present work can explain the reason why minor groove tetrads have been observed in these particular linear oligonucleotides and not in others. Based on the data shown in Table 3, and the strong similarity between the structures, we can assume that the stability of the dimeric quadruplexes formed by linear oligonucleotides would be similar for both G:C:G:C tetrads or C:G:G:C tetrad described in this work. However, the former ones have sequences like 5′-NGCNNGCN-3′ or 5′-NCGNNCGN-3′ (where N is any nucleotide). These sequences are susceptible of forming an antiparallel duplex with four GC base pairs and a central bulge, which may compete with quadruplex formation. Observation of this quadruplex DNA motif as a single well-defined structure in solution has been possible because, in these particular sequences, the competing Watson–Crick duplex cannot have more than two GC base pairs.
In previous studies, it has been shown that quadruplexes stabilized by minor groove tetrads are easily observed in cyclic oligonucleotides, where antiparallel Watson–Crick duplexes cannot be formed due to the conformational constraint induced by the cyclization. The fact that there are very few sequence requirements for their formation leads us to propose that we might be observing a rather general DNA motif, which might exist in cellular DNA. The thermodynamic and structural data obtained in this study add further support to the hypothesis that quadruplex structures stabilized by minor groove tetrads, at least those formed by GC base pairs, can also be a common motif in linear oligonucleotides. These structures can be observed in vitro, provided that the competing antiparallel duplexes are impeded. In short oligonucleotides, this impediment can be a conformational constraint (like in cyclic oligonucleotides), crystal packing forces [like in d(GCATGCT)], or sequences with very little propensity to form duplexes (as in the oligonucleotides presented in this study). In cellular DNA, this motif might stabilize the association of longer DNA hairpins or slipped structures.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We gratefully acknowledge Dr D.V. Laurents for revision of the manuscript and his useful comments.
FUNDING
Spanish Ministerio de Ciencia e Innovación (CTQ2007-68014-C02-01/02); Generalitat de Catalunya (2005SGR693 and Xarxa de Referència en Biotecnologia). Funding for open access charge: Spanish Ministerio de Ciencia e Innovación (CTQ2007-68014-C02-01/02).
Conflict of interest statement. None declared.
REFERENCES
- 1.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 2.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 5.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 8.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel PK, Koti AS, Hosur RV. NMR studies on truncated sequences of human telomeric DNA: observation of a novel A-tetrad. Nucleic Acids Res. 1999;27:3836–3843. doi: 10.1093/nar/27.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel PK, Hosur RV. NMR observation of T-tetrads in a parallel stranded DNA quadruplex formed by Saccharomyces cerevisiae telomere repeats. Nucleic Acids Res. 1999;27:2457–2464. doi: 10.1093/nar/27.12.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cáceres C, Wright G, Gouyette C, Parkinson G, Subirana JA. A thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+/Na+ ions. Nucleic Acids Res. 2004;32:1097–1102. doi: 10.1093/nar/gkh269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel PK, Bhavesh NS, Hosur RV. NMR observation of a novel C-tetrad in the structure of the SV40 repeat sequence GGGCGG. Biochem. Biophys. Res. Commun. 2000;270:967–971. doi: 10.1006/bbrc.2000.2479. [DOI] [PubMed] [Google Scholar]
- 13.Webba da Silva M. Association of DNA quadruplexes through G:C:G:C tetrads. Solution structure of d(GCGGTGGAT) Biochemistry. 2003;42:14356–14365. doi: 10.1021/bi0355185. [DOI] [PubMed] [Google Scholar]
- 14.Webba da Silva M. Experimental demonstration of T:(G:G:G:G):T hexad and T:A:A:T tetrad alignments within a DNA quadruplex stem. Biochemistry. 2005;44:3754–3764. doi: 10.1021/bi0478190. [DOI] [PubMed] [Google Scholar]
- 15.Kettani A, Kumar RA, Patel DJ. Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 1995;254:638–656. doi: 10.1006/jmbi.1995.0644. [DOI] [PubMed] [Google Scholar]
- 16.Kettani A, Bouaziz S, Gorin A, Zhao H, Jones RA, Patel DJ. Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 1998;282:619–636. doi: 10.1006/jmbi.1998.2030. [DOI] [PubMed] [Google Scholar]
- 17.Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. Dimeric DNA quadruplex containing major groove-aligned A-T-A-T and G-C-G-C tetrads stabilized by inter-subunit Watson-Crick A-T and G-C pairs. J. Mol. Biol. 2001;312:1073–1088. doi: 10.1006/jmbi.2001.5002. [DOI] [PubMed] [Google Scholar]
- 18.Bouaziz S, Kettani A, Patel DJ. A K cation-induced conformational switch within a loop spanning segment of a DNA quadruplex containing G-G-G-C repeats. J. Mol. Biol. 1998;282:637–652. doi: 10.1006/jmbi.1998.2031. [DOI] [PubMed] [Google Scholar]
- 19.Gaillard C, Strauss F. Association of poly(CA).poly(TG) DNA fragments into four-stranded complexes bound by HMG1 and 2. Science. 1994;264:433–436. doi: 10.1126/science.8153633. [DOI] [PubMed] [Google Scholar]
- 20.McGavin S. Models of specifically paired like (homologous) nucleic acid structures. J. Mol. Biol. 1971;55:293–298. doi: 10.1016/0022-2836(71)90201-4. [DOI] [PubMed] [Google Scholar]
- 21.Wilson JH. Nick-free formation of reciprocal heteroduplexes: a simple solution to the topological problem. Proc. Natl Acad. Sci. USA. 1979;76:3641–3645. doi: 10.1073/pnas.76.8.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonard GA, Zhang S, Peterson MR, Harrop SJ, Helliwell JR, Cruse WB, d’Estaintot BL, Kennard O, Brown T, Hunter WN. Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution. Structure. 1995;3:335–340. doi: 10.1016/s0969-2126(01)00165-4. [DOI] [PubMed] [Google Scholar]
- 24.Salisbury SA, Wilson SE, Powell HR, Kennard O, Lubini P, Sheldrick GM, Escaja N, Alazzouzi E, Grandas A, Pedroso E. The bi-loop, a new general four-stranded DNA motif. Proc. Natl Acad. Sci. USA. 1997;94:5515–5518. doi: 10.1073/pnas.94.11.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escaja N, Pedroso E, Rico M, González C. Dimeric solution structure of two cyclic octamers. A new four-stranded motif of DNA entirely formed by A:T:A:T and G:C:G:C tetrads. J. Am. Chem. Soc. 2000;122:12732–12742. [Google Scholar]
- 26.Escaja N, Gelpi JL, Orozco M, Rico M, Pedroso E, González C. A four-stranded DNA structure stabilized by a novel G:C:A:T tetrad. J. Am. Chem. Soc. 2003;125:5654–5662. doi: 10.1021/ja0344157. [DOI] [PubMed] [Google Scholar]
- 27.Escaja N, Gómez-Pinto I, Pedroso E, González C. Four-stranded DNA structures can be stabilized by two different types of minor groove G:C:G:C tetrads. J. Am. Chem. Soc. 2007;129:2004–2014. doi: 10.1021/ja066172z. [DOI] [PubMed] [Google Scholar]
- 28.Escaja N, Gómez-Pinto I, Viladoms J, Rico M, Pedroso E, González C. Induced-fit recognition of DNA by small circular oligonucleotides. Chem. Eur. J. 2006;12:4035–4042. doi: 10.1002/chem.200600050. [DOI] [PubMed] [Google Scholar]
- 29.Wahl MC, Sundaralingam M. A-DNA duplexes in the crystal. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Sructures. New York: Oxford University Press; 1999. pp. 117–144. [Google Scholar]
- 30.Tereshko V, Subirana JA. Influence of packing interactions on the average conformation of B-DNA in crystalline structures. Acta Crystallogr. D Biol. Crystallogr. 1999;55:810–819. doi: 10.1107/s0907444999000591. [DOI] [PubMed] [Google Scholar]
- 31.Adams A, Guss JM, Denny WA, Wakelin LP. Structure of 9-amino-[N-(2-dimethylamino)propyl]acridine-4-carboxamide bound to d(CGTACG)(2): a comparison of structures of d(CGTACG)(2) complexed with intercalators in the presence of cobalt. Acta Crystallogr. D Biol. Crystallogr. 2004;60:823–828. doi: 10.1107/S0907444904003907. [DOI] [PubMed] [Google Scholar]
- 32.Adams A, Guss JM, Collyer CA, Denny WA, Wakelin LP. A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)(2) Nucleic Acids Res. 2000;28:4244–4253. doi: 10.1093/nar/28.21.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorpe JH, Hobbs JR, Todd AK, Denny WA, Charlton P, Cardin CJ. Guanine specific binding at a DNA junction formed by d[CG(5-BrU)ACG](2) with a topoisomerase poison in the presence of Co(2+) ions. Biochemistry. 2000;39:15055–15061. doi: 10.1021/bi001749p. [DOI] [PubMed] [Google Scholar]
- 34.Teixeira SC, Thorpe JH, Todd AK, Powell HR, Adams A, Wakelin LP, Denny WA, Cardin CJ. Structural characterisation of bisintercalation in higher-order DNA at a junction-like quadruplex. J. Mol. Biol. 2002;323:167–171. doi: 10.1016/s0022-2836(02)00923-3. [DOI] [PubMed] [Google Scholar]
- 35.Yang XL, Robinson H, Gao YG, Wang AH. Binding of a macrocyclic bisacridine and ametantrone to CGTACG involves similar unusual intercalation platforms. Biochemistry. 2000;39:10950–10957. [PubMed] [Google Scholar]
- 36.Xu S, Dong M, Rauls E, Otero R, Linderoth TR, Besenbacher F. Coadsorption of guanine and cytosine on graphite: ordered structure based on GC pairing. Nano Lett. 2006;6:1434–1438. doi: 10.1021/nl060563u. [DOI] [PubMed] [Google Scholar]
- 37.Mamdouh W, Dong M, Xu S, Rauls E, Besenbacher F. Supramolecular nanopatterns self-assembled by adenine-thymine quartets at the liquid/solid interface. J. Am. Chem. Soc. 2006;128:13305–13311. doi: 10.1021/ja064988u. [DOI] [PubMed] [Google Scholar]
- 38.Alazzouzi E, Escaja N, Grandas A, Pedroso E. A straightforward solid-phase synthesis of cyclic oligodeoxyribonucleotides. Angew. Chem. Int. Ed. Engl. 1997;36:1506–1508. [Google Scholar]
- 39.Plateau P, Güeron M. Exchangeable proton NMR without base-line distortions, using new strong pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 40.Kumar A, Ernst RR, Wüthrich K. A two-dimensional nuclear overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 41.Bax A, Davies DJ. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 42.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 43.Goddard DT, Kneller G. San Francisco: University of California; SPARKY 3. [Google Scholar]
- 44.Borgias BA, James TL. MARDIGRAS, a procedure for atrix analysis of relaxation for discerning geometry of an aqueous structure. J. Magn. Reson. 1990;87:475–487. [Google Scholar]
- 45.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 46.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, et al. San Francisco: University of California; 2002. AMBER 7. [Google Scholar]
- 47.Soliva R, Monaco V, Gómez-Pinto I, Meeuwenoord NJ, Marel GA, Boom JH, González C, Orozco M. Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety. Nucleic Acids Res. 2001;29:2973–2985. doi: 10.1093/nar/29.14.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz K, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd generation force field for the simulation of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 49.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of single potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 50.Lavery R, Sklenar H. Paris: Laboratory of Theoretical Biochemistry CNRS; 1990. Curves 5.1. [Google Scholar]
- 51.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 52.González C, Escaja N, Rico M, Pedroso E. NMR Structure of two cyclic oligonucleotides. A monomer-dimer equilibrium between dumbbell and quadruplex structures. J. Am. Chem. Soc. 1998;120:2176–2177. [Google Scholar]
- 53.Breslauer KJ. Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transitions. Methods Enzymol. 1995;259:221–242. doi: 10.1016/0076-6879(95)59046-3. [DOI] [PubMed] [Google Scholar]
- 54.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.