Abstract
More than one-third of the RAG1 protein can be truncated from the N-terminus with only subtle effects on the products of V(D)J recombination in vitro or in a mouse. What, then, is the function of the N-terminal domain? We believe it to be regulatory. We determined, several years ago, that an included RING motif could function as an ubiquitin E3 ligase. Whether this activity is limited to automodification, or may alter other proteins in the cell, remains an open question. We revisited the issue of additional protein–protein interactions between RAG1 and other proteins by means of the yeast two-hybrid assay. We confirmed the interaction already described with KPNA2/RCH1/SRP1α and found two others—to the transcription factor GMEB1/PIF p96 and the splicing factor SF3A2/SF3a66. A luciferase reporter assay demonstrates that a protein complex containing RAG proteins and the transcription factor can assemble in cells. Further mapping identified a region within the N-terminal domain resembling a WW motif. Point mutation directed at residues conserved in WW motifs eliminated binding to one of the partners. Phylogenetic analysis shows the WW-like module to be highly conserved. The module contributes to protein–protein interactions that may also influence how RAG1 binds DNA targets.
INTRODUCTION
The adaptive immune response of vertebrate animals depends upon site-specific DNA rearrangement in a process termed ‘V(D)J recombination’. The combinatorial nature of this gene assembly explains a large part of the diversity witnessed in the resulting receptors, and permits a flexible response (1,2). The mechanism of the recombination reaction is known at the most general level. Recombination is targeted to the sequences encoding the V, D or J elements by the presence of adjacent recombination signal sequences (RSSs). Two proteins, called RAG1 and RAG2 (3,4), form the nuclease that recognizes these targets and cuts the DNA into ends that are processed and rejoined in a new configuration. Aside from the nuclease activity, there exists evidence that the RAG proteins function at a structural level in the synapsis of the two DNA targets and organization of the cut ends after the cleavage reaction. It is also possible that RAG1 functions at yet another level, as a regulatory molecule controlling V(D)J recombination and coordinating other aspects of cell physiology (5,6).
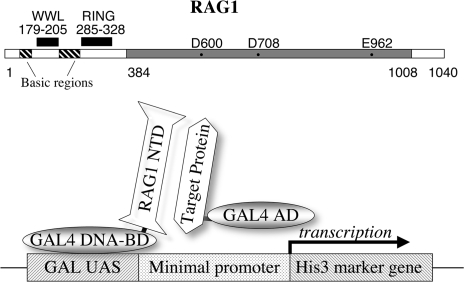
RAG1 protein is large, with 1040 amino acid residues in the mouse (Figure 1).
Figure 1.
(Top) Linear representation of the mouse RAG1 protein. The central core region (384–1008) is marked in gray, with essential acidic residues. The N-terminal region (residues 1–383) includes a previously identified RING motif, several clusters of basic residues, and (in this report) the WW-like domain. (Bottom) Representation of the two-hybrid assay. The GAL4 DNA-binding domain (GAL4 DNA-BD) is fused to the RAG1 N-terminal domain (NTD) or smaller peptides. A library of target proteins is obtained with each member fused to the GAL4 activation domain (GAL4 AD). In yeast, the GAL4 DNA-BD binds to the UAS element on a reporter construct. A protein–protein interaction between a target protein and the RAG1 NTD protein positions the GAL4 AD to activate transcription of the His3 marker gene.
Deletion analysis indicated that a sizable segment, residues 1–383 at the N-terminus, could be removed, along with a shorter segment at the C-terminus, to yield a core region that is active in V(D)J recombination in vitro and in cells (7–9). Despite being recognized for years, the function of the N-terminal region remains mysterious. Its existence is conserved through the evolution of RAG1, yet it clearly is not needed for the central enzymatic role as a recombinase. Sequence alignments show that this region exhibits a greater divergence through evolution than the enzymatic core (6). Notable in such alignments is the absolute conservation of a cluster of cystine and histidine residues recognized as a special zinc-binding motif termed a ‘RING finger’. RING structures have been found in enzymes (E3 ligases) that help modify other proteins through the covalent addition of small modifier proteins (10). In biochemical assays, the RAG1 N-terminus can act as an E3 ubiquitin ligase (11,12).
There is also phylogenetic evidence that the N-terminal domain of RAG1 may have a separate origin, independent of the nuclease-bearing core region (13).
These considerations suggest that RAG1, and specifically the N-terminal domain, may act through protein–protein interactions in ways that may modulate or complement the nuclease activity of the core region (14–16).
What other proteins interact with this region of RAG1? To date, only the single protein Karyopherin α2 (also known as KPNA2/RCH1/SRP1α/IPOA1/QIP2) has been reported (17–19).
Here we explore the binding of the RAG1 N-terminal domain with other proteins using the yeast two-hybrid assay (20). We describe binding to two additional proteins and find that a critical interaction maps to a distinct peptide motif in the RAG1 NTD.
MATERIALS AND METHODS
A yeast two-hybrid assay was conducted using Matchmaker Two-Hybrid System 3 (Clontech, Mountain View, CA), following the manufacturer's instructions. A Mouse T-cell lymphoma cDNA library (Clontech ML4001AE) was obtained, comprised of 3 million independent clones of 1.7 kb average size, cloned in the XhoI site of the pACT vector. The vector provides a fusion to the GAL4-AD and selectable markers [GAL4(768–881) AD, LEU2, Ampr]. The library was amplified once in Escherichia coli DH5α.
The RAG1 NTD was amplified by PCR from existing mouse RAG1 templates using primers Forward-1aa and Reverse-383aa (below), and cloned in pGBKT7 [GAL4(1–147) DNA-BD, TRP1, Kanr] using the introduced NcoI and XhoI sites. (NcoI and SalI in vector).
Simultaneous transformation of the RAG1 construct and the library was performed into Saccharomyces cerevisiae AH109 cells using the lithium acetate (0.1 M LiAc pH 7.5/40% polyethylene glycol/TE) method. Nine million yeast transformants (representing a 3-fold oversampling) were screened on 500 plates of 15-cm diameter at an average of 20 000 transformants per plate. Cotransformants were selected on minimal SD agar under permissive ‘double drop out’ (DDO: -LEU/-TRP) conditions or restrictive ‘triple drop out’ (TDO: -LEU/-TRP/-HIS) conditions requiring two-hybrid interaction, as well as TDO containing 2.5 mM, 5 mM and 7.5 mM 3-AT (3-amino1,2,4 triazole) (Sigma, St. Louis, MO, USA). DNA was isolated from positive clones, sequenced using a primer within the vector and recloned in pGADT7 [GAL4(768–881) AD, LEU2, Ampr] for confirmation.
Mammalian two-hybrid assay
Selected interacting proteins were tested for interaction with RAG1 peptides in mammalian cells using the mammalian two-hybrid assay (Clontech Matchmaker). RAG1 peptides were recloned into the pM vector (Clontech #K16021-1). Restriction sites in this vector were chosen to preserve the reading frame from pGBKT7. Proteins derived from the yeast screen were cloned in pVP16 (Clontech #K16021-1) with restriction sites in the same reading frame as in the pGADT7 vector. The protein–protein interaction was measured as Luciferase activity utilizing plasmid pFR-LUC essentially as described (21).
Mammalian one-hybrid assay
The interaction of GMEB1 to RAG proteins at the 12RSS was examined using the assay of Difilippantonio et al. (22). Reporter substrates were constructed by deleting the Gal UAS from plasmid pFR-Luc, and inserting 12RSS sequences. The eight copies of 12RSS were obtained from the original plasmid pMJD112 (6). Details will be shared upon request. Plasmid GMEB1-VP16 was a gift of Stoney Simons (NIH, Bethesda MD, USA). Full-length RAG1-VP16 was derived from the core RAG1-VP16 plasmid pCJM199 (6) by substitution. RAG2 was expressed from the full-length T7 tagged plasmid used previously (23).
Luciferase assay
Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) as per the manufacturer's protocol. COS7 cells, grown to 70% confluence on 6-cm culture plates, were transfected (Fugene, Roche, Indianapolis, IN, USA) using 3 µl reagent per microgram DNA. Typically, up to 6 µg DNA was used per dish and extracted 48–72 h post-transfection in 400 µl passive lysis buffer (30 min on ice). The supernatant measuring 100 µl (following two freeze–thaw cycles and centrifugation at 10 000 r.p.m. for 5 min) was assayed in 96-well format using a LUMIstar Galaxy luminometer and lumistar galaxy software (BMG LABTECH, Durham, NC, USA).
Primers used for construction of RAG1 plasmids expressing segments of the N-terminal domain are listed below. Included restriction sites are underlined
| PCR Primer | SEQUENCE |
|---|---|
| Forward-1aa | 5′ AAGCAACCATGGCTGCCTCCTTGCCGTCTACCCTG 3′ |
| Forward-104aa | 5′ CACCCCGGATCCGCCATGGCCAGGCTTAGACAC 3′ |
| Forward-139aa | 5′ CACCCCGGATCCGCCATGGCGCTTTTCCGAAAGAAG 3′ |
| Forward-173aa | 5′ CACCCCACTGAATTCTGCCAT 3′ |
| Forward-208aa | 5′ CAAAAAAGGATCCACACACCGTCCTGTGACATC 3′ |
| Reverse-224aa | 5′ CACCAAGTCGACTTATCAACTCTTCCTCTTGAGTCCCCG 3′ |
| Reverse-250aa | 5′ CACCAAGTCGACTTATCAAGTTCTCTTGCGACG 3′ |
| Reverse-383aa | 5′ AAGCCCCTCGAGTCATTACAAAGTCTCTTTAGATTCTTTGTG 3′ |
| Forward-250aa | 5′ AAAGTTCCATGGCAACTCAGGCTAGGGTCAGCAGCAAG 3′ |
| Reverse-350aa | 5′ AAAGGGCTCGAGTCACTAAGAATTCAAGATGTTCAGAAAGGACTTCAC 3′ |
Primers used for QuickChange (Strategene, La Jolla, CA, USA) mutagenesis of specific residues within the WWL motif of RAG1
| Name | Sequence |
|---|---|
| W179A forward | 5′ TTCTGCCATGACTGTGCGAGCATCATGCACAGA 3′ |
| W179A reverse | 5′ TCTGTGCATGATGCTCGCACAGTCATGGCAGAA 3′ |
| VYF193AAA forward | 5′ AGTTCCCACAGTCAGGCCGCCGCCCCAAGGAAAGTGACC 3′ |
| VYF193AAA reverse | 5′ GGTCACTTTCCTTGGGGCGGCGGCCTGACTGTGGGAACT 3′ |
| W203A forward | 5′ AAAGTGACCGTGGAGGCGCACCCCCACACACCG 3′ |
| W203A reverse | 5′ CGGTGTGTGGGGGTGCGCCTCCACGGTCACTTT 3′ |
| WHP203AHA forward | 5′ AAAGTGACCGTGGAGGCGCACGCCCACACACACCGTCCTGT 3′ |
| WHP203AHA reverse | 5′ ACAGGACGGTGTGTGGGCGTGCGCCTCCACGGTCACTTT 3′ |
Construct 1–173 was generated by restriction enzyme digestion of existing constructs without PCR amplification using the natural EcoRI site at residue 173.
Alignments
Over 1000 RAG1 sequences were collected using the NCBI PSI-Blast service (24) after five iterations seeded with the coelacanth peptide spanning the WWL domain. These sequences were then reformatted by filtering based on the taxonomic ID. The raw data sets are presented in supplemental material. Selected sequences were aligned using the CLC Sequence Viewer software (CLC Bio USA, Cambridge, MA, USA) as implemented for the Macintosh platform.
RESULTS
The N-terminal domain (NTD) of RAG1 comprises more than one-third of the protein (Figure 1, top). Known features within the NTD are the RING motif (residues 285–328) and clusters of basic residues. The core region contains the acidic residues essential for the nuclease activity (25,26) and known DNA-binding behavior. In an effort to explore whether the NTD alone was capable of interacting with other proteins, we performed a yeast two-hybrid screen (20). The RAG1 NTD was fused to the yeast GAL4 DNA-binding domain (DNA-BD) (Figure 1, bottom) and assayed for interaction with a library of proteins each fused to the GAL4 transcriptional activation domain (AD). We screened a library derived from a mouse T-cell lymphoma, at a level (>9 million transfected cells) calculated to exceed the representation of proteins by several fold. The combination of plasmids and yeast strains employed allowed screening and selection based on several criteria. Selection for the presence of the DNA-BD plasmid and an AD-expressing plasmid was provided by two separate nutritional markers (leucine and tryptophan). The interaction of the proteins activated a third nutritional selection, for histidine through the GAL1 UAS. The stringency of the histidine selection was modulated by the level of an additional agent, 3-AT, (3-Amino-1,2,4-triazole), which is a competitive inhibitor of the His3 protein. The concentration of this inhibitor determines a minimal level of His3 expression needed for survival. In addition, two-hybrid interaction also activates three other reporters; ADE2, driven by the GAL2 UAS promoter, β-galactosidase and MEL1 driven by the MEL1 promoter, the activity of which can be used to confirm the protein–protein interaction (data not shown).
Eight isolates emerged from the screen, representing three target proteins. Characterization of the initial plasmids is portrayed (in part) in Figure 2A. Each of the candidate target proteins is coexpressed with the empty DNA-BD vector (sectors labeled 177) or with the RAG1-NTD fusion (sectors labeled 179). All grow, as expected, on the control double-drop out plates lacking leucine and tryptophan (panel A, plate 4) but only candidates that are capable of interaction with the NTD survive on plates additionally lacking histidine, in the presence of 7.5 mM 3-AT (panel A, plates 1–3). One selected target is the protein Karyopherin α2 (also known as KPNA2/RCH1/SRP1α/IPOA1/QIP2), previously recognized by others using two-hybrid selection with full-length RAG1 as bait (17–19). This interaction served as a control in the analysis that follows. The remaining two target proteins have not been reported. These are splicing factor 3A2 (SF3A2) and glucocorticoid modulatory element binding protein 1 (GMEB1). The selected plasmids represented (almost) the full-length GMEB1 protein. In contrast, the several independent isolates of SF3A2 each were composed of the identical C-terminal 145 residues of that protein (Figure 2C). It is possible that this particular clone was over-represented in the library. We note that the residues selected are extraordinarily proline rich, being composed largely of imperfect repeats of the heptad PPAPGVH. The full SF3A2 protein is 475 amino acids long, of which the C-terminal 269 is 43% proline and is largely composed of this same proline-rich repeat (see supplement 1 in Supplementary Data). Potential physiologic relevance of these interactions will be discussed later.
Figure 2.
(A) Yeast two-hybrid assay. Positive interaction between the RAG1 N-terminal domain and any member of the library of fusion proteins allows growth under histidine-restrictive conditions (plates 1–3). In contrast, all cells grow on control double drop-out plates (plate 4). (B) Three proteins were isolated, including the previously known Karyopherin α2. (C) The isolated fragment of SF3A2 is shown. It is composed largely of imperfect repeats of a proline-rich heptad PPAPGVH.
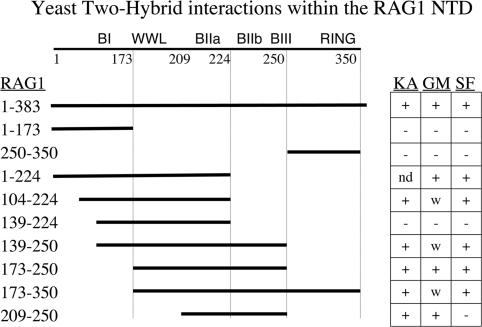
We employed variations of the same two-hybrid assay to help map the interactions shown above to smaller regions of RAG1. Shorter peptides, designed to capture or exclude known features in the RAG1-NTD, were cloned in the same DNA-binding domain vector as used above for the full NTD. Each of these constructs was tested in the yeast two-hybrid assay with the three binding partners. The results are presented in Figure 3. The basic regions are labeled using the nomenclature of McMahan et al. (16) as follows: BI 141-146, BIIa 218-224, BIIb 233-236, BIII 243-254. Dividing the NTD in half yielded no growth with any target protein when tested with residues 1–173, and gave growth with all targets using the segment 173–350. We note that the GMEB1 interaction appears to be weaker in the assay of this last construct as cell growth was observed only when the 3-AT concentration was reduced to 5 mM. Similar growth, marked W in the figure, was observed for GMEB1 with other constructs as well. It is worth remembering, however, that the qualitative measure of growth employed here does not translate reliably into a quantitative measure of protein interaction. Other factors, such as the relative protein stability of the various constructs, or structural effects that differ between constructs, also contribute to the behavior of the system.
Figure 3.
Mapping interactions within the NTD by yeast two-hybrid. Segments of RAG1 (solid lines) fused to GAL4 DNA-BD were tested with the three binding targets. Ka = KPNA2, GM = GMEB1, SF = SF3A2. (+) represents growth on histidine drop-out plates in the presence of 7.5 mM 3-AT. Weak growth (W) represents growth with 5 mM 3-AT. (–) represents no growth. ND is not determined.
The RING motif was not required for the interaction with these targets. Truncating the NTD at residue 250 (middle of BIII) was compatible with interaction with all three targets, and the small fragment from 173 to 250 was positive in each case. Further deletion of the basic residues between 224 and 250 (BIIb and BIII) interfered with binding (construct containing RAG1 139–224), but this could be restored by providing the basic region BI (construct 104–224).
In yeast, the peptide 209–250 was sufficient to interact with KPNA2 and GMEB1 but not with SF3A2 and suggests that, for SF3A2 at least, a structural contribution is provided by the RAG1 peptide within residues 173–209. We will return to this observation following further confirmation of the binding behavior of these target proteins using other assays.
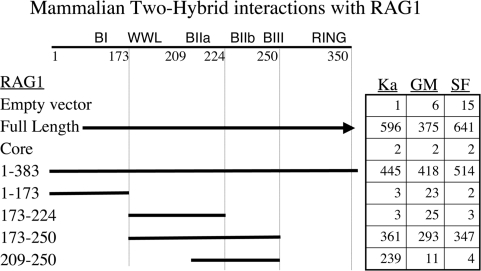
Translating the two-hybrid assay from yeast into mammalian cells seemed an important step in confirming the central observations. Two additional advantages are the more physiologic environment for RAG1 function and better quantitation of activity afforded by the luciferase-based assay. We employed a system that is conceptually similar to that used above, but uses the herpes viral VP16 transactivation domain rather than the GAL4-AD. Plasmids expressing the RAG1 fragments fused to the GAL4 DNA-BD, and the target proteins fused to VP16, were cotransfected into COS7 cells, along with the pFRLuc reporter. Extracts of these cells were assayed for luciferase activity and the results tabulated in Figure 4. For all three proteins, full-length RAG1 and the NTD gave parallel positive evidence of interaction, while the core region of RAG1 exhibited background levels of activity. The constructs further truncated within the NTD were largely consistent with the equivalent assays performed in yeast. Most important, the short segment of RAG1 173–250 was again positive while truncation into that segment from either side had striking consequences. Removal of the basic region BIIb and BIII (plasmid 173–224) prevented interaction with all three target proteins. Truncation from the other direction, (plasmid 209–250) still bound KPNA2 but lost interaction with GMEB1 and SF3A2. We note that the behavior of GMEB1 in this context differs from the parallel experiment in yeast in Figure 3. While we cannot fully account for the difference, we feel the mammalian two-hybrid result better reflects RAG1 behavior in its appropriate context. These data therefore also indicate that the region between 173 and 209 contains sequence that is essential for the protein binding assayed here.
Figure 4.
Mammalian two-hybrid results. The table presents luciferase units (in thousands). DNA-BD RAG1 fusions interact with the target proteins (as VP16 fusions) to activate a luciferase gene. Nomenclature as in Figure 3.
The luciferase assay was also used in a second context to measure GMEB1 interaction with the RAG protein complex. We adapted the one-hybrid assay (22) that had been previously used to detect RAG1 and RAG2 binding to the RSS. The authors of the original study used VP16 transactivator fusions to RAG1 and RAG2 to address the occupancy of the RAG protein complex on DNA. Here we extend that assay to demonstrate that GMEB1 fused to VP16 can be recruited to the RSS through interaction with the RAG1-NTD. Figure 5A shows the original scheme. Three reporter plasmids encode luciferase driven by a minimal promoter region and differ by the number of 12RSS elements in the intervening spacer (zero, three or eight RSS repeats). The previous study (22) showed that binding of RAG proteins to the RSS depended on both RAG1 and RAG2 coexpression. Panel 5B presents a variation on this assay, where only GMEB1 carries the VP16 transactivator. GMEB1 binding to RAG1 localized to the RSS activates transcription of the luciferase reporter. The results are tabulated in Figure 5C. We see, using full-length or core RAG1-VP16, that the RAG complex binds to the reporter plasmids, with luciferase activity increasing with the number of RSS copies (lines 1 and 2). GMEB1-VP16, in the absence of the RAG protein complex, does not transactivate the luciferase (line 3). GMEB-VP16 strongly transactivates the reporter when full-length RAG1 protein (absent VP-16), coupled with RAG2, binds the RSS elements (line 4). In contrast, core RAG1 plus RAG2 do not recruit GMEB1-VP16 (line 5). These data extend the results of the mammalian two-hybrid by showing that GMEB1 is capable of binding to RAG1 in the context of the RAG1/RAG2 complex situated on an RSS. One is struck by the magnitude by which the GMEB-VP16 fusion drives transcription compared to the relatively weak signal elicited by RAG1-VP16. Alternatives include the possibility that the RAG1-VP16 fusion is intrinsically a poor transcriptional activator for structural reasons, while the GMEB1-VP16 bound to RAG1 is better able to activate the promoter. It is also possible that the complex including GMEB1 binds DNA better than the RAG complex alone. These questions are under further study.
Figure 5.
One-hybrid assay adapted to show GMEB binding to RAG proteins at the RSS. (A) The original one-hybrid assay (ref. 22). The RAG protein complex, fused to VP16, activates luciferase when bound to the 12RSS. (B) Modified one-hybrid assay is portrayed in which VP-16 is only present fused to GMEB1. Activation of luciferase requires protein–protein interaction between GMEB1 and the RAG complex on the 12RSS. (C) Luciferase activity (machine units) obtained from the listed combinations of proteins transfected into COS7 cells.
We return now to the observation (Figures 2 and 3), that deletion of the RAG1 peptide residues 173–209 rendered it unable to bind the SF3A2 fragment in yeast cells, and both GMEB1 and the SF3A2 fragment in mammalian cells. We had not chosen the endpoints for that deletion arbitrarily. Owing to the striking proline-rich character of the SF3A2 fragment, we examined the sequence of the RAG1-NTD for motifs that would correspond to known proline-binding domains. Such a correspondence can be found with the domain called WW, for its two conserved tryptophans (W).
Figure 6A shows an alignment between the RAG1 NTD and one well-studied WW domain, the fourth WW domain in the mouse protein Itch (Genebank NM_008395). The larger type emphasizes the important conserved features. These include the two tryptophan residues (W) as well as an internal hydrophobic cluster often containing a tyrosine (Y) and commonly, a proline two residues C-terminal to the second tryptophan. The spacing of the aligned residues within the RAG1 sequence does not correspond precisely to the current definition so we suggest calling the motif in RAG1 WW-like or WWL for now. This sequence is highly conserved across all mammalian RAG1 sequences, as summarized in Figure 6B (an alignment to 343 mammalian sequences is provided in supplement 2 of the Supplementary Data). The consensus is set at 90% stringency and shows complete conservation of the defining elements (although the tyrosine residue is replaced by cystine in the dog). The conservation is less absolute across the full vertebrate phylogeny. Figure 6C shows representatives of the major vertebrate radiations. Additional alignments are also presented in Supplementary Data.
Figure 6.
Alignments over the WW-L motif. (A) The mouse RAG1 WWL is aligned to Itch, an E3 ligase (genebank NM_008395, fourth WW domain). Residues that contribute to the definition of the WW domain are highlighted in larger type. (B) Alignment of a wide selection of representative mammals over the WWL region. (C) Alignment of selected RAG1 sequences from all vertebrates. Boxed residues in (B) and (C) correspond to the defining features in (A). Positions with a consensus that exceeds 90% are listed. Dots represent identities to the first sequence; dashes represent gaps. Genebank accession numbers for each sequence are listed on the right.
All mammals, amphibians, reptiles, turtles and crocodilians preserve perfectly the two tryptophans and the VYF hydrophobic core. The proline (mouse 206, boxed in Figure 6B) is universally conserved among mammals and crocodilians, but in reptiles, amphibians and turtles, is often replaced by a serine.
Lizards show somewhat less conservation. Among lizards, the blast alignment provided hits for 366 organisms, of which 14% replaced the first tryptophan with arginine. One organism replaced the second tryptophan. The hydrophobic core was frequently mutated with conservative replacements, and three organisms failed to conserve the proline (mouse 206).
Similarly with birds, we find high conservation of all residues, with specific replacement of the first tryptophan by arginine in 2% (15/741), replacement of the VYF with VCF in 4% (33/741) of the species (as was seen with the dog among mammals) and with six other changes in this cluster. The second tryptophan is perfectly conserved, and the proline (mouse 206) largely conserved, with roughly 4% (28/741) arginines and two cysteines substituting.
Fish show a distinct difference, among catfish and boney fish, most importantly owing to the deletion of residues between the first tryptophan and the hydrophobic core (VYF in mouse). While the final phenylalanine (F) is conserved, the tyrosine (Y) is lost, and it is not clear whether this suffices for the hydrophobic core. The remaining positions remain largely conserved. We have no data addressing binding activity. It is possible that function has been lost in these fish, although retained in the Bull Shark.
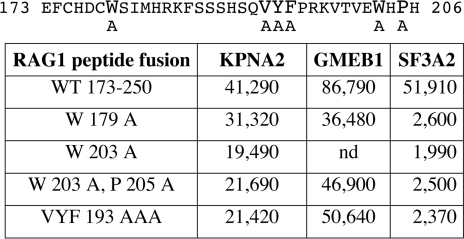
Given the strong conservation, especially across mammals, we chose to test whether mutation could confirm a functional role of these residues in binding interactions. We prepared several mutated versions of the minimal RAG1 construct used in Figure 4, spanning the region 173–250. We created single alanine substitutions at each of the two tryptophan residues (W179A or W203A), the double mutant containing W203A and P205A, and the clustered replacement of three alanines for the three hydrophobic residues (VYF193AAA). The constructs are presented in Figure 7, along with the luciferase assays reflecting the binding to each of the three interacting proteins using the mammalian two-hybrid system. KPNA2 and GMEB1 each show only a modest reduction in binding (∼2-fold), which may be attributed to subtle effects on protein structure or stability within the RAG1 peptide. In striking contrast, binding to the SF3A2 peptide shows a 20-fold reduction in binding to each of the mutant RAG1 proteins. These data are in complete accord with the prediction that a WW-like domain is binding to a proline-containing peptide. They suggest that the WW-like domain plays a necessary role in stabilizing the interaction with SF3A2 but is not as important for KPNA2 and GMEB1. For these latter proteins, the interaction with the basic region appears to be dominant.
Figure 7.
The effect of site specific mutations within the WWL motif using the mammalian two-hybrid assay. (Top) Mouse RAG1 from 173-206, spanning the WWL motif. Residues in larger font are variously mutated. (Bottom) Mammalian two-hybrid assay revealing interaction between the RAG1 peptide and the three other proteins. Data are raw luciferase units. The interaction with SF3A2 is very sensitive to all mutations.
DISCUSSION
The RAG proteins remain the only identified cell-specific factors required for V(D)J recombination. Biochemical studies indicate that the RAG proteins alone are sufficient to recognize the RSS in vitro, and perform the initial DNA cleavages. However, the recombination reaction requires many additional steps, and the extent to which the RAG proteins participate before and following cleavage remains largely speculative. On natural chromatin, the initial binding is constrained by accessibility and there is evidence of protein–protein interactions with nucleosomes (27–29). We believe that additional protein–protein interactions remain to be described. These are likely to shape the distribution of recombination events, the consequences of recombination, and the state of the cell during the recombination reaction. One avenue by which RAG1 could mediate effects beyond its role as a nuclease is through an ubiquitin ligase activity mapped to the N-terminal domain (11,12).
This report reveals additional protein–protein interactions by means of a yeast two-hybrid screen using the RAG1 N-terminal domain as one binding partner. Previous yeast two-hybrid screens, by others, used the entire RAG1 protein. Three groups reported interaction with the protein Karyopherin α2, also called Srp1α and Rch1 (17–19). In our hands, this protein binds only to the NTD, but the core protein still localizes to the nucleus. Our screen reiterated the binding to KPNA2, and revealed binding to two others; a C-terminal segment of Splicing Factor 3A subunit 2 (SF3A2, also known as SF3a66) and the GMEB1 also called PIF p96 (30,31).
Only a brief description of the activities of these proteins will be provided here, until we have more evidence of function in V(D)J recombination. KPNA2 is recognized as a nuclear transport protein. As a complex with β-karyopherin, it has a second role as an activator of the proteasome (32,33).
Although the interaction with an RNA splicing protein seems far afield for RAG1, a previous report found that RAG1, when overexpressed, localized to the nucleolus (19). This was interpreted as reflecting a direct RNA-binding behavior, but it may actually reflect binding to this RNA-processing protein. Two other binding interactions with SF3A2 appear to be independent of a role in splicing. One group finds this protein tethers to the microtubules in the nucleus (34). A second finds that a developmental transcription factor (nuclear FGF-2) binds this factor in neurons (35).
GMEB1 is a transcription factor initially described as a cofactor of genes controlled by the glucocorticoid receptor. Subsequently, it has been found to contribute to the regulation of many genes (36), and bind the CREB-binding protein CBP (30). GMEB1 binding is regulated by DNA methylation. GMEB1 is already known to bind a RING-containing ubiquitin E3 ligase and play a regulatory role in muscle (37). GMEB1 also interacts with the SUMO pathway. One important function seems to be recruiting Ubc9, the SUMO-specific E2 enzyme, to transcription complexes (38). The GMEB1 heterodimer with GMEB2 is associated with a DNA nicking activity. It is a host factor, which binds and activates a mouse parvovirus viral DNA nickase during viral replication (31). GMEB1 also blocks pro-apoptosis signals induced by a variety of stresses (39).
We mapped the binding interaction of each of the three proteins onto RAG1 and defined small peptides that were able to mediate this interaction using the two-hybrid assay. Two different transactivation domains were used between the yeast and mammalian assays. The segment of RAG1 including residues 173–250 was sufficient to recapitulate the interactions with all three targets analyzed here (Figures 3 and 4). The luciferase assay yielded roughly 50% of the activity using this short peptide, compared to the full NTD, indicating that we have isolated a major component to this interaction. Better quantitation of the binding will require in vitro analysis, not subject to confounding effects of differing protein expression or stability, or structural constraints contributed by the fusion partners.
The segment 173–250 contains clusters of basic residues previously designated as the BIIa, BIIb, and (most of) BIII. It also contains the motif we are designating WWL (for WW-like). The basic clusters, by themselves, appear sufficient for interaction with KPNA2 (Figures 3 and 4; RAG1 209–250). In the mammalian two-hybrid assay, the other two proteins appear to require both an interaction within the WWL motif and a basic region. Truncation from the C-terminal side to residue 224, removing the BIIb and BIII clusters, eliminated binding to all three interacting proteins in both yeast and mammalian assays (RAG1 139–224 in Figure 3, and RAG1 173–224 in Figure 4). Curiously, adding additional sequence to the N-terminal side restored binding to all three proteins, as seen in Figure 3 RAG1 104–224. Two explanations are consistent with these data. Either the segment from 104 to 139 provides an alternate binding motif, or the change in spacing allows the basic region BI to function better as a substitute than when it was positioned at the junction with the GAL4-BD fusion partner. In summary, a small cluster of basic residues alone appears to be sufficient for interaction with KPNA2. In contrast, the proteins GMEB1 and SF3A2 bound best to constructs that provided the WWL motif accompanied by a basic cluster, which could be located on either side.
The motif we consider WW-like shares several properties with canonical WW domains. As described by the Conserved Domain Database (40) http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=119576 the ‘two conserved tryptophans’ (WW) domain is composed of around 40 residues and functions as an interaction module in a diverse set of signaling proteins. It binds specific proline-rich sequences at low affinities. We note that several ubiquitin ligases bind their substrates through the action or one or several associated WW domains. To date, these are all HECT type E3 ligases and include the Smurf proteins, the NEDD4 family, Itch and Suppressor of Deltex.
Canonical WW domains share a protein structure in which the first tryptophan contributes to the folding of the domain, while the internal hydrophobic core (frequently a tyrosine) and the second tryptophan form hydrophobic stacks with a proline of the binding ligand (41). None of these positions needs to be absolutely conserved but the spacing between these features does not match the spacing observed in RAG1. Specifically, the canonical motif [calculated from the 100 most diverse members of the conserved domain database for motif cd00201, (40)] is spaced W (X9–11) Y (X9–14) W XX P. The RAG1 (mammalian alignment in Figure 6B) sequence is W (X14) Y (X8) W X P. Our mutational data using the mammalian two-hybrid assay (Figure 7) support our proposal of a structure that mimics the canonical WW domain. Mutation of the defining features of the WW motif, single-point mutations of the tryptophans, or of the hydrophobic cluster including the tyrosine, reduced binding activity to the SP3A2 proline-rich ligand by over 20-fold as determined by the mammalian two-hybrid assay.
Also noteworthy are the two potential metal-binding motifs CxxC that bracket the WWL (conserved in Figure 6B and C). Were these to cooperatively bind a metal ion, they would structurally constrain and approximate the ends of the WWL motif. We have not yet examined their function in isolation.
Examination of the phylogenetic alignment (Figure 6 and Supplementary Data) reinforces the significance of the WWL motif. The relatively infrequent mutations at the signature residues are highly constrained in identity.
We conclude by noting that, although we detected new binding interactions with the RAG1 NTD, we did not detect interaction with the RING motif. Such an interaction is expected between proteins behaving as an ubiquitin E3 and their necessary partners, the E2 proteins. It is the case, however, that two-hybrid assays have not been successful in detecting other such E3–E2 interactions. This may reflect the weak signal that is likely to be generated by a relatively transient interaction. The two-hybrid approach is most sensitive to stable binding. The new interacting partners we did detect reveal a previously uncharacterized motif within RAG1 that seems to behave similarly to the WW family. We suspect that protein–protein interactions with RAG1 could influence DNA binding and signaling as the recombination reaction progresses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health grant (AI072055). Funding for open access charge: National Institutes of Health (AI072055).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for materials and discussion provided by the laboratories of Dr David Schatz and Dr Stoney Simons.
REFERENCES
- 1.Gellert M. Molecular analysis of V(D)J recombination. Ann. Rev. Genet. 1992;22:425–446. doi: 10.1146/annurev.ge.26.120192.002233. [DOI] [PubMed] [Google Scholar]
- 2.Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 3.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 4.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 5.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 6.Sadofsky MJ. The RAG proteins in V(D)J recombination: more than just a nuclease. Nucleic Acids Res. 2001;29:1399–1409. doi: 10.1093/nar/29.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl Acad. Sci. USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirch SA, Sudarsanam P, Oettinger MA. Regions of RAG1 protein critical for V(D)J recombination. Eur. J. Immunol. 1996;26:886–891. doi: 10.1002/eji.1830260425. [DOI] [PubMed] [Google Scholar]
- 10.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yurchenko V, Xue Z, Sadofsky M. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 2003;17:581–585. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones JM, Gellert M. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc. Natl Acad. Sci. USA. 2003;100:15446–15451. doi: 10.1073/pnas.2637012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudley DD, Sekiguchi J, Zhu C, Sadofsky MJ, Whitlow S, DeVido J, Monroe RJ, Bassing CH, Alt FW. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J. Exp. Med. 2003;198:1439–1450. doi: 10.1084/jem.20030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roman CA, Cherry SR, Baltimore D. Complementation of V(D)J recombination deficiency in RAG-1(–/–) B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity. 1997;7:13–24. doi: 10.1016/s1074-7613(00)80506-3. [DOI] [PubMed] [Google Scholar]
- 16.McMahan CJ, Difilippantonio MJ, Rao N, Spanopoulou E, Schatz DG. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol. Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuomo CA, Kirch SA, Gyuris J, Brent R, Oettinger MA. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl Acad. Sci. USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortes P, Ye ZS, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl Acad. Sci. USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spanopoulou E, Cortes P, Shih C, Huang CM, Silver DP, Svec P, Baltimore D. Localization, interaction, and RNA-binding properties of the V(D)J recombination-activating proteins Rag1 and Rag2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 20.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, He Y, Simons SS., Jr. Structure/activity relationships for GMEB-2: the second member of the glucocorticoid modulatory element-binding complex. Biochemistry. 2004;43:245–255. doi: 10.1021/bi035311b. [DOI] [PubMed] [Google Scholar]
- 22.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 23.Yurchenko V, Xue Z, Sadofsky MJ. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol. Cell Biol. 2006;26:1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl Acad. Sci. USA. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaul S, Blackford JA, Jr, Chen J, Ogryzko VV, Simons SS., Jr. Properties of the glucocorticoid modulatory element binding proteins GMEB-1 and -2: potential new modifiers of glucocorticoid receptor transactivation and members of the family of KDWK proteins. Mol. Endocrinol. 2000;14:1010–1027. doi: 10.1210/mend.14.7.0494. [DOI] [PubMed] [Google Scholar]
- 31.Christensen J, Cotmore SF, Tattersall P. Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur. J. Virol. 2001;75:7009–7017. doi: 10.1128/JVI.75.15.7009-7017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glickman MH, Raveh D. Proteasome plasticity. FEBS Lett. 2005;579:3214–3223. doi: 10.1016/j.febslet.2005.04.048. [DOI] [PubMed] [Google Scholar]
- 33.Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J. Struct. Biol. 2004;146:425–430. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Takenaka K, Nakagawa H, Miyamoto S, Miki H. The pre-mRNA-splicing factor SF3a66 functions as a microtubule-binding and -bundling protein. Biochem. J. 2004;382:223–230. doi: 10.1042/BJ20040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gringel S, van Bergeijk J, Haastert K, Grothe C, Claus P. Nuclear fibroblast growth factor-2 interacts specifically with splicing factor SF3a66. Biol. Chem. 2004;385:1203–1208. doi: 10.1515/BC.2004.156. [DOI] [PubMed] [Google Scholar]
- 36.Burnett E, Christensen J, Tattersall P. A consensus DNA recognition motif for two KDWK transcription factors identifies flexible-length, CpG-methylation sensitive cognate binding sites in the majority of human promoters. J. Mol. Biol. 2001;314:1029–1039. doi: 10.1006/jmbi.2000.5198. [DOI] [PubMed] [Google Scholar]
- 37.McElhinny AS, Kakinuma K, Sorimachi H, Labeit S, Gregorio CC. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J. Cell Biol. 2002;157:125–136. doi: 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaul S, Blackford JA, Jr, Cho S, Simons SS., Jr. Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J. Biol. Chem. 2002;277:12541–12549. doi: 10.1074/jbc.M112330200. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa T, Tsuruma K, Uehara T, Nomura Y. GMEB1, a novel endogenous caspase inhibitor, prevents hypoxia- and oxidative stress-induced neuronal apoptosis. Neurosci Lett. 2008;438:34–37. doi: 10.1016/j.neulet.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudol M. In: Modular Protein Domains. Cesareni G, Gimona M, Sudol M, Yaffe M, editors. Weinheim, FRG: Wiley-VCH Verlag GmbH; 2005. pp. 59–72. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.