Abstract
Apoptosis is essential for the maintenance of inherited genomic integrity. During DNA damage-induced apoptosis, mechanisms of cell survival, such as DNA repair are inactivated to allow cell death to proceed. Here, we describe a role for the mammalian DNA repair enzyme Exonuclease 1 (Exo1) in DNA damage-induced apoptosis. Depletion of Exo1 in human fibroblasts, or mouse embryonic fibroblasts led to a delay in DNA damage-induced apoptosis. Furthermore, we show that Exo1 acts upstream of caspase-3, DNA fragmentation and cytochrome c release. In addition, induction of apoptosis with DNA-damaging agents led to cleavage of both isoforms of Exo1. The cleavage of Exo1 was mapped to Asp514, and shown to be mediated by caspase-3. Expression of a caspase-3 cleavage site mutant form of Exo1, Asp514Ala, prevented formation of the previously observed fragment without any affect on the onset of apoptosis. We conclude that Exo1 has a role in the timely induction of apoptosis and that it is subsequently cleaved and degraded during apoptosis, potentially inhibiting DNA damage repair.
INTRODUCTION
DNA is constantly damaged by endogenous factors (e.g. free radicals generated during normal cellular metabolism) and exogenous factors [e.g. ultraviolet (UV) light]. In order for genomic stability to be maintained, it is essential that this damage is repaired. The repair of DNA damage involves a highly coordinated series of events: first, the cell must signal to halt cell cycle progression at precise cell cycle checkpoints, following this, DNA damage-specific repair pathways are activated (1). These pathways lead to repair of the damaged DNA and their composition is dependant on the type of damage. Following repair, cell cycle checkpoints are released and the cell cycle can progress normally. However large amounts of DNA damage can trigger another pathway called apoptosis, this initiates signals which ultimately result in controlled cell death. Apoptosis is essential for the removal of damaged cells, which would have the potential to carry deleterious mutations onto daughter cells. If such cells were allowed to continue dividing in an organism, this could potentially lead to tumour development (1).
Caspases are the major proteases involved in apoptosis. This family of proteins contribute to cellular disintegration via targeted cleavage of a collection of proteins involved in many processes within the cell, including DNA repair and checkpoint activation (2). Of the proteins in the caspase family, caspase-3, caspase-6 and caspase-7 have been shown to be the major effector caspases in apoptosis (3). In order to completely understand the role of caspases in apoptosis, it is essential to identify their downstream targets. The cleavage of proteins by caspases is not a random event and appears to target proteins involved in maintenance of cellular integrity in a highly specific manner. Caspases do not completely degrade their targets, but rather cleave proteins at a few specific sites. In general, caspase substrates become inactivated upon cleavage, however, a subset become activated (4) and contribute to apoptosis. A comprehensive list of caspase substrates can be found on the CASBAH web site (http://www.casbah.ie). The major apoptotic nuclease Caspase-activated DNase (CAD) is cleaved by caspase-3 during apoptosis, this results in the translocation of CAD into the nucleus and induction of CAD-mediated DNA fragmentation (5,6). Two major kinases involved in DNA damage signalling events; Ataxia Telangiectasia mutated (ATM) (7) and the catalytic subunit of DNA-dependent protein kinase (DNA-PK) (8) are also cleaved by caspase-3 during apoptosis. Cleavage of these two proteins is suggested to prevent DNA repair during apoptosis. Interestingly, ATM is also required to induce apoptosis in response to some DNA-damaging agents (9).
The present study provides support for a role for the DNA damage repair nuclease Exonuclease 1 (Exo1) in the induction of apoptosis. Exo1 was first identified as a nuclease required for meiosis in fission yeast (10). Exo1 belongs to the RAD2 family of nucleases and possesses 5′-3′ nuclease activity and 5′-flap endonuclease activity (11,12). There are two isoforms of Exo1 (a and b), which result from alternate splicing. The isoforms differ at the C-terminus, with Exo1b having an additional 48 amino acids. Several proteins involved in replication and DNA repair including PCNA and mismatch repair (MMR) proteins interact with Exo1 (13). Exo1 has a role in several DNA repair pathways including MMR, post-replication repair, meiotic and mitotic recombination (14–16). Many DNA repair proteins have been implicated in tumourigenesis, for example mutations in MLH1, an essential component of MMR are linked to colorectal cancer (17). The involvement of Exo1 in DNA repair pathways including MMR suggests it may also be a target for mutation in tumourigenesis. Consistent with this, Exo1 deficient mice display a cancer-prone phenotype, including increased susceptibility to lymphoma development (18). In addition, germ-line variants of Exo1, which affect nuclease function and MMR protein interactions have been detected in patients with atypical human non-polyposis colon cancer and other forms of colorectal cancer (19,20).
In this study, we show that DNA damage-induced apoptosis is defective in cells depleted of Exo1, suggesting that Exo1 is required for the timely induction of apoptosis. In addition, we show that both Exo1a and Exo1b are cleaved during apoptosis induced by ultra-violet C (UVC) and etoposide (ETOP). Caspase-3 is shown to be the protease responsible for Exo1b cleavage at a specific site. We conclude that Exo1 plays a significant role in the induction of apoptosis and that the lack of apoptosis observed in Exo1-deficient cells may have significant implications for chemotherapy in cancer patients.
MATERIALS AND METHODS
Reagents, antibodies and cell lines
All cell lines were grown in DMEM supplemented with 10% FCS. Antibodies used were as follows: mouse anti-Flag M2 antibody (Sigma); mouse anti-PARP1 (Zymed); rabbit anti-caspase-3 (Biosource); rabbit anti-γ-tubulin and mouse anti-β-actin (Sigma); rabbit anti-GFP (Molecular Probes); rabbit anti-Chk1 S317 and rabbit anti-p53 S15 (Cell Signalling); goat anti-ATR and goat anti-Exo1 (Santa Cruz). The recombinant caspase-3 and caspase inhibitor, ZVAD-FMK were supplied by Biosource. Exo1-deficient and wild-type mouse embryonic fibroblasts (MEFs) have been described previously (21).
Cloning, site-directed mutagenesis and expression of Exo1 constructs
A full length Exo1b clone was purchased from Origene. The full length Exo1b, and fragment of Exo1 encoding amino acids 1–514 were cloned into the ECOR-V and BamHI sites of the Flag-2 plasmid (Fermentas). Full length Exo1b was cloned into the SalI and BamHI sites of pEGFP-C1 (Clontech). Flag-Exo1b was mutated via site-directed mutagenesis to create Flag-Exo1a, which was 46 amino acids shorter. Mutation of Asp514 and Asp78 to Ala was carried out by site-directed mutagenesis. Expression constructs were transfected into cells using Lipofectamine 2000™ (Invitrogen) as per manufacturers instructions, samples were assayed 24 h after transfection.
siRNA
Stealth™ Exo1 (UAGUGUUUCAGGAUCAACAUCAUCU) and control siRNA was purchased from Invitrogen. The siRNA was transfected into Hela cells using Lipofectamine 2000™ (Invitrogen) as per manufacturers instructions and samples were analysed 48 h after transfection.
Immunoblotting
The media was removed from Hela or MEF cells cultured in 10 cm tissue culture plates and replaced with 2 ml of PBS. Cells were then exposed to 40 J/m2 UVC (BioRad Stratalinker), 100 μM etoposide, 10 μM actinomycin D and incubated for the indicated times. Following incubation cells were scraped from tissue culture plates and washed once in PBS. Cells were lysed (lysis buffer: 20 mM Hepes pH 8, 150 mM KCl, 5% glycerol, 10 mM MgCl2, 0.5 mM EDTA, 0.02% NP-40, before use buffer was supplemented with NaF, NaVO4, PMSF and protease inhibitors) and sonicated. Lysates were cleared by centrifugation and protein concentrations were estimated using the standard Bradford assay (Bradford reagent supplied by Bio-Rad). Typically 50 μg of protein lysate was separated on a 4–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gel (Invitrogen) and immunoblotted with the indicated antibodies.
Immunoprecipitation
Cells were treated and lysed as for immunoblotting. Following cell lysis, lysates were diluted in 600 μl of lysis buffer and pre-cleared with 100 μl sepharose. The lysate was transferred to a new tube, to which 20 μl (total volume) of anti-FlagM2 beads (Sigma) were added. Lysates were incubated with beads for at least 2 h. Beads were then washed 3–4 times with lysis buffer. Loading buffer containing 10% β-mercaptoethanol was then added to bead pellets and samples were boiled for 5 min. Samples were analysed via immunoblotting (as above).
Cytochrome c staining (fluorescence activated cell sorting and immunofluorescence)
MEFs were grown at a density of 1 × 106, treated with 40 J/m2 UVC and incubated for 14 h. Cytochrome c release was measured using an InnoCyte™ Flow Cytometric Cytochrome c Release Kit (Calbiochem), according to the manufacturers instructions.
Annexin V assay
Hela or MEF cells were treated with UVC, 1 μM staurosporine or 10 μM actinomycin D as indicated to induce apoptosis. Following incubation, all cells (including floating ones) were trypsinized, washed once in PBS and resuspended in 100 μl 1X annexin V binding buffer (10 mM HEPES, 140 mM NaCl and 2.5 mM CaCl2 pH 7.4). 5 μl of anti-Annexin V antibody (Molecular Probes) and 1 μl propidium iodide (1 mg/ml) were added to cells and incubated for 20 min. Following incubation 400 μl of 1X Annexin V binding buffer was added and cells were analysed by fluorescence activated cell sorting (FACS). Cells positive for Annexin V were scored as apoptotic.
In vitro caspase assay
Hela cells were transfected with the Flag-Exo1b wild-type or Flag-Exo1b D514A constructs. Cell pellets were taken and lysed (as for immunoblotting) 24 h after transfection. The 100 μg of protein lysate was added to 20 μl of caspase activity buffer (6 mM Tris–Cl pH7.5, 1.2 mM CaCl2, 5 mM DTT, 1.5 mM MgCl2 and 1 mM KCl) containing three units of recombinant caspase-3 (cat # PH20‱014, Biosource). Where indicated 20 μM ZVAD-FMK was added. Samples were incubated for 1 h at 37°C. Following incubation samples were boiled in 10 μl SDS loading dye (Invitrogen). Samples were immunoblotted as described above.
Immunofluorescence
Hela cells were seeded onto coverslips the day before siRNA transfection. Following siRNA transfection, cells were allowed to grow for 48 h before treatment with 40 J/m2 UVC. After UVC treatment, cells were treated with an extraction buffer (22) for 10 min before fixation in 4% paraformaldehyde (PFA). Cells were permeabilized with 0.2% Triton-X for 5 min and blocked in 3% bovine serum albumin (BSA) for 30 min. Cells were incubated with monoclonal RPA antibodies (Neomarkers) and alexia-conjugated secondary antibodies for 1 h each at room temperature. Cells were stained with DAPI before mounting onto slides. Cells containing over 20 foci were scored as positive for RPA foci.
Terminal deoxynucleotidyl transferase dUTP nick end labeling assay
Hela cells were grown on coverslips and transfected with the indicated siRNA or plasmids. Cells were treated with 40 J/m2 UVC to induce apoptosis after 24 (plasmids) or 48 h (siRNA) transfection. At the indicated time-point, cells were fixed in 4% paraformaldehyde. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining was carried out using a Click-iT TUNEL Alexa Fluor Imaging Assay (Invitrogen) as per the manufacturers instructions. In each condition, 200 cells were scored for TUNEL staining.
RESULTS
Exo1 depletion confers resistance to apoptosis
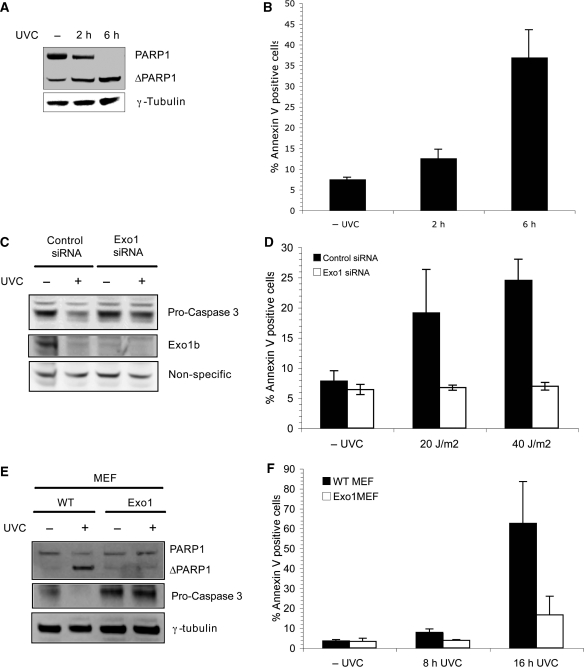
Exo1 is known to function in many types of DNA damage repair, however, the role of Exo1 in apoptosis remains to be elucidated. Since other MMR proteins are required for UV-induced apoptosis, we initially sought to investigate whether Exo1 has a role in apoptosis (23,24). Treatment of Hela cells with 40 J/m2 UVC was shown to induce apoptosis rapidly as measured by the cleavage of poly-ADP-polymerase (PARP1), a well characterized apoptotic substrate of caspase-3 (Figure 1A). It was further confirmed that these conditions induced apoptosis by annexin V analysis (Figure 1B). Hela cells were depleted of Exo1 using siRNA, which targets the mRNA of both isoforms of Exo1 (Exo1a and Exo1b) and exposed to UVC. The siRNA induced silencing of Exo1 reduced the protein level as evident by the disappearance of ∼100 kDa band recognized by the Exo1 antibody (Figure 1C). Interestingly, we also found significant reduction in Exo1 expression in control siRNA transfected cells after exposure of cells to UVC, suggesting that Exo1 might itself be targeted during apoptosis. The basis and significance of this degradation has been pursued in the latter part of the manuscript.
Figure 1.
Exo1 is required for the induction of apoptosis. (A) Hela cells were treated with 40 J/m2 UVC and lysates were taken at the indicated time-points for immunoblotting with the indicated antibodies. (B) Hela cells were treated as in (A), cells were harvested at the indicated times and analysed for annexin V staining via FACS. (C) Hela cells were transfected with control or Exo1 siRNA before treatment with 40 J/m2 UVC. Lysates were taken and immunoblotted with the indicated antibodies 6 h after treatment. (D) Hela cells were treated as in (C) and stained with annexin V antibodies 4 h after the indicated dose of UVC. Cells were analysed for annexin V staining via FACS. (E) Wild-type and Exo1 deficient MEFs were treated with 40 J/m2 UVC, incubated for 8 h and lysates immunoblotted with the indicated antibodies. (F) Wild-type and Exo1-deficient MEFs were treated with 40 J/m2 UVC and incubated for 8 or 16 h before staining with annexin V antibodies and FACS analysis. For annexin V experiments, the means and SD of three independent experiments are shown.
Next, we examined apoptosis induction in Exo1 deficient cells. Given that pro-caspase-3 cleavage is required for caspase-3 to become active and cleave its substrates, we examined pro-caspase-3 cleavage in Exo1 deficient cells. We observed that caspase-3 was cleaved within 4 h of exposure of control siRNA-transfected cells to UVC, however, caspase-3 cleavage and subsequent activation was suppressed in Exo1-deficient cells (Figure 1C). In order to further confirm the apoptotic defect in Exo1-deficient cells the annexin V assay was utilized. Cells transfected with control siRNA displayed around 25% apoptosis 4 h after treatment with 40 J/m2 UVC. In contrast, cells transfected with Exo1 siRNA were markedly resistant to UV-induced apoptosis and this resistance was characterized by a low percentage of annexin V positive cells that remained at around 6% following UVC exposure (Figure 1D). Camptothecin was also used to induce apoptosis and similarly to UVC showed that Exo1-deficient cells are resistant to DNA damage-induced apoptosis (Supplementary Figure 1).
Furthermore, we found that Exo1-deficient MEFs (21) showed a similar defect in pro-caspase-3 cleavage (Figure 1E) and apoptosis defect, as measured by accumulation of annexin V positive cells compared with wild-type MEFs (Figure 1F). It should be noted that the Exo1 deficient MEFs used were also heterozygote for hMSH6. MSH6 heterozygote MEFs do not display an apoptosis defect (17). Overall, these data provide strong evidence that Exo1 is required for the induction of DNA damage-induced cell death/apoptosis.
Exo1 is required for cytochrome c release and DNA fragmentation
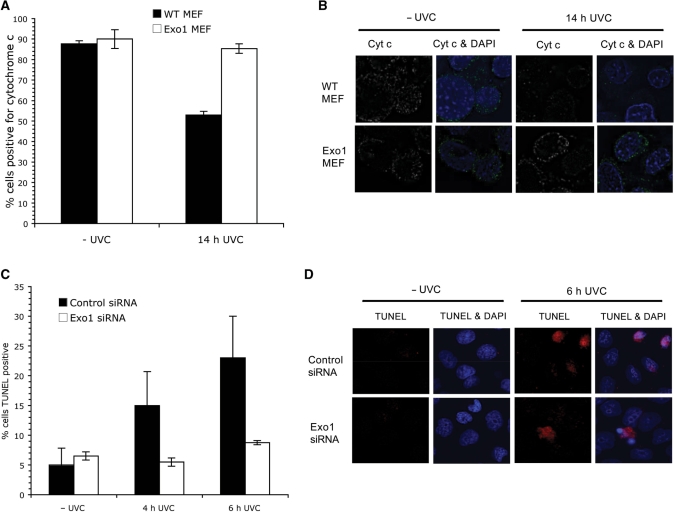
During apoptosis, cytochrome c is released from the mitochondria in response to a number of pro-apoptotic stimuli. The release of cytochrome c leads to the activation of caspases and progression of cell death (25). As depletion of Exo1 prevented the activation of caspase-3 (Figure 1C and E), we next examined whether cytochrome c was still released from the mitochondria in Exo1 deficient cells. Exo1 deficient MEFs were treated with 40 J/m2 UVC and cytochrome c levels were measured via FACS 14 h after treatment. In wild-type MEFs, the number of cytochrome c positive cells were reduced by around 40% following UVC treatment, whereas levels were only reduced by around 5% in Exo1 deficient MEFs (Figure 2A and B), indicating that Exo1's role in apoptosis is upstream of cytochrome c release and subsequent caspase activation.
Figure 2.
Exo1 is required for cytochrome c release. (A) Wild-type and Exo1 deficient MEFs were treated with 40 J/m2 UVC and incubated for 14 h. Cells were fixed and incubated with a cytochrome c antibody before FACS analysis was carried out. (B) Cells were treated as in (A) and subjected to immunofluorescence. Representative images are shown. (C) Hela cells were grown on coverslips and transfected with the indicated siRNAs. Cells were treated with 40 J/m2 UVC after 48 h transfection. Cells were fixed and subjected to TUNEL staining after 4 or 6 h treatment. For TUNEL staining, 200 cells were scored. The results presented are the average of three independent experiments and the error bars represent the SD. (D) Representative images of C.
During apoptosis, caspase-3 dependent DNA-degradation occurs, executed by the CAD nuclease (26). To examine whether Exo1 is required for DNA fragmentation during apoptosis, we utilized TUNEL staining, which specifically identifies cells undergoing DNA fragmentation. Hela cells depleted of Exo1 with siRNA were found to have less TUNEL positive cells, indicating less DNA fragmentation (Figure 2C and D). Similar results were also obtained in Exo1-deficient MEFs (Supplementary Figure 2).
Exo1 is dispensable for other forms of apoptosis
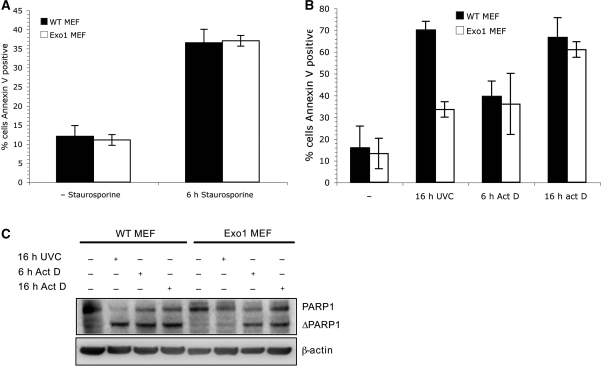
Given the role of Exo1 in DNA damage repair, we reasoned that Exo1 may be required for apoptosis induced specifically by DNA damage. We examined the extent of apoptosis induced by staurosporine and actinomycin D in Exo1 deficient cells. Staurosporine (27) is an inhibitor of protein kinase C and actinomycin D is an inhibitor of mRNA synthesis (28), but neither cause DNA damage. Like UVC, both staurosporine and actinomycin D induce apoptosis via the caspase-3 pathway. Exo1 deficient MEFs were found to be as sensitive as wild-type MEFs to treatment with staurosporine or actinomycin D as measured by annexin V assay (Figure 3A and B) and PARP1 cleavage (Figure 3C). These data indicate that Exo1 is required for DNA damage-induced apoptosis but is not universally required for caspase-3 activation.
Figure 3.
Exo1 is dispensable for other forms of apoptosis. (A) Wild-type and Exo1-deficient MEFs were treated with 1 μM staurosporine and incubated for 6 h before staining with annexin V antibodies and FACS analysis. The means and SD of two independent experiments are shown. (B) Wild-type and Exo1-deficient MEFs were treated with 10 μM actinomycin D or 40 J/m2 UVC and incubated for the indicated times before staining with annexin V antibodies and FACS analysis. The means and SD of three independent experiments are shown. (C) Wild-type and Exo1 deficient MEFs were treated with 40 J/m2 UVC or 10 μM actinomycin D, incubated for the indicated times and lysates were immunoblotted with the indicated antibodies.
Depletion of Exo1 does not disrupt UV-induced ATR-mediated pathways
The above data suggests Exo1 is a differential regulator of apoptosis depending on the type of cellular insult and may have a role upstream of both caspase-3 and PARP1 cleavage in response to UV-induced apoptosis This raises the possibility that Exo1 acts as a DNA damage sensor to induce apoptosis.
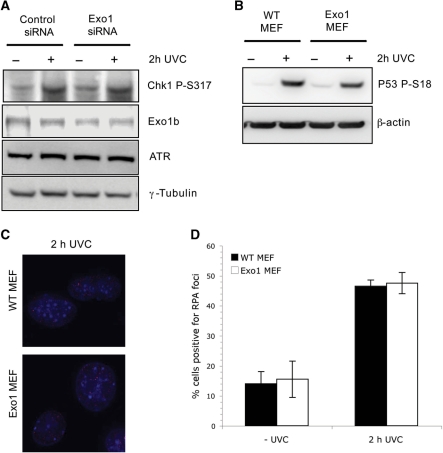
Following DNA damage primary lesions are often processed by nucleases that generate single-stranded DNA (ssDNA), which is believed to play a key function in the DNA damage response. The major kinase involved in the checkpoint response to UV-induced DNA damage is Ataxia Telangiectasia mutated and Rad3 related (ATR) (29). Since previous research has suggested that ATR has a role in UVC-induced apoptosis (9), we next examined whether Exo1b is required for the ATR-mediated UV-response. ATR mediated signalling preferentially triggers the phosphorylation and activation of the Chk1 kinase after UV. Exo1-depleted Hela cells were found to have a similar level of Chk1 phosphorylation as control cells following UVC treatment (Figure 4A). In addition, ATR levels were not altered in Exo1 depleted cells (Figure 4A). Since phosphorylation of p53 on serine 15 in humans (30) and serine 18 in mice (31) has been shown to be required for induction of apoptosis we next sought to examine p53 phosphorylation. Exo1 deficient MEFs were found to have comparable p53 serine 18 phosphorylation to wild-type MEFs following UVC treatment (Figure 4B), suggesting that the inhibition of apoptosis in Exo1-deficient cells is p53 independent.
Figure 4.
Depletion of Exo1 does not disrupt UV-induced ATR-mediated pathways. (A) Hela cells were transfected with the indicated siRNAs, treated with 40 J/m2 UVC and extracts taken 2 h after treatment for immunoblotting with the indicated antibodies. (B) Wild-type and Exo1 deficient MEFs were treated as in (A), and immunoblotted with p53 serine 15 antibodies (which detect phosphorylation on serine 18 in MEFs). (C) Wild-type and Exo1 deficient MEFs were treated as in (A), and stained with monoclonal RPA antibodies. (D) Graphical representation of the percentage of MEF cells positive for RPA foci in three independent experiments. The error bars represent the SD.
Following DNA damage by agents such as UVC many repair proteins localize to nuclear foci, marking the sites of DNA damage. RPA is a single stranded DNA binding protein involved in many forms of DNA repair and is known to coat long stretches of ssDNA generated after the processing of primary DNA lesions. RPA localizes to these sites of DNA damage after UVC treatment in an ATR-dependent manner, so we next decided to examine whether RPA localized correctly in the absence of Exo1. We observed that RPA foci formation was not affected by the lack of Exo1 in Exo1-deficient MEFs (Figure 4C and D), indicating an Exo1-independent mechanism processes primary UV-induced lesions and activates the ATR pathway.
Cleavage of Exo1 during apoptosis
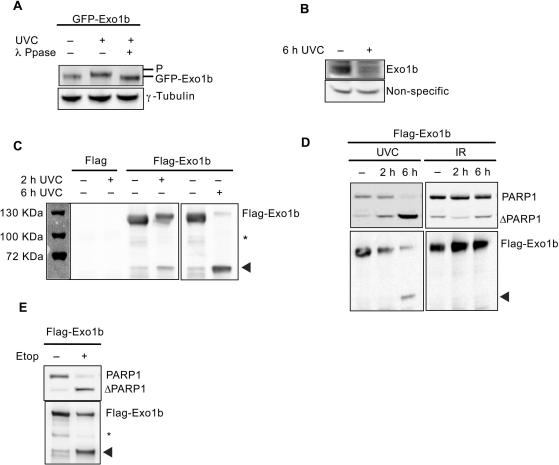
During the course of above studies, we noticed that Exo1 was degraded in normal/wild-type cells undergoing apoptosis (Figure 1C) and given the resistance of Exo1-deficient cells to apoptosis, we next investigated whether cleavage of Exo1 is required to activate the apoptotic pathway. To determine the fate of Exo1 during UV-induced apoptosis, we transfected Hela cells with GFP-Exo1b and treated with UVC. We detected a protein mobility shift of Exo1b within 1 h of exposure to UV, which was shown to be due to phosphorylation, as lambda phosphatase treatment prevented the shift (Figure 5A). As mentioned previously (32), Exo1b is difficult to detect in human cells due to a low abundance of protein and the lack of high affinity antibodies. We found that in 100 μg of nuclear lysates endogenous Exo1b could be detected via immunoblotting and at 6 h after UVC treatment Exo1b levels were significantly depleted (Figures 5B and 1C). In agreement with this, an exogenous Flag-tagged Exo1b driven by a heterologous promoter was reduced in levels in a time-dependent manner (Figure 5C). In parallel to the observed reductions in full length Exo1b, we observed the appearance of a 60 kDa polypeptide specifically recognized by anti-Flag antibody in cells expressing Flag-Exo1b (Figure 5C). Since the Flag-tag is localized on the N-terminus of this fusion protein, we postulated that the fragment generated comprised the N-terminus of Exo1. This band was subsequently analysed by Matrix Assisted Laser Desorption Ionisation-Time of Flight-mass spectrometry (MALDI-TOF-MS) and was confirmed to be Exo1b (data not shown). The other isoform of Exo1, Exo1a was also degraded following UVC treatment, producing an approximately 60 kDa fragment (Supplementary Figure 3). For the rest of our investigation, we have focused on the longer isoform, Exo1b as this is the more extensively studied and therefore best characterized isoform.
Figure 5.
Distinct fates of Exo1b following DNA damage. Unless otherwise stated, Hela cells were treated or mock-treated with 40 J/m2 UVC. (A) Hela cells were transiently transfected with GFP-Exo1b, treated with UVC and lysates taken 1 h after treatment for immunoblotting with the indicated antibodies. Lysates were incubated with llambda phosphatase where indicated. (B) Hela cells were treated with UVC and extracts taken for immunoblotting 6 h after treatment. (C) Hela cells were transfected with Flag or Flag-Exo1b, treated with UVC and extracts were taken at the indicated time-points. Immunoprecipitations were carried out using Flag M2 beads and immunoblotted with the Flag antibody. The panel to the left shows the protein molecular weight markers. (D) Hela cells were treated with 40 J/m2 UVC or 10Gy IR and lysates taken for immunoprecipitation and immunoblotting at the indicated times. (E) Hela cells were treated with 100 μM ETOP, extracts taken and immunoprecipitated with Flag M2. Immunoprecipitations were immunoblotted with the indicated antibodies. Arrowheads denote cleaved Exo1b. Asterisk indicates non-specific cross-reacting bands.
Since the doses of UVC used earlier in this investigation were shown to induce apoptosis (Figure 1A and B) we concluded that the degradation of Exo1 was a result of the apoptotic process. The degradation of Exo1 paralleled the extent of apoptosis induction as evident by annexin V positive cells, which increased in a time-dependent manner following UVC treatment. In addition, the cleavage of Exo1 coincided with the cleavage of PARP1 (33), a well-characterized substrate of caspase-3, in a time-dependent manner following UVC treatment (Figures 1A and 5D).
The cleavage of Exo1 was specific to apoptosis as no degradation was observed in Hela cells treated with ionising radiation (IR) (10Gy) (Figure 5D). IR fails to induce apoptosis in Hela cells at this dose as demonstrated by the lack of cleavage of PARP1 for up to 6 h post-IR treatment. In addition, the specific cleavage of both Exo1 and PARP1 was evident in Hela cells exposed to other agents, including etoposide, shown previously to induce apoptosis in these cells (Figure 5E), indicating that Exo1 cleavage is correlated with apoptosis.
Cleavage of Exo1b is mediated by caspase-3 during apoptosis
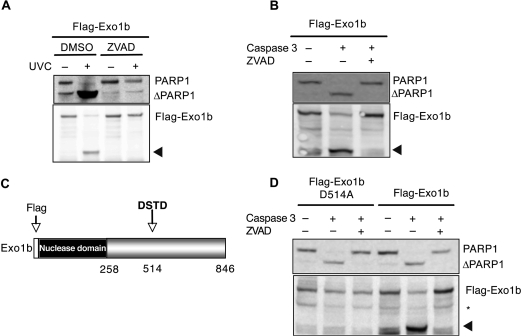
Given that caspases are the main proteases responsible for proteolysis during apoptosis, we next examined whether Exo1b cleavage is dependent upon this group of proteins. We initially employed a broad-spectrum caspase inhibitor, ZVAD-FMK, to study the requirement of caspases in Exo1 cleavage during apoptosis. The presence of ZVAD-FMK completely abrogated Exo1b cleavage in UVC-induced apoptosis (Figure 6A). The ZVAD-FMK inhibitor also protected the caspase-3 substrate PARP1 from cleavage following UVC-treatment, suggesting that caspase-3 may be the protease required for Exo1b cleavage.
Figure 6.
Caspase-3 cleaves Exo1 during apoptosis. Hela cells were transiently transfected with the indicated constructs. (A) Hela cells were mock-treated or pre-treated with ZVAD-FMK for 1 h, before being treated with 40 J/m2 UVC. Lysates were taken for immunoprecipitation with Flag M2 beads and immunoblotting 6 h after UVC treatment. (B) Hela lysates were incubated with 3 U of recombinant caspase-3 at 37°C in the presence or absence of ZVAD-FMK before immunoblotting with the indicated antibodies. (C) Schematic diagram of Exo1b showing putative caspase-3 cleavage site, D514A. (D) Hela lysates containing either Flag-Exo1b wild-type or D514A were incubated with 3 U of recombinant caspase-3 at 37°C in the presence or absence of ZVAD-FMK before immunoblotting with the indicated antibodies. Arrowheads denote cleaved Exo1b. Asterisk indicates non-specific cross-reacting bands.
To provide direct evidence for the involvement of caspase-3 in Exo1b cleavage, recombinant caspase-3 was added to lysates containing Flag-Exo1b. Following incubation with caspase-3, lysates were immunoblotted with anti-Flag antibodies. While predominantly full length Exo1b could be detected in the absence of caspase-3, the lysates incubated with recombinant caspase-3 contained predominantly cleaved Exo1b (Figure 6B). PARP1 was also cleaved in these lysates showing caspase-3 cleavage specificity. As anticipated, the presence of the ZVAD-FMK caspase inhibitor prevented the caspase-mediated cleavage of both Exo1b and PARP1.
Identification of the caspase-3 cleavage site in Exo1b
Caspase-3 recognizes the DXXD motif in its substrates and cleaves immediately following this site (34). Analysis of the Exo1b amino acid sequence led to identification of a putative caspase-3 cleavage site, DSTD514 (Figure 6C). We predicted that cleavage of Exo1b at this site would give rise to a N-terminal fragment of around 60 kDa, which is consistent with the size of the cleaved Exo1b fragment observed during apoptosis. In order to confirm this motif as a genuine caspase-3 cleavage site we generated a Flag-Exo1b fusion protein containing a mutation of the aspartic acid amino acid at position 514 to alanine (11). Plasmids expressing the mutant and wild-type proteins were transiently transfected into Hela cells. Lysates containing the mutant protein were prepared and incubated with recombinant caspase-3 and immunoblotted. In contrast to the wild-type protein, the D514A mutant was not degraded by recombinant caspase-3 (Figure 6D). This confirms the Asp514 site to be a likely in-vivo caspase-3 target site. PARP1 cleavage was also analysed as a positive control and was still cleaved in the lysates containing D514A.
The effect of over-expression of an Exo1 fragment or cleavage-resistant Exo1 on apoptosis
Cleavage of proteins during apoptosis sometimes leads, as in the case of other DNA repair proteins such as hMLH1 (24) and Claspin (35) to fragments that have dominant-negative activity. Such fragments appear to have pro-apoptotic roles in the progression of this type of cell death. Since the N-terminal fragment generated during apoptosis contains the nuclease domain of Exo1, this prompted us to speculate that the nuclease activity of cleaved-Exo1 may have a role in DNA degradation during apoptosis. This hypothesis is based on an earlier report that demonstrated that a fragment of Exo1 containing amino acids 1–391 was proficient in 5′-3′ nuclease activity (11). The fragment generated during apoptosis in the current study contains amino acids 1–514 of Exo1 and in light of the above it seems likely that this fragment will retain nuclease and DNA binding activity. Therefore, we tested the effect of over-expression of this fragment (1–514) and degradation-resistant (D514A) full length Exo1b on DNA damage-induced apoptosis in Hela cells in comparison with wild-type and nuclease-deficient (D78A) Exo1b. Expression of these constructs in Hela cells in the absence or presence of UVC had very little effect on the levels of apoptosis as assessed by cleavage of PARP1 (Supplementary Figure 4) or annexin V staining (Supplementary Figure 5B). Expression of these constructs also had no significant effect on DNA fragmentation, as measured by TUNEL staining (Supplementary Figure 5C).
DISCUSSION
Apoptosis is a fundamentally important cellular process, which orchestrates a series of controlled events, which ultimately leads to cell death. Apoptosis exists solely in multi-cellular organisms where it protects the organism from the accumulation of deleterious DNA mutations, which could result in loss of cellular programming. Specific DNA repair proteins such as, MLH1 (24), MSH2 (36) ATR and ATM (9) have been shown to have a role in the induction of apoptosis. Since Exo1 is known to be involved in DNA repair and interacts with other MMR proteins we examined whether Exo1 itself has a role in apoptosis. Here, we provide for the first time a role for the evolutionary conserved exonuclease, Exo1b, in UV-induced apoptosis.
Using Exo1-deficient MEFs and Hela cells, we found that Exo1 was required for timely, UVC-induced apoptosis. We further show that Exo1 deficient cells do not release cytochrome c from the mitochondria and both caspase-3 activation and PARP1 cleavage are absent following the induction of apoptosis by UVC. To elucidate the function of Exo1 in this process, we examined the potential roles of the DNA damage response kinase, ATR (9). It has been previously shown that ATR is required for apoptosis following UVC-treatment (9). We have shown here that Exo1 must function downstream or in a different pathway from ATR as depletion of Exo1 did not affect the phosphorylation or the localization of key ATR target proteins. This is consistent with a recent study in which Exo1 depletion had little impact on ATR signalling (37). Furthermore, we have shown here that p53 is unlikely to be involved in Exo1-mediated apoptosis as Exo1-deficient MEF cells can still phosphorylate p53 on serine 18. The Exo1-mediated apoptosis documented here is therefore downstream of or independent of ATR and p53 and suggests the involvement of other signalling pathways.
There is increasing evidence that polymorphisms in Exo1 may be associated with colorectal and lung cancer occurrence (19,20,38). Some of these polymorphisms have been shown to disrupt the interactions between Exo1, hMLH1 and hMSH2 and therefore inhibit MMR (20). It is possible that these polymorphisms may also disrupt the role of Exo1 and other MMR proteins in apoptosis. Thus such polymorphisms may not only compromise DNA repair but promote tumour cell survival via an apoptotic defect, especially in environments where DNA damage may accumulate, such as the colon or lungs.
The key effectors in apoptosis are the caspase proteases, a group of proteins that function by cleaving specific proteins at conserved sites this results in either the activation or inactivation of their substrates. In order to dissect the downstream events of apoptosis, it is of fundamental importance to identify the substrates of the effector caspases. In addition to Exo1 acting upstream of caspases in the induction of UV-induced apoptosis, we also noticed that Exo1 was cleaved during apoptosis. The fate of the Exo1b isoform, following DNA damage-induced apoptosis, was found to be time-dependant. Initially, Exo1b is phosphorylated following UVC treatment, however 2 h after treatment the levels of full length Exo1b decreased with the concomitant generation of a smaller, stable 60 kDa Exo1b fragment. This cleavage of Exo1b correlated with the induction of apoptosis. Previous work in both lower and higher eukaryotic systems has provided evidence for the involvement of Exo1 in several types of DNA repair (11,12,15,39). In light of the above, it is notable that the cleavage of Exo1 mimics that of other DNA repair proteins including PARP1 (33), ATM (7), DNAPKcs (8), BRCA1 (40), MLH1 (24) and Rad51 (41). The low abundance of Exo1b protein in mammalian cells may provide an explanation as to why Exo1 has remained undiscovered as a caspase substrate for so long.
Since Exo1 was cleaved during apoptosis, it was predicted that caspases would be the most likely effector of such an event and indeed we have mapped the site of Exo1 cleavage to Asp514 (DXXD caspase 3 consensus site) and found this event to be directly mediated by the caspase 3 protease.
Since DNA is fragmented during apoptosis and Exo1 is a nuclease it is tempting to speculate that Exo1 has a role in degrading DNA during apoptosis. We consider this implication unlikely when we compare it with the major apoptotic nuclease CAD for a number of reasons. First, CAD is expressed in the cytoplasm and once cleaved by caspase-3 translocates into the nuclease and degrades DNA (5,6). In contrast Exo1b is predominantly found in the nucleus and its localization does not change during apoptosis (Bolderson and Khanna, unpublished data). In addition, CAD has been shown to function solely downstream of caspase-3 activation, while we have shown Exo1 to have an essential apoptotic function upstream of caspase-3 (5,6). Although we have shown that Exo1-deficient cells are resistant to apoptotic DNA-fragmentation, this is not unexpected as we have also shown that Exo1 inhibits caspase-3 activation and caspase-3 activity is required for DNA fragmentation (26).
Over-expression of the full length Exo1b, cleavage-resistant Exo1b (D514A), nuclease deficient Exo1b (D78A) or the caspase-3 cleaved Exo1 fragment (amino acids 1–514) did not affect the induction of apoptosis as measured by PARP1 cleavage or annexin V. In addition, expression of these constructs had no significant affect on the induction of DNA fragmentation during apoptosis, as measured by TUNEL staining. Taken together, these data suggest that depletion of Exo1 inhibits DNA fragmentation during apoptosis via a caspase-3 dependent mechanism. In addition, we conclude that the fragment of Exo1b produced during apoptosis is not likely to be pro-apoptotic. However, it is possible that unlike other DNA repair proteins (24,40) the presence of the Exo1 cleavage fragment alone is not sufficient to stimulate apoptosis and that the proteolysis of other apoptotic factors is also required.
Given the upstream role for Exo1 in the induction of apoptosis, it is thus likely that the presence of the full length wild-type Exo1 protein is required for the induction of apoptosis, but only in the presence of DNA damage stimuli such as UVC. Exo1 is then subsequently cleaved to allow cells to undergo apoptosis efficiently by preventing the repair of fragmented DNA.
The potential function of Exo1 in DNA repair suggests that its cleavage may compromise its repair function by preventing it from participating in the repair of DNA damage such as DNA breaks generated by CAD (5,6). Following Caspase-3 cleavage PARP1 (33) and ATM (7) also retain their DNA-binding ability and it has been suggested that these inactive proteins may bind to the fragmented DNA and prevent DNA damage repair signaling. Together, these findings led us to speculate that Exo1 plays a crucial role in initially contributing to the induction of apoptosis and once apoptosis commences it is functionally inactivated by caspase 3 cleavage.
In summary, these results suggest that the full length Exo1 is required for induction of apoptosis, possibly via a role in a DNA damage-sensing complex. Our finding that Exo1 is specifically cleaved in cells undergoing apoptosis supports the hypothesis that a function of the effector caspases is to disassemble proteins that can repair the DNA and thus allow apoptosis to proceed.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online
FUNDING
National Health and Medical Research Council of Australia (241917).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Nigel Waterhouse for critical reading of the manuscript and helpful suggestions.
REFERENCES
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 4.Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14:44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- 5.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 6.Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 7.Smith GC, d’Adda di Fagagna F, Lakin ND, Jackson SP. Cleavage and inactivation of ATM during apoptosis. Mol. Cell Biol. 1999;19:6076–6084. doi: 10.1128/mcb.19.9.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Q, Lees-Miller SP, Kumar S, Zhang Z, Chan DW, Smith GC, Jackson SP, Alnemri ES, Litwack G, Khanna KK, et al. DNA-dependent protein kinase catalytic subunit: a target for an ICE-like protease in apoptosis. EMBO J. 1996;15:3238–3246. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Ma WY, Kaji A, Bode AM, Dong Z. Requirement of ATM in UVA-induced signaling and apoptosis. J. Biol. Chem. 2002;277:3124–3131. doi: 10.1074/jbc.M110245200. [DOI] [PubMed] [Google Scholar]
- 10.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J. Biol. Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 11.Lee BI, Wilson DM., 3rd The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 12.Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J. Biol. Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 13.Lee SD, Alani E. Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. J. Mol. Biol. 2006;355:175–184. doi: 10.1016/j.jmb.2005.10.059. [DOI] [PubMed] [Google Scholar]
- 14.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 18.Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Jr, Kneitz B, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Berends MJ, Post JG, Mensink RG, Verlind E, Van Der Sluis T, Kempinga C, Sijmons RH, van der Zee AG, Hollema H, et al. Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology. 2001;120:1580–1587. doi: 10.1053/gast.2001.25117. [DOI] [PubMed] [Google Scholar]
- 20.Sun X, Zheng L, Shen B. Functional alterations of human exonuclease 1 mutants identified in atypical hereditary nonpolyposis colorectal cancer syndrome. Cancer Res. 2002;62:6026–6030. [PubMed] [Google Scholar]
- 21.Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, et al. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young DB, Jonnalagadda J, Gatei M, Jans DA, Meyn S, Khanna KK. Identification of domains of ataxia-telangiectasia mutated required for nuclear localization and chromatin association. J. Biol. Chem. 2005;280:27587–27594. doi: 10.1074/jbc.M411689200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Richards B, Wilson T, Lloyd M, Cranston A, Thorburn A, Fishel R, Meuth M. Apoptosis induced by overexpression of hMSH2 or hMLH1. Cancer Res. 1999;59:3021–3027. [PubMed] [Google Scholar]
- 24.Chen F, Arseven OK, Cryns VL. Proteolysis of the mismatch repair protein MLH1 by caspase-3 promotes DNA damage-induced apoptosis. J. Biol. Chem. 2004;279:27542–27548. doi: 10.1074/jbc.M400971200. [DOI] [PubMed] [Google Scholar]
- 25.Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 26.Janicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 27.Tamaoki T, Nakano H. Potent and specific inhibitors of protein kinase C of microbial origin. Biotechnology. 1990;8:732–735. doi: 10.1038/nbt0890-732. [DOI] [PubMed] [Google Scholar]
- 28.Sobell HM. Actinomycin and DNA transcription. Proc. Natl Acad. Sci. USA. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair. 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Unger T, Sionov RV, Moallem E, Yee CL, Howley PM, Oren M, Haupt Y. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 31.Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol. Cell Biol. 2004;24:976–984. doi: 10.1128/MCB.24.3.976-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari S. Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res. 2005;65:3604–3609. doi: 10.1158/0008-5472.CAN-04-4069. [DOI] [PubMed] [Google Scholar]
- 33.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 34.Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 35.Semple JI, Smits VA, Fernaud JR, Mamely I, Freire R. Cleavage and degradation of Claspin during apoptosis by caspases and the proteasome. Cell Death Differ. 2007;14:1433–1442. doi: 10.1038/sj.cdd.4402134. [DOI] [PubMed] [Google Scholar]
- 36.Toft NJ, Winton DJ, Kelly J, Howard LA, Dekker M, te Riele H, Arends MJ, Wyllie AH, Margison GP, Clarke AR. Msh2 status modulates both apoptosis and mutation frequency in the murine small intestine. Proc. Natl Acad. Sci. USA. 1999;96:3911–3915. doi: 10.1073/pnas.96.7.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin G, Wang H, Hu Z, Liu H, Sun W, Ma H, Chen D, Miao R, Tian T, Jin L, et al. Potentially functional polymorphisms of EXO1 and risk of lung cancer in a Chinese population: a case-control analysis. Lung Cancer. 2008;60:340–346. doi: 10.1016/j.lungcan.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 40.Zhan Q, Jin S, Ng B, Plisket J, Shangary S, Rathi A, Brown KD, Baskaran R. Caspase-3 mediated cleavage of BRCA1 during UV-induced apoptosis. Oncogene. 2002;21:5335–5345. doi: 10.1038/sj.onc.1205665. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y, Nakada S, Ishiko T, Utsugisawa T, Datta R, Kharbanda S, Yoshida K, Talanian RV, Weichselbaum R, Kufe D, et al. Role for caspase-mediated cleavage of Rad51 in induction of apoptosis by DNA damage. Mol. Cell Biol. 1999;19:2986–2997. doi: 10.1128/mcb.19.4.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.