Abstract
Nur, a member of the Fur family, is a nickel-responsive transcription factor that controls nickel homeostasis and anti-oxidative response in Streptomyces coelicolor. Here we report the 2.4-Å resolution crystal structure of Nur. It contains a unique nickel-specific metal site in addition to a nonspecific common metal site. The identification of the 6-5-6 motif of the Nur recognition box and a Nur/DNA complex model reveals that Nur mainly interacts with terminal bases of the palindrome on complex formation. This contrasts with more distributed contacts between Fur and the n-1-n type of the Fur-binding motif. The disparity between Nur and Fur in the conformation of the S1-S2 sheet in the DNA-binding domain can explain their different DNA-recognition patterns. Furthermore, the fact that the specificity of Nur in metal sensing and DNA recognition is conferred by the specific metal site suggests that its introduction drives the evolution of Nur orthologs in the Fur family.
INTRODUCTION
Transition metal ions are essential nutrients for cells since they stabilize the folded conformations or mediate chemical reactions of metalloproteins which constitute about one-third of all proteins (1). Nickel is required to maintain the structures and functions of microbial and plant enzymes including urease, hydrogenase, carbon monoxide dehydrogenase, acetyl-CoA decarbonylase/synthase, methyl-CoM reductase, glyoxylase I, acireductone dioxygenase, methylenediurease and superoxide dismutase (SOD) (2). Despite the structural and functional role in some enzymes, however, the intracellular level of nickel should be tightly regulated since nickel inflicts toxic effect on cells when in excess (3). Therefore, it is critical for cells to keep a delicate balance of nickel level through sensing and transporting systems. The NikR proteins of Gram-negative bacteria are well-known transcription factors that repress and/or activate specific genes implicated in nickel import and utilization in response to nickel availability (4). Recently, we have identified a new type nickel-uptake regulator (Nur) that is distinct from NikR. Nur belongs to the ferric-uptake regulator (Fur) family and controls nickel homeostasis and antioxidative response in Streptomyces coelicolor (5,6). In the presence of nickel, nickel-bound Nur directly represses the expression of nikABCDE operon encoding components of ABC-type transporter and sodF gene encoding Fe-containing SOD. It activates the expression of sodN gene encoding Ni-containing SOD, possibly in an indirect way (5).
Many of the Fur family members regulate metal-ion homeostasis and oxidative stress responses in prokaryotes at the level of transcription (7). In most cases, the binding of Fur regulators to the promoter regions represses the expression of target genes by blocking the access of transcription machinery. Among these, Fur subfamily orthologs, as respresented by Escherichia coli Fur (8), function as iron-responsive regulators for iron homeostasis in a wide range of bacteria (7). Zur orthologs as represented by E. coli Zur (9) and Rhizobium leguminosarum Mur (10), are regulated by zinc and manganese, respectively, and maintain homeostasis of their specific metals. Bacillus subtilis PerR (BsPerR) (11) is different from other members: it is a peroxide sensor that regulates inducible peroxide defense genes rather than having a primary role in controlling metal homeostasis. In spite of diverse metal selectivity and physiological functions, the above-mentioned Fur family regulators are common in that only the specific metal-bound forms bind tightly to their target sites.
According to the crystal structures of Pseudomonas aeruginosa Fur (PaFur) with two zinc ions (12), BsPerR with the regulatory metal site empty (apo-BsPerR) (13), and Mycobacterium tuberculosis FurB (MtFurB or MtZur) with three zinc ions (14), the Fur family members are homodimeric and each monomer consists of an N-terminal DNA-binding domain (DB-domain), a C-terminal dimerization domain (D-domain) and a hinge region that connects the two domains. According to the structural comparison between PaFur and apo-BsPerR, which represent the DNA-binding competent and the DNA-binding incompetent conformations, respectively, DB-domain seems to swing around the hinge region with respect to D-domain in response to regulatory metal association (13). The metal-induced hinge motion that brings two DB-domains in close proximity is the key to understand the metal-mediated activation mechanism of the Fur family members since the spatial orientation of two DB-domains should match with that of interacting tandem subsites along the target DNA molecule for complex formation (15).
Nur is likely to have a modular dimeric structure like other Fur family members. However, nickel-binding sites in Nur could not be deduced from the previous structural studies that revealed only zinc-binding sites. In addition, among the published crystal structures of the Fur family members, only PaFur is considered to adopt a DNA-binding competent conformation (12). Due to the restricted structural information, the question on how members in the Fur family with highly similar structural architecture select different metals and recognize distinct DNA sequences still remains to be addressed. Here, we report the crystal structure of DNA-binding competent Nur to reveal atomic details of nickel-coordination and to provide insights into the structural mechanism that underlies the divergence of Fur family members.
MATERIALS AND METHODS
Crystallization, structure determination and refinement
We have previously reported a 3.0-Å resolution MAD data set (16). However, we could not complete model building because electron density maps for some regions were poor. Thus, we found a new crystallization condition with Nur proteins of ∼12 mg ml–1. Crystals of Nur were grown at 23°C with mother liquors of 14% PEG3350, 0.4 M sodium malonate (pH 7), 0.1 mM benzamidine hydrochloride and 0.1 mM NiCl2. The crystals belonged to the space group P31 with cell parameters a = 79.16, b = 79.16 and c = 49.73 Å. For data collection crystals were frozen at 100 K using a cryostream cooler after they were briefly immersed in a cryoprotectant solution containing 15% ethylene glycol in the same mother liquors. A 2.4-Å MAD data set (Supplementary Table 2) was collected at beamline 5A of Photon Factory, Japan. Data were integrated and scaled with HKL and SCALEPACK (17). The four nickel positions in crystals of Nur were located, and phases were calculated using the program SOLVE. The subsequent solvent flattening by RESOLVE gave rise to an interpretable map, based on which the de novo model building of Nur was completed. The subsequent refinement and manual refitting of the initial model reduced R and Rfree values to 22.4% and 27.0%, respectively. The ideality of the stereochemistry of the final model containing residues 3–135 was verified by PROCHECK. The Ramachandran plot indicates 83.3% of non-glycine residues are in the most favored regions and the remaining 14.8% residues are in the allowed region. The model building was performed using QUANTA and refinement was done with maximum-likelihood algorithm implemented in CNS program (Supplementary Table 2).
Preparation of Nur mutants and electrophoretic mobility shift assay
Site-directed mutagenesis of residues in Nur was carried out with QuikChange™ Site-directed Mutagenesis Kit (Stratagene) according to the manufacturer's instruction. A recombinant pET3a-based plasmid (Novagen) that contains the cloned nur gene (3) was used as a template. Mutated clones were selected and confirmed by nucleotide sequencing. Electrophoretic mobility shift assay between sodF promoter DNA fragment and cell extracts containing recombinant Nur proteins was done as described previously (3). In order to assess the amount of Nur proteins in each binding reaction, we performed western blot analysis of cell extracts used for binding assay with polyclonal antibody raised against wild type Nur protein in mice. Following SDS–PAGE (15%), blots were prepared and detected by ECL system (Amersham) using anti-mouse goat antibody conjugated with peroxidase (Cappell).
RESULTS AND DISCUSSION
Overall structure of Nur
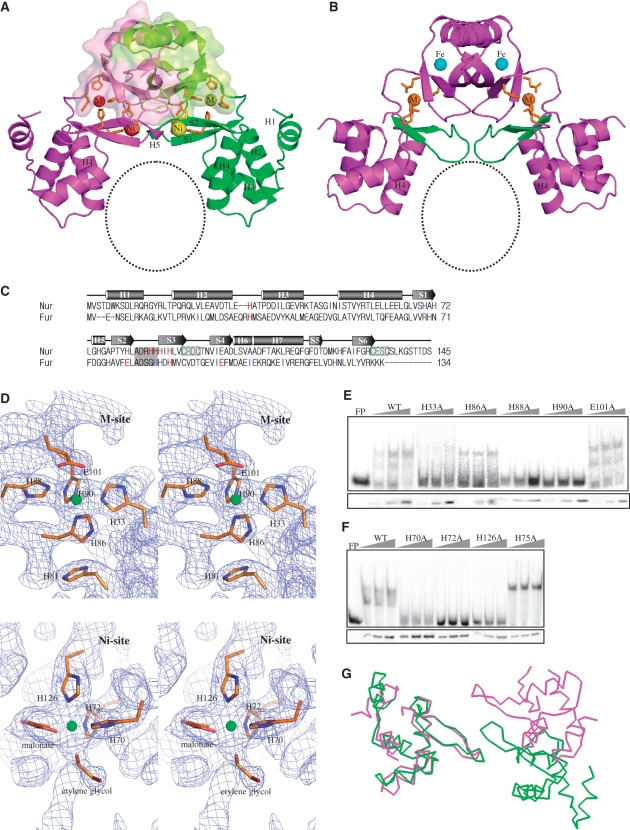
Nur is a homodimer with a modular architecture: two DB-domains are attached to the dimeric core constructed by two D-domains (Figure 1A). The triangular dimeric conformation of Nur with two closely located DB-domains resembles that of PaFur (Figure 1B), and most likely represents the DNA-binding competent conformation. DB-domain (residues 1–82) consists of four helices and two strands (H1-4 and S1-2; Figure 1A–C). Except an N-terminal helix (H1) extending outward, three helices (H2-H4) form the typical type of the winged helix motif. In this classical motif, the last helix (H4 in Nur), which could make specific contacts with bases in the major groove of DNA, is called a ‘recognition helix’ (Figure 1A). D-domain (residues 88–145) folds as a simple α/β structure (Figures 1A and 2) and a loop of residues 83–87 is the hinge region connecting the two domains (Figure 1C). The domain structures of Nur are similar to those of PaFur, BsPerR and MtFurB as predicted from their sequence homology (data not shown).
Figure 1.
Structure of Nur and metal sites. (A) Ribbon diagram of Nur with the dimeric core veiled by transparent surface. Nickel ions and metal-coordinating residues are represented by spheres and sticks, respectively. M and Ni indicate M- and Ni-sites, respectively. A black circle indicates the plausible DNA-binding site. For clarity, secondary structure elements only for DB-domain are labeled. (B) Ribbon diagram of PaFur. M and Fe indicate M- and Fe-sites, respectively. The S1–S2 sheets are in green. Zinc ions and residues of M-site are shown by spheres and sticks, respectively. A black circle indicates the plausible DNA-binding site. (C) A structure-based sequence alignment of Nur with PaFur. Red and blue letters in the Nur (PaFur) sequence indicate residues of M- and Ni (Fe)-site, respectively. The hinge region is dark shaded. Two Cys-X-X-Cys motifs are boxed. (D) Stereo view of the final 2Fo – Fc electron density maps contoured at 1σ, showing M- and Ni-sites. Ni and Ni-coordinating residues are shown in spheres and sticks, respectively. (E) DNA-binding activity of Nur variants with substitution mutations of M-site residues. E101A mutant was examined in parallel for comparison. Electrophoretic mobility shift assay was performed for binding between the sodF promoter DNA fragment (–60 to +30 nt from transcription start site) and cell extracts containing either wild-type or mutant Nur proteins with H33A, H86A, H88A and H90A mutations. For each Nur variant, three separate reactions with increasing amounts of cell extracts were examined, along with the assessment of Nur protein levels by western blot analysis (lower panel). FP indicates a sample with free probe only. (F) DNA-binding activity of Nur variants with substitution mutations of Ni-site residues. Nur variants with H70A, H72A and H126A mutations were examined as described above. H75A mutant was examined in parallel for comparison. (G) Cα-tracings of DB-domains from Nur (green) and PaFur (violet) that are superposed using only one DB-domain.
Figure 2.
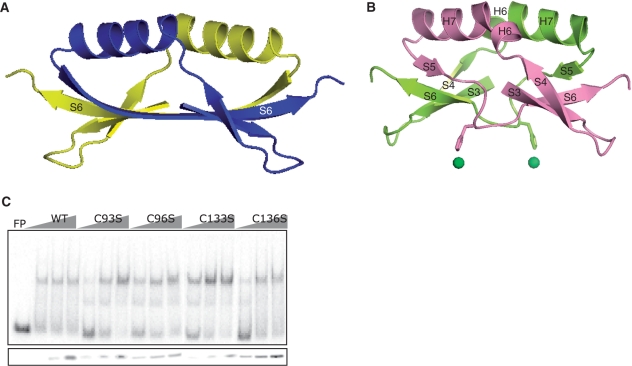
Structure of the dimeric core and insignificant contribution of conserved cysteines to DNA association. (A) Ribbon diagram of the dimeric core of PaFur. Each monomer is represented by different colors. For clarity, only S6 is labeled. (B) Ribbon diagram of the dimeric core of Nur with secondary structural elements labeled. Each monomer is represented by different colors. His126 and a nickel ion at Ni-site are presented in sticks and spheres, respectively. (C) DNA-binding activity of C93S, C96S, C133S and C136S mutants of Nur.
A nonspecific metal site commonly observed in Nur and other Fur family members
The anomalous scattering from nickel was exploited for the structure determination of Nur, which allowed us to determine the number and the position of metal sites. Two metal sites, M-site and Ni-site, exist in each monomer of Nur. M-site at the domain interface is constructed by His33, His86, His88, and His90 (Figure 1A and D). Here nickel is coordinated by four nitrogen atoms of the four histidines with a square-planar geometry, one of the preferred coordination geometries for nickel (18) (Figure 1D). Interestingly, M-site seems to be able to accommodate zinc, too. Nur contains near stoichiometric amount of zinc and nickel ions per each monomer, according to an ICP-AES analysis (data not shown). Therefore, we tried to determine the position of zinc using a zinc MAD data. Although we could not solve the structure due to the low occupancy of zinc, it was possible to identify the zinc position. Unexpectedly, the zinc position was nearly identical to the nickel position at M-site, which is compatible with a relatively low nickel-occupancy at M-site. It would appear that among Nur proteins in crystals, most proteins have nickel but some have zinc at this site. It is probable that the addition of NiCl2 to crystallization conditions might have rendered nickel predominance at M-site in Nur crystals.
It is interesting to note that other Fur family members have a metal site with affinity for various divalent cations whose location is similar to M-site (19). Especially, in the crystal structure of PaFur (12), zinc is located at a position that is nearly identical to the nickel position in M-site of Nur (Figure 1A–C). Remarkably, the four residues of PaFur that coordinate zinc are conserved in Nur: His32, Glu80, His89 and Glu100 in PaFur correspond to His33, His81, His90 and Glu101 in Nur, respectively (Figure 1C). Since histidine is the most common metal-binding residue, the only replacement of a glutamate residue (Glu80 in Fur) with a histidine residue (His81 in Nur) is not likely to affect zinc binding. In Nur, therefore, zinc could be coordinated by His33, His81, His90 and Glu101 at M-site (Figure 1D). Consequently, the conservation of zinc-coordinating residues in Nur might be the basis for the accommodation of zinc at M-site.
A nickel-specific metal site in Nur
In contrast to M-site, Ni-site has never been observed in structures of other Fur family members, suggesting that this site is the unique metal site of Nur. Ni-site resides at the domain interface: His70 and His72 in DB-domain and His126 in D-domain participate in metal coordination at this site (Figure 1A and D). Three nitrogen atoms from the histidines, together with three oxygen atoms from malonate and ethylene glycol, coordinate a nickel ion with an octahedral geometry (Figure 1D). The oxygen contributors would be water molecules in vivo. We grew crystals in another condition without malonate (16). For the data collection, we dehydrated these crystals instead of using ethylene glycol as a cryoprotectant (16). Although we failed to build the complete Nur model with this data, we confirmed that a nickel ion is coordinated by His70, His72 and His126 as shown in Figure 1D. Although due to the low resolution we could not identify electron densities for water molecules, water molecules are highly likely to be involved in nickel coordination.
Nickel coordination with three protein ligands and three water molecules indicates that Ni-binding at the surface exposed Ni-site is probably weak. However, it is worthy of notice that NikR also has a low-affinity nickel site that is occupied at high nickel concentration (21,22). The occupation of this weak nickel site is critical for the maintenance of the closed conformation of NikR, that is primed for DNA-binding (21,22).
Compared with M-site with an affinity for zinc, this site probably prefers nickel, as indicated by high occupancy of nickel at this site. The preference of Ni-site for nickel is further supported by its octahedral environment, the most favorable geometry for nickel binding in proteins (18). The nickel specificity of Ni-site was also crystallographically confirmed. A crystal of Nur was transferred to a crystallization solution containing ZnCl2 instead of NiCl2. We repeated this procedure five times for 4 h, collected a Ni-MAD data set with the transferred crystal and the position of nickel ions was identified using the SOLVE program. Interestingly, Ni-site still contains a nickel ion. Nur is exquisitely selective for nickel in vitro and in vivo as revealed by no DNA-binding activity of Nur in the presence of other divalent cations (5,19). Nickel specificity of Ni-site, coupled with the affinity of M-site for zinc, strongly suggests that Ni-site determines the nickel-responsive activation of Nur.
Evaluation of metal-binding residues by site-directed mutagenesis
To evaluate the role of the two metal sites in DNA binding, we mutated all the histidine residues into alanine and then performed gel-shift assay. As shown in Figure 1E, H88A and H90A mutants exhibited nearly no DNA-binding activity, indicating that each of these residues is critical in constructing M-site. H33A exhibited greatly reduced binding, whereas H86A did not affect DNA binding significantly. It is conceivable that the absence of one histidine residue like His86 cannot disrupt M-site. In such a mutant protein, a water molecule or a nearby residue such as His81 could possibly participate in metal-coordination. At Ni-site, His70, His72 and His126 are all essential for the maintenance of Ni-site since their respective mutant proteins showed nearly no DNA-binding activity (Figure 1F).
To investigate the relationship between metal binding and DNA-binding activity, we purified the wild-type Nur and two mutant Nur proteins (H72A and H90A), performed gel-shift assay, and analyzed their metal contents by ICP-AES (Supplementary Table 1). The gel-shift patterns of the three purified proteins (data not shown) resembled those of corresponding cell extracts (Figure 1E) with H72A mutation caused more drastic loss of DNA-binding activity than H90A. Moreover, the metal content of the purified proteins was revealed to be correlated with their DNA-binding activity. The nickel content of H72A mutant was just half of that of the wild type whereas its zinc content is comparable to the wild type (Supplementary Table 1), strongly suggesting that the drastically reduced nickel content led to impaired DNA-binding activity of H72A mutant. In addition, the ignorable effect of His72Ala mutation on zinc content supports the nickel specificity of Ni-site. In the case of H90A mutant, the content of both nickel and zinc was ∼20% reduced compared with the wild type. This coincides with our crystallographic observation that M-site can accommodate both metal ions.
The mutational study reveals that the occupation of both metal sites is required to maintain the DNA-binding competent conformation of Nur since the loss of single metal site can lead to the loss of DNA-binding activity. Based on the mutational study and the location of the two metal sites at the domain interface (Figure 1A), it can be safely assumed that both metal sites play regulatory roles to determine interdomain arrangement that is key to activate Nur to bind DNA. It deserves attention that PaFur with an active dimeric conformation also contains a specific metal site (Fe-site) where iron bind (12) in addition to the common M-site (Figure 1B). M-, Ni-, Fe-sites are all located at the domain interface or near the hinge region (Figure 1A and B), suggesting their contribution in arranging DB-domains to bind DNA. In this view, Nur and PaFur of a similar conformation would have different spatial arrangement of DB-domains since metal binding to Ni-site and Fe-site is sure to induce different hinge motions. The disparity in the spatial arrangement becomes clear when DB-domains of Nur and PaFur are superposed (Figure 1G). In functional aspect, it is natural that Nur and Fur adopt similar but distinct dimeric conformations since they are activated by different metals and recognize different DNA sequences despite their overall homology in sequence and fold. Consequently, the development of both a common metal site and a unique metal site in Nur and PaFur appears to be a molecular strategy to insure both the general similarity and the functional disparity to be encrypted in their DNA-binding competent structures.
Structural features of the dimeric core in Nur
Dimeric cores in other Fur family members are commonly featured by an intermolecular sheet binding up the two dimerization domains. For example, two three-stranded sheets of D-domains from each monomer in PaFur are combined to form a six-stranded antiparallel sheet (Figure 2A). In contrast, in the dimeric core of Nur, two sheets from each monomer are not combined into a stable intermolecular sheet (Figure 2B), which is most likely due to nickel binding to Ni-site. To coordinate a nickel ion, His126 stretches out its imidazole group toward Ni-site, resulting in the concomitant shift of the segment harboring His126 which otherwise forms the N-terminal part of S6 as in the corresponding region of PaFur (Figure 2B). In fact, the Cα atom of H126 in Nur is shifted down ∼3.7 Å compared with that of the corresponding residue (N126) in PaFur. The segment shift prevents the formation of interstrand hydrogen bonds between two S6s from each monomer, which is essential to combine two sheets into one, by hindering the side-by-side arrangement of the two strands.
The existence of two Cys-X-X-Cys motifs in the primary structure (Figure 1C) has been related to the existence of zinc (Cys4-Zn) that are coordinated by four cystein residues (11,13). In the case of BsPerR, the zinc coordination by four cysteines in the motifs was suggested to be critical to stabilize their dimerization domain and hence their dimeric structures, indicating the structural role of Cys4-Zn (13). However, there is no Cys4-Zn coordinated by four cysteines of two Cys-X-X-Cys motifs in Nur. The absence of Cys4-Zn is not related to the oxidation of cystein residues since crystals of Nur were grown in reducing conditions. Despite the absence of Cys4-Zn, Nur maintains the dimeric conformation (Figure 1A). Furthermore, the Cys→Ser mutations did not affect the DNA-binding activity of Nur (Figure 2C), which contrasts with the loss of repressor function of BsPerR mutants with the Cys→Ser substitutions (11). Taken together, Cys4-Zn appears not to be essential for maintaining the DNA-competent conformation and hence for the DNA-binding activity of Nur. Our structural and biochemical data do not necessarily mean that Nur has no zinc ions at this site in vivo. Considering the zinc avidity of the Cys-X-X-Cys motif and the abundance of zinc ions, Nur could have zinc ions bound at this site in vivo. Both the bridging sheet and Cys4-Zn appears to result in the loose packing of the dimeric core although it does not lead to the break of the dimeric core.
The Nur box and the Nur/DNA complex model
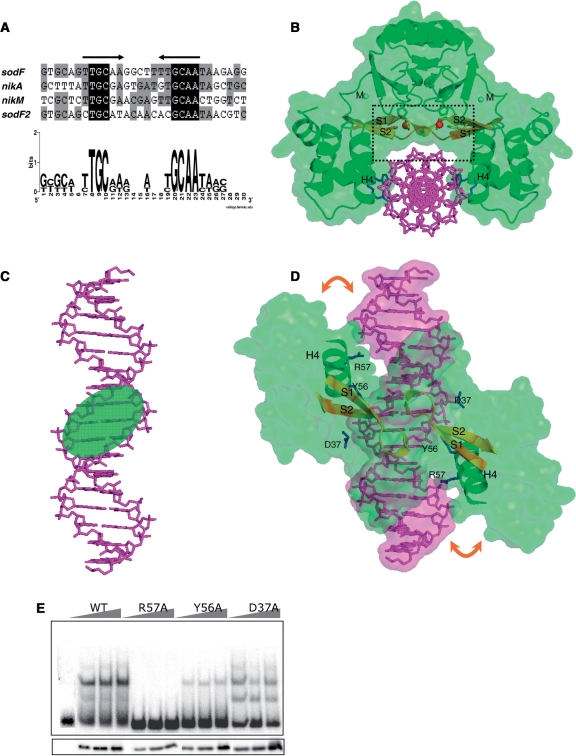
To elucidate the regulatory mechanism of Nur at the transcription level, it is essential to characterize the Nur–DNA recognition mechanism. For this purpose, the consensus DNA sequences were searched among Nur-binding promoter regions. In addition to previously identified genes (sodF, sodF2 and nikABCDE) (5), a new type of nickel transporter (nikMNOQ) has been suggested in S. coelicolor (23) and we determined that Nur also binds to nikM promoter DNA in a nickel-dependent manner (see Supplementary Figure S1). The Nur-binding sites in each promoter region were analyzed by DNase I footprinting (Supplementary Figure S1), and the nucleotide sequences of primary protected regions were aligned. A consensus Nur box emerged with a palindromic structure of six residues with 5-bp spacing (tTGCaa-N5-ttGCAA). The 6-5-6 motif implies that five nonconserved bases in the middle of the motif may contribute little to interactions with Nur (Figure 3A).
Figure 3.
The proposed Nur box sequence and a model for Nur/DNA complex. (A) Palindromic sequences conserved within Nur-binding sites. The primary DNase I- protected regions in sodF, sodF2, nikA and nikM promoters by Nur binding (Supplementary Figure S1) were aligned, and a conserved palindromic pattern was proposed for Nur-binding consensus. (B) The front view of a Nur/DNA complex model. Nickel ions at Ni-site are shown by red spheres. The S1–S2 sheets are colored in yellow for emphasis. The 17-bp Nur box DNA in sodF promoter and DNA-contacting residues (Tyr56, Arg57 and Asp37) are shown in violet and blue sticks, respectively. Recognition helices (H4) are labeled. A black rectangle highlights the location of S1–S2 sheets on top of the central minor groove [see also (D)]. (C) The contacting face of the Nur box DNA in the Nur/DNA complex model. A green ellipsoid indicates the central minor groove. (D) The top view of the Nur/DNA complex model. For clarity, Nur is represented by a green surface with recognition helices and the S1–S2 sheets shown by ribbon drawing. DNA is shown in sticks covered with violet surface. The orange arrows indicate the deduced induced fit on complex formation. Tyr56, Arg57 and Asp37 are shown in blue sticks. Recognition helices (H4) are labeled. (E) DNA-binding activity of Nur variants with mutations of residues contacting DNA. Nur variants with mutations for R57, Y56 and D37 were examined as in Figure 1E.
To verify the interaction mode deduced from the 6-5-6 motif, we built a Nur/DNA complex model based on complex structures between winged helix motifs and DNAs. First, one DB-domain of Nur was superposed onto that of a template structure with a recognition helix as a reference, and then the dimeric Nur was superposed onto the previously superposed DB-domain. Among many complex structures that were used as a template, the complex structure between Mu repressor and DNA (24) gave us a plausible Nur/DNA model with little steric clashes. Second, the DNA molecule in the template structure was replaced by a 17-bp DNA molecule with the sodF sequence. Third, we manually tuned the position of Nur relative to DNA, keeping it in mind that recognition helices of Nur should face toward DNA major grooves. Finally, the position of DNA was refined by the ‘solid docking’ module in QUANTA (Molecular Simulation, Inc.). The module allows a successful docking only when an electrostatic and geometric complementarity is accomplished between a host and a guest molecule. In the final complex model that was completed without any structural alteration in both Nur and DNA, Nur takes a grip on DNA with its recognition helices facing major grooves. Interestingly, our complex model, where DNA is accommodated in the space below the dimeric core with two DB-domains on both sides (Figure 3B), is similar to a Fur/DNA complex model that best fits experimental observations and allows good insertion of recognition helices into major grooves (25).
The contact pattern between Nur and DNA in the Nur/DNA complex model is examined to assess whether it supports the interaction mode deduced from the 6-5-6 motif of the Nur box. In structural aspect, on the contacting face of the Nur box, there are three grooves in an order of a major groove, a minor groove and a major groove (Figure 3C). According to this arrangement of grooves, six conserved bases at the terminals of the motif, which are thought to mainly interact with Nur, are located in the major grooves on both ends, while five nonconserved bases in the middle of the Nur box are in the central minor groove. In the Nur/DNA complex model, although recognition helices do not nestle down in major grooves, a slight induced fit on complex formation would be enough to elicit direct contact between residues in recognition helices and bases in major grooves (Figure 3D). In contrast, the S1–S2 sheets on top of the central minor groove are not likely to contact with bases in the central minor groove (Figure 3B and D). To make contact, the horizontal sheet should sink down toward DNA on complex formation. However, this is impossible since the sheets are tied to the dimeric core through metal-coordination (see below) (Figure 3B). No contact between the horizontal sheets and bases in the central minor groove is compatible with the deduced little contribution of five nonconserved bases to complex formation.
Mutational analyses of Nur–DNA interactions based on the complex model
Arg57 in recognition helices extends its side chain toward the major groove of DNA in our complex model (Figure 3B and D), indicating that its guanidino group presumably interacts with conserved palindromic bases in the Nur box. Specific contact between bases and the guanidino group of arginine residues is prevalent in protein–DNA interactions (20). Therefore, the importance of Arg57 in DNA recognition was assessed by a mutational analysis. As expected, the R57A mutant did not display DNA-binding activity at all, exhibiting the critical role of Arg57 in DNA recognition (Figure 3E). In addition to Arg57, Tyr56 and Asp37 make contact with the phosphorous backbone of DNA in the complex model (Figure 3D), indicating their potential role in DNA binding. The Y56A mutation resulted in drastic loss of DNA-binding activity and the D37A mutant exhibited slightly reduced DNA-binding activity, pointing to their relative contribution to DNA binding (Figure 3E). The side chains of Glu63 and Glu64 in the recognition helix face major grooves in the complex model. Although their electrostatic property and distance from DNA indicates no direct contact with DNA, water-mediated interactions between the two residues and DNA cannot be excluded. Therefore, we mutated the two residues into alanine to investigate their role in DNA binding and found that the two negatively charged residues appear to contribute little to DNA binding since the mutant proteins efficiently bound to DNA (data not shown).
Structural correspondence of Nur and Fur to their respective DNA boxes
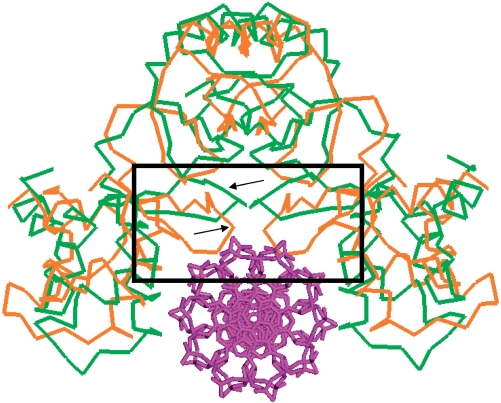
The n-1-n motif of the Fur box, where n is 6, 7 or 9 depending on different interpretations of the box (26–28), is distinctly different from the 6-5-6 motif of the Nur box in that the conserved palindromic half sequences are separated only by a single base. In contrast to the Nur box, the n-1-n motif suggests that the middle bases in the central minor groove of the Fur box are involved in interactions with Fur. The different configurations of the Nur and Fur boxes indicate different conformations of DNA-binding sites in Nur and Fur. To disclose the structural correspondence of Nur and PaFur [the PaFur box is also n-1-n type (27)] to their respective DNA boxes, therefore, we performed a detailed structural investigation and discovered that they have a striking disparity in the conformation of the S1–S2 sheets that face the central minor groove (Figures 1A and B, and 4). Compared with the turn regions of the horizontal S1–S2 sheets folding into a short 310-helix in Nur, the turn regions in PaFur are just loops, slanting down toward the DNA-binding site (Figures 1A and B, and 4). Considering both the proximity to minor grooves and the flexibility, the turn regions in PaFur would protrude into the central minor groove, making contact with bases on complex formation (Figure 4). In the winged-helix motif, the turn region connecting strand S1 and S2 is called a wing. Noticeably, the wing region is known to interact with bases in the minor groove adjacent to major grooves harboring recognition helices (29). In summary, the S1–S2 sheets of Nur and Fur appear to be well adapted for the specific recognition of their respective DNA boxes.
Figure 4.
Superposition of PaFur to Nur in the Nur/DNA complex. Dimeric structure of PaFur is superposed onto Nur in the Nur/DNA complex model. Cα atoms of Nur (green) and PaFur (orange) are shown with DNA in sticks (violet). Arrows point out S1–S2 sheets. A black rectangle highlights the protrusion of slant S1–S2 sheets toward DNA in PaFur in comparison with horizontal S1–S2 sheets of Nur.
Introduction of Ni-site for the evolution of Nur orthologs
Metal binding generates the disparity between Nur and Fur in the conformation of the S1–S2 sheets. In Nur, His70 and His72 in the sheets, together with His126 in the dimeric core, coordinate a nickel ion at Ni-site (Figures 1A and D, and 3B). Therefore, metal binding to Ni-site has an effect to tie the sheets up to the dimeric core through metal coordination. As a result, the sheets just run horizontally without sinking toward the DNA-binding site (Figures 1A and 3B). In contrast, the S1–S2 sheets in Fur, which are not involved in metal coordination, are not fixed to the dimeric core. This allows their slant conformation and confers conformational flexibility, facilitating interactions with bases in the central minor groove on complex formation (Figure 4).
In the Fur superfamily, only Nur orthologs appear to have the horizontal S1–S2 sheets. The three histidine residues constituting Ni-site are conserved only in Nur orthologs (Supplementary Figure S2), indicating that other Fur family members do not contain metal-binding sites that correspond to Ni-site in Nur. Therefore, the S1–S2 sheets in other Fur subfamilies possibly adopt the slant conformation, which allows contact with bases in the central minor groove on complex formation. Consistently, the DNA boxes of other Fur family members such as the Per box and the Zur box (27,30) have a single base between conserved palindromic sequences just as with the Fur box.
Nur is outstanding among the Fur family members in that (i) it is exquisitely selective for nickel in vitro and in vivo (5); (ii) it has no bridging sheet in the dimeric core (Figure 2B); and (iii) it has a horizontal S1–S2 sheet that fits recognition of palindromic sequences separated by a long interval (Figures 1 and 4). The exclusive activation of Nur by nickel is associated with the nickel-specific Ni-site. Moreover, both the absence of the intermolecular bridging sheet and the horizontal conformation of the S1–S2 sheet are consequences of nickel-binding to Ni-site. Taken together, all the unique features of Nur regarding the specificity in metal sensing and DNA recognition have to do with Ni-site which is present exclusively in Nur orthologs (subfamily) in the Fur family. Therefore, the introduction of Ni-site can be assumed to be the origin of specialization of Nur orthologs in the evolution of the Fur superfamily. Based on this perspective, development of new metal-binding sites could be a working mechanism to evolve subfamilies of Fur that respond to different metals and recognize different patterns of DNA sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Marine & Extreme Genome Research Center Program from the Ministry of Land, Transport, and Maritime Affairs, the KORDI in-house programs (PE98202 and PE98402); and NRL research grant from KOSEF. Funding for open access charge: The Marine & Extreme Genome Research Center Program from the Ministry of Land, Transport, and Maritime Affairs.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Jin-Won Lee for critical reading of the manuscript. Data deposition: The atomic coordinates have been deposited in the Protein Data Bank with code 3EYY.
REFERENCES
- 1.Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Structural elements of metal selectivity in metal sensor proteins. Proc. Natl Acad. Sci. USA. 2003;100:3713–3718. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulrooney SB, Hausinger RP. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 3.Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 4.Dosanjh NS, Michel SL. Microbial nickel metalloregulation: NikRs for nickel ions. Curr. Opin. Chem. Biol. 2006;10:123–130. doi: 10.1016/j.cbpa.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 2006;59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- 6.Kim EJ, Chung HJ, Suh B, Hah YC, Roe JH. Transcriptional and post-transcriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Müller. Mol. Microbiol. 1998;27:187–195. doi: 10.1046/j.1365-2958.1998.00674.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 8.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 9.Patzer SI, Hantke K. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 2000;275:24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, Johnston AW. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology. 2004;150:1447–1456. doi: 10.1099/mic.0.26961-0. [DOI] [PubMed] [Google Scholar]
- 11.Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 12.Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- 13.Traore DA, El Ghazouani A, Ilango S, Dupuy J, Jacquamet L, Ferrer JL, Caux-Thang C, Duarte V, Latour JM. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol. Microbiol. 2006;61:1211–1219. doi: 10.1111/j.1365-2958.2006.05313.x. [DOI] [PubMed] [Google Scholar]
- 14.Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, Pohl E. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- 15.Nam TW, Jung HI, An YJ, Park YH, Lee SH, Seok YJ, Cha SS. Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration. Proc. Natl Acad. Sci. USA. 2008;105:3751–3756. doi: 10.1073/pnas.0709295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An YJ, Ahn BE, Roe JH, Cha SS. Crystallization and preliminary X-ray crystallographic analyses of Nur, a nickel-responsive transcription regulator from Streptomyces coelicolor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008;64:130–132. doi: 10.1107/S1744309108001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Rulisek L, Vondrasek J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J. Inorg. Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 19.Giedroc DP, Arunkumar AI. Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Trans. 2007:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 20.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chivers PT, Tahirov TH. Structure of Pyrococcus horikoshii NikR: nickel sensing and implications for the regulation of DNA recognition. J. Mol. Biol. 2005;348:597–607. doi: 10.1016/j.jmb.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Schreiter ER, Wang SC, Zamble DB, Drennan CL. NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc. Natl Acad. Sci. USA. 2006;103:13676–13681. doi: 10.1073/pnas.0606247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 2006;188:317–327. doi: 10.1128/JB.188.1.317-327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wojciak JM, Iwahara J, Clubb RT. The Mu repressor-DNA complex contains an immobilized ‘wing’ within the minor groove. Nat. Struct. Biol. 2001;8:84–90. doi: 10.1038/83103. [DOI] [PubMed] [Google Scholar]
- 25.Tiss A, Barre O, Michaud-Soret I, Forest E. Characterization of the DNA-binding site in the ferric uptake regulator protein from Escherichia coli by UV crosslinking and mass spectrometry. FEBS Lett. 2005;579:5454–5460. doi: 10.1016/j.febslet.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 26.Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 2002;184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, Storz G, Schneider TD. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 2007;35:6762–6777. doi: 10.1093/nar/gkm631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavrrar JL, McIntosh MA. Architecture of a fur binding site: a comparative analysis. J. Bacteriol. 2003;185:2194–2202. doi: 10.1128/JB.185.7.2194-2202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huffman JL, Brennan RG. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 30.Fuangthong M, Helmann JD. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 2003;185:6348–6357. doi: 10.1128/JB.185.21.6348-6357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.