Abstract
Synthesis of the pre-mRNA poly(A) tail in the nucleus has important consequences on the translational activity of the mature mRNA in the cytoplasm. In most eukaryotes, nuclear polyadenylation of pre-mRNAs is thought to require the nuclear poly(A)-binding protein (PABP2/PABPN1) for poly(A) tail synthesis and ultimate length control. As yet, however, the extent of the association between PABP2 and the exported mRNA remains poorly understood. Here, we used chromatin immunoprecipitation (ChIP) assays to show that the fission yeast ortholog of mammalian PABP2 (Pab2) is cotranscriptionally recruited to active genes. Notably, the association of Pab2 to genes precedes that of a typical 3′-processing/polyadenylation factor, suggesting that Pab2 recruitment during the transcription cycle precedes polyadenylation. The inclusion of an RNase step in our ChIP and immunoprecipitation assays suggests that Pab2 is cotranscriptionally recruited via nascent mRNA ribonucleoprotein (mRNPs). Tandem affinity purification coupled with mass spectrometry also revealed that Pab2 associates with several ribosomal proteins as well as general translation factors. Importantly, whereas previous results suggest that the nuclear poly(A)-binding protein is not present on cytoplasmic mRNAs, we show that fission yeast Pab2 is associated with polysomes. Our findings suggest that Pab2 is recruited to nascent mRNPs during transcription and remains associated with translated mRNPs after nuclear export.
INTRODUCTION
Two evolutionarily conserved poly(A)-binding proteins (PABPs) have been characterized with some details: PAPBC in the cytoplasm and PABP2/PABPN1 in the nucleus (1,2). Consistent with its cytosolic localization, PABPC (Pab1 in yeast) stimulates translation initiation by mediating contacts between the mRNA 5′- and 3′-ends via interactions between PABPC and components of the translational machinery (3,4). PABPC also appears to act as an antagonist of nonsense-mediated decay (5–7), a pathway of mRNA surveillance that targets transcripts with premature termination codons. Studies in budding yeast and mammals indicate that Pab1 and PABPC, respectively, shuttle between the nucleus and cytoplasm (8–10) and that Pab1 facilitates the biogenesis and the export of mRNAs (9–11). Consistent with an evolutionarily conserved nuclear function for the cytosolic PABP, intron-containing RNAs can be copurified with mammalian PABPC (12).
The nuclear counterpart of PABPC, PABP2, is structurally different from PABPC and thought to function during polyadenylation of pre-mRNAs. Polyadenylation of most eukaryotic pre-mRNAs consists of a two-step reaction involving endonucleolytic cleavage and poly(A) tail addition. An exhaustive list of evolutionarily conserved proteins responsible for specific and efficient 3′-end processing have been characterized (13–15). These conserved proteins form large multisubunit complexes that bind different cis-acting elements in the 3′-end of pre-mRNAs and determine the site of endonucleolytic cleavage. It has now become clear that 3′-end processing events are tightly integrated to the transcription cycle [reviewed in (13,14,16,17)] through the carboxy-terminal domain (CTD) of RNA polymerase II (Pol II). The Pol II CTD consists of evolutionarily conserved heptad repeats that are thought to act as a platform for the recruitment of various trans-acting factors required for pre-mRNA maturation (18). Notably, this includes many factors involved in 3′-end processing/polyadenylation that are recruited late during the transcription cycle and near the polyadenylation site of yeast (19–21) and human (22,23) genes.
Following cleavage, the nuclear poly(A) polymerase is responsible for the synthesis of poly(A) tails with average length of 70 and 300 nt in yeast and mammals, respectively (2,24). Experiments using in vitro polyadenylation assays suggest that PABP2 has a dual role in 3′-end formation: (i) PABP2 stimulates processive poly(A) synthesis by direct and simultaneous interactions with the growing poly(A) tail and the poly(A) polymerase (25) and (ii) PABP2 promotes the transition from processive to distributive poly(A) synthesis once a specific length is reached (26). Whereas the genome of the yeast Saccharomyces cerevisiae does not encode for an ortholog of mammalian PABP2, we have recently reported the identification of the PABP2 ortholog in the yeast Schizosaccharomyces pombe (27). Deletion of S. pombe PAB2 results in the expression of RNAs with hyperadenylated tails, indicating that factors other than Pab2 stimulate poly(A) polymerase processivity. Therefore, the precise role of the nuclear poly(A)-binding protein in pre-mRNA polyadenylation in vivo remains unclear.
Translocation of the mRNA ribonucleoprotein (mRNP) complex from the nucleus to the cytoplasm is linked to remodeling events mediated by a myriad of different proteins (28,29). As yet, the status of the association between PABP2 and nascent mRNPs during and after transit from the nuclear pore complex remains poorly understood. Although mammalian PABP2 shuttles between the nucleus and the cytoplasm (30), earlier results suggest that PABP2 is restricted to nuclear transcripts. Specifically, it has been shown that human PABP2 copurifies with a subunit of the nuclear cap-binding complex, but not with the general translation initiation factor, eIF4E (31). On the basis of these results and the different steady-state distribution of PABPC and PABP2, it has been suggested that poly(A)-bound PABP2 is replaced by PABPC upon transit of the mRNP to the cytosol. The mechanism and cellular compartment of such a substitution between PABP2 and PABPC remain elusive, however.
To further characterize the role of the nuclear poly(A)-binding protein during mRNA synthesis, we performed a comprehensive analysis of Pab2 during mRNP formation in fission yeast. Using chromatin immunoprecipitation (ChIP) assays, our results suggest that Pab2 associates with pre-mRNAs cotranscriptionally prior to 3′-end processing/polyadenylation. Furthermore, tandem affinity purification and mass spectrometry revealed that Pab2 associates with proteins involved in diverse RNA-related functions, including several proteins involved in cytoplasmic translation. Notably, we show that Pab2 is a shuttling protein and present strong evidence that Pab2 associates with translating mRNPs. Our data suggest that Pab2 is recruited early during the transcription cycle and remains associated with translated mRNPs after nuclear export.
MATERIALS AND METHODS
Strains, growth media and genetic methods
The strains used in this study are listed in Table 1. Schizosaccharomyces pombe was grown at 30°C in yeast extract medium with amino acid supplements (YES) and Edinburgh minimum medium (EMM) containing appropriate amino acid supplements. Schizosaccharomyces pombe cells were transformed with plasmids and PCR products by the lithium acetate method. The strains expressing TAP-tagged version of Pab2 and Pcf11 were constructed by PCR-based gene targeting as described previously (32,33). The oligonucleotides sequences used for the construction of these strains are available upon request. Appropriate tagging of strain was confirmed by PCR and immunoblotting.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| FBY13 | h+ ade6M210 leu1-32 ura4Δ18 his3Δ1 | 39 |
| FBY101 | h+ ade6M210 leu1-32 ura4Δ18 his3Δ1 Pab2-TAP::kanMX6 | This study |
| FBY152 | MATa ura3 leu2 lys NUP49::TRP1 (pUN100 nup49-313-LEU2) | 40 |
| FBY153 | h+ ade6M210 leu1-32 ura4Δ18 his3Δ1 Pcf11-TAP::kanMX6 | This study |
| FBY187 | h+ ade6M210 leu1-32 ura4Δ18 his3Δ1 PAB2::URA4 | This study |
Plasmids
Cloning of Pab2 in fusion with the green fluorescent protein (GFP) in a S. pombe-based plasmid was previously described (27). To generate a S. cerevisiae-based plasmid that expresses GFP-Pab2, the DNA encoding the GFP-Pab2 cassette was amplified by PCR using oligonucleotide sequences containing XhoI and EcoRI restriction sites. The resulting PCR product was then cloned into p416ADH (34) that was previously digested with EcoRI and XhoI restriction enzymes to create plasmid pFB197. The ADH1 promoter of pFB197 was replaced by the CYC1 promoter (406 bp) via PCR amplification from S. cerevisiae genomic DNA using oligonucleotide sequences containing SacI and SpeI restriction sites. Following digestion of the PCR product, the DNA was cloned into the SacI and SpeI site of pFB197 to create pFB235. The open reading frame encoding for S. pombe Pab1 was amplified by PCR from genomic DNA using oligonucleotide sequence containing NotI and BglII restriction sites. Following digestion of the PCR product with NotI and BglII, the DNA was cloned into the NotI and BglII site of pSLF273 (35) to create plasmid pFB243.
Antibodies
Mouse monoclonal antibody specific to recognize the carboxy-terminal heptapeptide repeat present on the largest subunit of RNA Pol II (Rpb1; 8WG16) was from Covance Research Products. Rabbit anti-Protein A was from Sigma. Mouse monoclonal antibody specific to HA (clone 12CA5) was from Roche Applied Science. Rabbit polyclonal antibodies specific to fission yeast 40S ribosomal protein S2 (Rps2) were raised at Covance Research Products against GST fusion proteins purified from Escherichia coli.
ChIP assays
ChIP assays were performed as previously described (36). For RNA Pol II ChIPs, a mouse monoclonal antibody specific for the C-terminal domain of Pol II (8WG16) was used. For the analysis of the NMT1 gene, thiamine was added to the culture medium at a final concentration of 30 μM for 3 h. Determination of RNA-dependent ChIP signals was based on a previously described procedure (37). Briefly, the cross-linking time was reduced to 5 min and the lysis buffer was adjusted to a final SDS concentration of 0.05%. Chromatin preparation were then treated or not treated with a cocktail of RNases to obtain 7.5 U of RNase A and 300 U of RNase T1. Subsequent steps were as previously described (36).
Quantification of the immunoprecipitated DNA was done by quantitative real-time PCR (Rotor-gene 3000, Corbett life sciences) using gene-specific primer sets. Specific dilutions of coimmunoprecipitated DNA as well as of sonicated and reverse cross-linked input DNA were used to determine the percentage of input DNA in each immunoprecipitate. To calculate the increase in signal for the different gene regions, the percentage input values obtained by quantitative PCR were normalized to the percentage input value obtained with the nontranscribed intergenic region, arbitrarily set to 1. The 104-bp PCR amplicon corresponding to the intergenic region lies on the left arm of chromosome I (nucleotides 3009380 to 3009484). Each PCR was run in triplicate, and all ChIPs were repeated at least three times using independent chromatin extracts.
Protein purification
Coimmunoprecipitation assays of RNA Pol II were based on a previously described procedure (38). Briefly, cells were grown in YES at 30°C to mid-log phase and lyzed in ice-cold lysis buffer (25 mM NaPO4 at pH 6.8, 0.1 M KOAc, 2 mM MgOAc, 10% glycerol, 1 mM PMSF, 3 ng/ml pepstatin, 3 ng/ml leupeptin, 3ng/ml aprotinin, 3 ng/ml chymostatin, 0.2 mM Na3VO4, 5 mM β-glycerophosphate, 1 mM NaF) with a Fastprep FP120 using 0.5 mm glass beads. The protein concentration of lysates was determined by the Bradford protein assay. Lysates were precleaned overnight at 4°C with Glutathione-Sepharose beads that had been previously pre-equilibrated in lysis buffer. The lysate was transferred to IgG-Sepharose beads that had been previously pre-equilibrated in IP buffer (Lysis buffer with 0.5 mM NH4OAc and 0.1% Tween 20) for 4 h at 4°C. The beads were then washed five times with 1 ml IP buffer, and the bound proteins eluted by incubating for 5 min at 95°C in 1× SDS–PAGE sample buffer. For coimmunoprecipitation with RNase treatment, the IgG-Sepharose beads were washed four times and incubated for 30 min at room temperature with 7.5 U of RNase A and 300 U of RNase T1. After the RNase treatment, the beads were washed two times and the bound proteins were eluded with 1× SDS–PAGE at 95°C for 5 min. Samples were subjected to a 7% SDS–PAGE gel, transferred to nitrocellulose in 10 mM CAPS (pH 11) and 1% MeOH. Following incubation with specific primary antibodies, membranes were probed with goat anti-rabbit and anti-mouse secondary antibodies conjugated to IRdye 800 (LI-COR Odyssey) and Alexa fluor 680 (Invitrogen), respectively. Linear detection of the proteins was performed and quantified using the Odyssey infrared imaging system (LI-COR).
Purification of the carboxy-terminal TAP-tagged Pab2 was performed as previously described (39) from 8 l of fission yeast cells. Mass spectrometry analyses were done at the Southern Alberta Mass Spectrometry (SAMS) Centre for Proteomics (University of Calgary).
Nuclear export assays
Nuclear export assays in S. cerevisiae were done as previously described (40) with some modifications. Briefly, GFP-Pab2 (pFB235), NLS-LacZ-GFP (pFB145) and Nab2-GFP (pFB147) expression constructs were transformed into S. cerevisiae strain FBY152 that harbors the nup49-313 temperature-sensitive allele. Cells were grown to OD600 0.1 at 25°C in synthetic medium and separated into two different cultures that were grown for 5 h at the permissive (25°C) or nonpermissive (37°C) temperature. To prevent the synthesis of new GFP-tagged proteins in the cytoplasm, cycloheximide (100 μg/ml) was added to each sample for the last hour to inhibit translation. Cells were examined for GFP fluorescence signal by live fluorescence.
Polysome assay
Polysome profiles were generated from S. pombe as previously described (39). To disrupt ribosomes by chelating Mg2+ ions, EDTA was added to final concentration of 20 mM to the lysis buffer. The lysate was loaded onto 5–45% (w/w) sucrose gradient containing 20 mM EDTA. For RNase disruption of polysomes, the lysate was incubated with 22 U/ml RNase A for 10 min at room temperature before ultracentrifugation onto 5–45% (w/w) sucrose gradients. KCl treatment of extracts was performed by the addition of salt to a final concentration of 500 mM in polysome lysis buffer and the sucrose gradient. Analysis of formaldehyde cross-linked ribosome profiles was done as previously described (41). Briefly, shaved ice and formaldehyde were added to cultures at final concentrations of 25% and 1%, respectively. After gentle mixing, cells were left on ice for 15 min and 0.1 M glycine was added to stop the cross-linking reaction. Cell lysis and ultracentrifugation were as previously described (39) except that KCl was adjusted to 500 mM in lysis buffer and sucrose gradients. For puromycin treatment experiments, a mixture of 1 mM puromycin/2 mM GTP was added to the culture for 15 min at 30°C before cell lysis. Lysis buffer was also adjusted to 1 mM puromycin/2 mM GTP. Sucrose gradients were fractioned by upward displacement with 55% (w/w) sucrose using a gradient fractionator (Brandel Inc.) connected to a UA-6 UV monitor (Teledyne Isco) for continuous measurement of the absorbance at 254 nm. Twenty fractions of 600 μl were collected and the proteins were TCA-precipitated. Proteins from same amounts of each fraction were separated onto 12% SDS–PAGE, and analyzed by immunoblotting.
RESULTS
Cotranscriptional recruitment of Pab2 to active genes precedes 3′-end processing/polyadenylation signals
Electron microscopy has previously detected PABP2 in the vicinity of transcription complexes using salivary glands from the insect Chironomus tentans (42). To investigate further the mechanism by which the nuclear poly(A)-binding protein associates with the transcriptional machinery, we used ChIPs to examine whether S. pombe Pab2 is recruited during the transcription cycle of RNA Pol II. ChIP assays have been used extensively to determine the position of mRNA processing factors along genes and infer the steps at which they are recruited during the transcription cycle. Three Pol II-transcribed genes (ADH1, PYK1 and PGK1) were first examined because of their relatively strong transcription levels (43). To perform ChIP assays, we used a strain in which a Pab2-TAP fusion protein was expressed from the endogenous PAB2 promoter. The Pab2-TAP strain did not confer cold sensitivity in contrast to PAB2-null cells (27) and showed doubling times similar to a wild-type control (data not shown), suggesting that TAP-tagged Pab2 is functional. The amount of genomic DNA associated with a TAP-tagged version of Pab2 was determined by real-time PCR using primer sets located at the 5′-end, the middle and the 3′-end of the ADH1, PYK1 and PGK1 genes (Figure 1A).
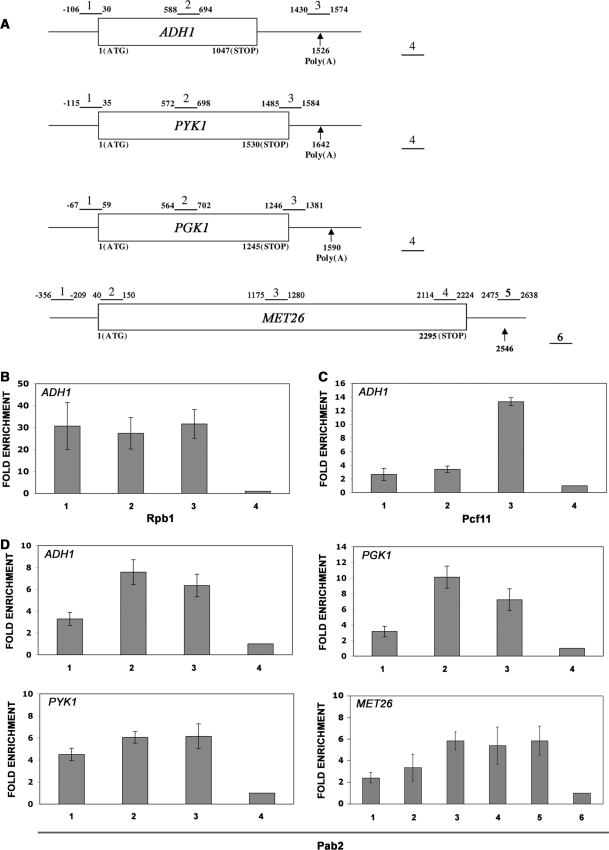
Figure 1.
Association of Pab2 to active genes precedes that of the characterized 3′-processing factor, Pcf11. (A) Schematic diagram of genes used for ChIP assays. Boxes represent open reading frames and nucleotides numbers are relative to the initiation codon. Arrows indicate the position of the predicted polyadenylation site based on the 3′-UTR sequences of the ADH1, PYK1, PGK1 and MET26 genes (74). Bars above each gene show the position of PCR products used for analyses in the ChIP assays. The number above each bar is used for identification in subsequent figures. PCR product number 4 (ADH1, PYK1 and PGK1) and number 6 (MET26) corresponds to the nontranscribed intergenic region (see Materials and methods section) and served as the internal background control. Cross-linked and sonicated extracts from wild-type cells (B) as well as from cells that express Pcf11-TAP (C) and Pab2-TAP (D) were subjected to affinity purification using IgG-sepharose (C and D) or antibodies specific for the large subunit of RNA Pol II (Rpb1) (B). The coprecipitating DNA was amplified by real-time PCR using gene-specific primer pairs as indicated in (A). The abundance of the different gene segments from ADH1 (B–D) as well as from the PYK1, PGK1 and MET26 genes (D) in each immunoprecipitate was expressed as fold enrichment relative to the nontranscribed intergenic region value, arbitrarily set to 1. The values correspond to the means of at least three independent experiments.
ChIP experiments using antibodies specific for the large subunit of RNA Pol II (Rpb1) demonstrated similar cross-linking levels across the ADH1-coding region (Figure 1B). In contrast, ChIP assays using extracts of cells that expressed a TAP-tagged version of the cleavage/polyadenylation factor Pcf11 showed robust cross-linking near the polyadenylation site of the ADH1 gene (Figure 1C). The ChIP patterns for fission yeast Rpb1 and Pcf11 are consistent with similar analyses reported in the budding yeast S. cerevisiae (20,37,44) and demonstrate the validity of our ChIP assays. Pab2 associated mainly with the middle and the 3′-end of the ADH1, PYK1 and PGK1 genes, although some levels of cross-linking were also detected at the 5′-end of these genes (Figure 1D). To confirm and better visualize the gradient of association of Pab2 from 5′ to 3′, we used the 2.3-kb long MET26 gene (Figure 1A). MET26 was chosen because the larger size of this gene allows for a better resolution of gene segments. Consistent with the data obtained for the ADH1, PYK1 and PGK1 genes, the enrichment of Pab2 at the 5′-end of the MET26 gene (Figure 1D, regions 1 and 2) clearly increased in the middle and at the 3′-end. Further analysis of Pab2 cross-linking using intron-containing genes demonstrated ChIP profiles similar to intronless genes (Figure 2). The results of these ChIP experiments using six different genes indicate a general 5′ to 3′ enrichment of Pab2.
Figure 2.
Association of Pab2 to intron-containing genes. (A) Schematic diagram of genes used for ChIP assays. Boxes represent exons and nucleotides numbers are relative to the initiation codon. The number above each bar is used for identification in (B). PCR product number 6 (CPC2) and number 5 (RPP0) corresponds to the nontranscribed intergenic region and served as the internal background control. (B) ChIPs were performed on cells that express TAP-tagged Pab2 using methods similar to those described in Figure 1.
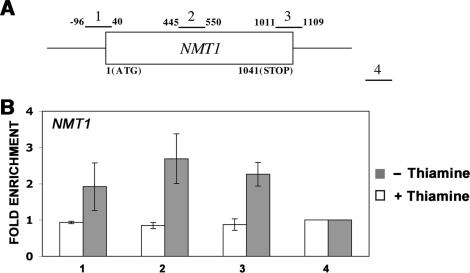
To determine whether the association of Pab2 to Pol II-specific genes is dependent on active transcription, real-time PCR was performed on Pab2-enriched genomic DNA using primer sets spanning the NMT1-coding region (Figure 3A). Expression of the S. pombe NMT1 gene is strongly repressed following addition of thiamine (45). As can be seen in Figure 3B, Pab2 immunoprecipitates prepared from extracts of cells that were previously grown in the absence of thiamine (active conditions) showed a ChIP pattern on the NMT1 gene consistent with the cross-linking profiles detected for the ADH1, PYK1, PGK1 and MET26 genes. In contrast, the levels of Pab2 cross-linking across the NMT1 gene were similar to the intergenic control region after repression of the NMT1 promoter (with thiamine). These results indicate that recruitment of Pab2 to genes requires active transcription by RNA Pol II.
Figure 3.
Transcription-dependent recruitment of Pab2 to the NMT1 gene. (A) The schematic diagram shows the positions of the PCR products used to analyze the NMT1 gene in the ChIP assays. (B) ChIP assays were performed on cells that express TAP-tagged Pab2 and that were treated (white bars) or not treated (gray bars) with 30 μM thiamine for 3 h. The coprecipitating DNA was amplified by real-time PCR using gene-specific primer pairs as indicated in (A). The abundance of the different NMT1 gene segments in each immunoprecipitate was expressed as fold enrichment relative to the nontranscribed intergenic region value, arbitrarily set to 1. The values correspond to the means of at least three independent experiments.
Pab2 interacts with the nascent mRNP in the vicinity of the transcription complex
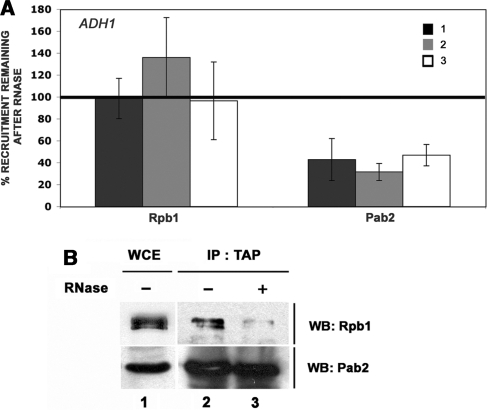
The requirement for active transcription to detect Pab2 cross-linking along the coding region of the NMT1 gene suggested that Pab2 might physically interact with components of the transcriptional machinery. We therefore affinity purified TAP-tagged Pab2 from cell extracts using IgG-sepharose and analyzed the precipitated proteins by immunoblotting. Consistent with a previous study in which the coimmunoprecipitation of PABP2 and RNA Pol II from insect cells was demonstrated (42), our experiments also indicated that the large subunit of yeast RNA Pol II (Rpb1) copurifies with TAP-tagged Pab2, but not with a control purification (data not shown). Importantly, we further examined whether the association between Pab2 and the transcriptional machinery is mediated by the nascent mRNP. To test this possibility, extracts were treated with a cocktail of RNases before ChIP analysis. Whereas the ChIP signals for RNA Pol II were not perturbed upon RNase treatment, the level of Pab2 cross-linking over the entire ADH1 coding region was reduced after the samples were treated with RNases (Figure 4A). These results indicate that the nascent transcript is important for the association between Pab2 and genomic DNA in the ChIP assays. Furthermore, the level of Rpb1 that copurified with Pab2-TAP was reduced after RNase treatment (Figure 4B, lanes 2–3), consistent with the importance of RNA for the association between Pab2 and the transcription machinery. Taken together, these immunoprecipitation experiments suggest that recruitment of Pab2 to transcribed genes is mediated by nascent mRNPs.
Figure 4.
Pab2 interacts with the nascent mRNP in the vicinity of the transcription complex. (A) Rpb1 and Pab2-TAP ChIPs with and without an RNase treatment step were performed using wild-type cells (for Rpb1) as well as using cells that express TAP-tagged Pab2 using methods similar to those described in Figure 1. The coprecipitating DNA was quantified by real-time PCR using ADH1-specific primer pairs as shown in Figure 1. The percentage signal remaining after RNase treatment was calculated as the ratio of the fold enrichment for the RNase-treated sample over the fold enrichment of the sample not treated with RNases. Values correspond to the means of at least three independent experiments. (B) Equal amounts of a whole-cell extract (WCE, lane 1) prepared from Pab2-TAP cells were subjected to affinity purification using IgG-sepharose (lanes 2 and 3). The amount of extract loaded in lane 1 represents 2% of the protein used in the immunoprecipitation. Following extensive washing steps, the beads were treated (lane 3) or not treated (lane 2) with RNases. The eluted proteins were analyzed by western blotting (WB) using antibodies specific to Rpb1 (upper panel) and to the TAP epitope (lower panel).
Pab2 associates with cellular components involved in diverse RNA-related functions
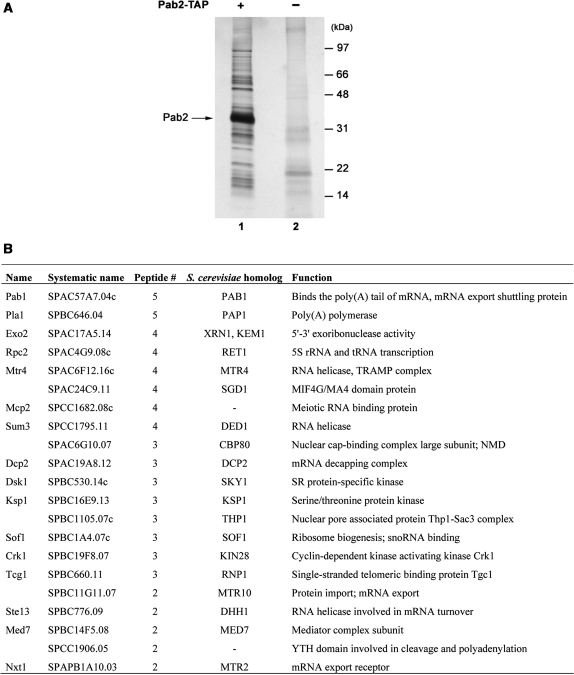
To date, the protein interaction network of the nuclear poly(A)-binding protein remains largely unknown. To get further insights into the mechanism by which Pab2 is cotranscriptionally recruited to nascent mRNPs, a tandem affinity purification approach was used to identify proteins that associate with fission yeast Pab2. Extracts were prepared from 8 l of cells that expressed a TAP-tagged version of Pab2 as well as from untagged control cells. Following two rounds of purification over IgG-sepharose and calmodulin-bound resins, the eluted proteins were resolved by SDS–PAGE and visualized by silver staining. As can be seen in Figure 5A, all of the associated proteins were in substoichiometric amounts relative to Pab2. Analysis of the eluted proteins by mass spectrometry identified unique peptides that corresponded to gene products that are involved in a wide range of RNA-related functions (Figure 5B and Table 2). Notably, peptides from 32 ribosomal proteins, 4 general translation factors, as well as from the cytosolic poly(A)-binding protein (Pab1) were identified (Table 2 and Figure 5B). Peptides that correspond to the evolutionarily conserved nuclear poly(A) polymerase were also detected (Figure 5B), consistent with the role of Pab2 in polyadenylation (27). Other proteins identified that participate in RNA metabolism included the RNA helicase Mtr4, the 5′-to-3′ exonuclease Exo2, the mRNA decapping subunit Dcp2 and the nuclear cap-binding protein Cbp80 (Figure 5B). Protein factors for which the S. cerevisiae orthologs are functionally implicated in mRNA export were also coprecipitated with Pab2; these included the orthologs of the S. cerevisiae proteins Thp1, Mtr10 and Mtr2. Consistent with the aforementioned results (Figure 4), RNA could be responsible for the copurification of some of these proteins. Importantly, with the exception of one ribosomal protein for which some peptides were detected in the control purification, no peptides were detected in the control for any of the proteins indicated in Figure 5B and Table 2. In conclusion, our proteomic analysis suggests that Pab2 is associated with factors involved in several steps of the mRNA life cycle.
Figure 5.
Purification of proteins associated with TAP-tagged Pab2. (A) Proteins copurified with Pab2 by tandem affinity purification (lane 1) were resolved using a Bis–Tris 4–12% gradient SDS–PAGE and analyzed by silver staining. The result for an identically treated extract from control S. pombe is shown in lane 2. Molecular weight markers are shown on the right in kilodaltons (kDa). The position of Pab2 is indicated on the left. (B) Summary of the nonribosomal proteins identified by mass spectrometry from the Pab2-TAP purification.
Table 2.
Ribosomal proteins and translation factors identified in the Pab2-TAP purification
| Name | Systematic name | Function |
|---|---|---|
| Ribosomal proteins | ||
| Rps2 | SPCC576.08c | 40S ribosomal subunit S2 |
| Rps7 | SPAC18G6.14c | 40S ribosomal subunit S7 |
| Rps801 | SPAC2C4.16c | 40S ribosomal subunit S8 |
| Rps802 | SPAC521.05 | 40S ribosomal subunit S8 |
| Rps901 | SPAC24H6.07 | 40S ribosomal subunit S9 |
| Rps902 | SPBC29A3.12 | 40S ribosomal subunit S9 |
| Rps1101 | SPAC31G5.03 | 40S ribosomal subunit S11 |
| Rps13 | SPAC6F6.07c | 40S ribosomal subunit S13 |
| Rps1501 | SPCC1393.03 | 40S ribosomal subunit S15 |
| Rps2202 | SPAC5D6.01 | 40S ribosomal subunit S15a |
| Rps3002 | SPBC19G7.03c | 40S ribosomal subunit S30 |
| Rpl401 | SPBC1711.06 | 60S ribosomal subunit L2 |
| Rpl402 | SPBP8B7.03c | 60S ribosomal subunit L2 |
| Rpl301 | SPAC17A5.03 | 60S ribosomal subunit L3 |
| Rpl302 | SPAPB8E5.06c | 60S ribosomal subunit L3 |
| Rpl6 | SPCC622.18 | 60S ribosomal subunit L6 |
| Rpl701 | SPBC18H10.12c | 60S ribosomal subunit L7 |
| Rpl8 | SPBC29A3.04 | 60S ribosomal subunit L7a (L8) |
| Rpl1101 | SPAC26A3.07c | 60S ribosomal subunit L11 |
| Rpl1201 | SPCC16C4.13c | 60S ribosomal subunit L12A |
| Rpl13 | SPAC664.05 | 60S ribosomal subunit L13 |
| Rpl1801 | SPBC11C11.07 | 60S ribosomal subunit L18 |
| Rpl2001 | SPAC3A12.10 | 60S ribosomal subunit L20a |
| Rpl2101 | SPBC365.03c | 60S ribosomal subunit L21 |
| Rpl2102 | SPAC959.08 | 60S ribosomal subunit L21 |
| Rpl2501 | SPBC106.18 | 60S ribosomal subunit L25 |
| Rpl2802 | SPCC5E4.07 | 60S ribosomal subunit L27a.2/L28A |
| Rpl29 | SPBC776.01 | 60S ribosomal subunit L29 |
| Rpl3001 | SPAC9G1.03c | 60S ribosomal subunit L30 |
| Rpl31 | SPAC890.08 | 60S ribosomal subunit L31 |
| Rpl3601 | SPCC970.05 | 60S ribosomal subunit L36 |
| Rpl3602 | SPBC405.07 | 60S ribosomal subunit L36 |
| Translation factors | ||
| Ef1a-a | SPCC794.09c | Translation elongation factor |
| Moe1 | SPAC637.07 | Translation initiation factor eIF3d |
| Sup35 | SPCC584.04 | Translation release factor class II |
| Tif11 | SPBC25H2.07 | Translation initiation factor eIF1A |
Pab2 is a shuttling protein that associates with translating mRNPs
Fission yeast and human nuclear PABPs are predominantly nuclear at steady state (27,46). Yet, PABP2 was detected in the cytoplasm of human cells by electron microscopy (46) and has been shown to shuttle between the nucleus and cytoplasm (30). To test the capacity of fission yeast Pab2 to shuttle, we used a NUP49-based assay in S. cerevisiae (47) as such an in vivo export assay does not exist in S. pombe. The nup49-313 allele of S. cerevisiae expresses a temperature-sensitive nucleoporin that exhibits defective nuclear import at the nonpermissive temperature without perturbing export of protein and RNA from the nucleus (48). Accordingly, the nup49-313 allele has been used to assay the export of several nuclear RNA-binding proteins (40,47,49).
Consistent with previous reports indicating that S. cerevisiae Nab2 is a shuttling protein (40,50), cytoplasmic accumulation of Nab2 was observed at the nonpermissive temperature (Figure 6G and H), whereas Nab2 was predominantly nuclear at the permissive temperature (Figure 6E and F). The expression of GFP-tagged Pab2 in S. cerevisiae resulted in nuclear localization (Figure 6I and J), consistent with the steady-state localization of Pab2 in fission yeast (27). In contrast, significant GFP signal was detected in the cytoplasm of Pab2-expressing cells that were shifted to the nonpermissive temperature (Figure 6K and L). As a control, a GFP-tagged LacZ protein that includes a strong nuclear localization signal remained in the nucleus at both permissive and nonpermissive temperatures (Figure 6A–D). This control also demonstrated that the cycloheximide treatment before imaging prevented de novo synthesis of GFP-tagged proteins in the cytoplasm. The results of the nuclear export assay indicate that Pab2 can shuttle between the nucleus and the cytoplasm.
Figure 6.
Pab2 shuttles between the nucleus and cytoplasm. The nuclear export assay was performed using S. cerevisiae cells that express the nup49-313 allele as described in the Materials and methods section. The nup49-313 cells expressing either NLS-LacZ-GFP (A–D), Nab2-GFP (E–H) and GFP-Pab2 (I–L) were incubated at 25°C (A, B, E, F, I and J) or shifted to 37°C (C, D, G, H, K and L) before treatment with cycloheximide to block new protein synthesis. Phase contrast (A, C, E, G, I and K) and GFP fluorescence (B, D, F, H, J and L) are shown.
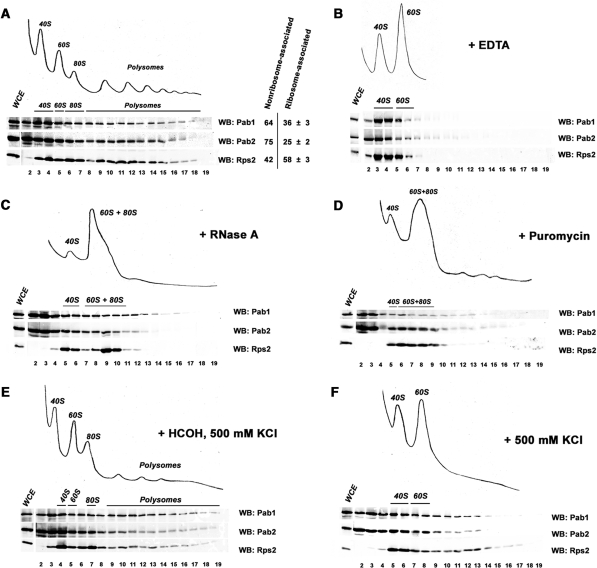
The aforementioned results indicating that Pab2 is a shuttling protein suggested that the detection of ribosomal proteins and translation factors in the eluate of the Pab2-TAP purification could be the result of the association between Pab2 and translating mRNPs. We therefore tested whether Pab2 cosedimented with polysomal mRNAs by examining the fractionation profile of Pab2 after ultracentrifugation using sucrose gradients. As can be seen in Figure 7A, western blot analysis indicated that significant amounts of Pab2 were detected in ribosome-containing fractions (lanes 6–19). We also determined the ratio of ribosome-associated proteins for Pab2, Pab1 and a protein of the small ribosomal subunit, Rps2 (Figure 7A). Roughly 36% of cellular Pab1 cosedimented with ribosomes, consistent with earlier studies (51–53). Notably, our sucrose gradient fractionation experiments indicated that 25% of cellular Pab2 was ribosome-associated (Figure 7A). As expected for a ribosomal protein, most of Rps2 was associated with ribosome-containing fractions (lanes 6–19) and free ribosomal subunits (lanes 3–5). These results indicate that Pab2 cosediments with polysomes.
Figure 7.
Pab2 associates with translating mRNPs. (A–F, top panels) Ribosomal profiles from S. pombe were determined by continuous measurement of rRNAs at 254 nm. (A–F, bottom panels) Pab1, Pab2, and Rps2 were visualized by western blotting using anti-HA, anti-protein A and anti-Rps2, respectively. (A) Sedimentation profile of extracts from Pab2-TAP cells on sucrose gradients. Quantification of the amount of Pab1, Pab2 and Rps2 in nonribosomal fractions (lanes 2–5) and in ribosomal fractions (lanes 6–19) is shown on the right. Values correspond to the means of at least three independent experiments. (B) Sedimentation profile of EDTA-treated extracts from Pab2-TAP cells on sucrose gradients. (C) Sedimentation profile of RNAse-treated extracts from Pab2-TAP cells on sucrose gradients. (D). Sedimentation profile of extracts from puromycin-treated Pab2-TAP cells on sucrose gradients. (E) Sedimentation profile of salt-treated extracts from Pab2-TAP cells that were previously treated with formaldehyde. (F) Sedimentation profile of salt-treated extracts from Pab2-TAP cells on sucrose gradients.
To validate that the sedimentation behavior of Pab2 reflects an association with polysomes, ribosomes were first disrupted into ribosomal subunits by chelating Mg2+ ions. As can be seen in Figure 7B, Pab2 was no longer detected in the heavy fractions of the gradient under these conditions, suggesting that the sedimentation of Pab2 in heavy fractions requires the presence of polysomes. Treating cellular extracts with RNase A prior to ultracentrifugation also perturbed the normal distribution of Pab2 after velocity sedimentation on sucrose gradients (Figure 7C). Specifically, RNase treatment resulted in the disruption of polysomal complexes and the concomitant redistribution of Pab2 to lighter fractions. These results are consistent with the association of Pab2 with translating mRNPs.
To confirm that Pab2 associates with polysomes and not with other large RNPs, we used the translation inhibitor, puromycin. Puromycin specifically disrupts polysomes by causing premature release of nascent peptides as well as of mRNAs from ribosomes (54). Accordingly, treatment of cells with puromycin resulted in the almost complete disappearance of polysomes and the concurrent accumulation of 80S monosomes (Figure 7D). Importantly, Pab1, Pab2 and Rps2 were removed from polysomes-containing fractions after puromycin treatment (Figure 7D). These results demonstrate that Pab2 is specifically associated with polysomal mRNPs.
Given that Pab2 shows nuclear localization at steady state [Figure 6; (27)], it was important to determine whether the association of Pab2 with polysomes occurs in vivo. To test this, cells were cross-linked with formaldehyde prior to lysis, extracts were prepared using high-salt conditions, and separated on sucrose gradients. As can been seen in Figure 7E, a significant amount of Pab2 cosedimented with polysomes under these experimental conditions. Conversely, Pab2 was redistributed to lighter fractions under similar high-salt conditions, but lacking the formaldehyde cross-linking step (Figure 7F). These experiments indicate that the high-salt conditions dissociated proteins that had not been cross-linked to the mRNPs before cell lysis, including Pab2. Our data thus indicate that a fraction of Pab2 is associated to polysomes in vivo.
DISCUSSION
Although experiments using purified versions of the nuclear PABPs (PABP2) have provided insights into its biochemical properties, the functions as well as the network of associations of PABP2 in a cellular context remain poorly understood. We have recently reported the identification of the ortholog of the mammalian nuclear poly(A)-binding protein in S. pombe (27). Consistent with a functional role in poly(A) tail synthesis, RNAs from PAB2-null cells display hyperadenylated 3′-ends (27). In this study, we investigated the recruitment of Pab2 during mRNP formation and the subsequent association between Pab2 and exported mRNPs. Based on our results, we propose that Pab2 is recruited early during the transcription cycle of RNA Pol II genes and remains associated with translating mRNPs after nuclear export. These findings provide novel insights into the events that govern mRNP remodeling as it transits to the cytoplasm.
During eukaryotic transcription, nascent pre-mRNAs are wrapped in ribonucleoprotein complexes that contain factors required for processing and export (55). It is now established that most factors required for processing and export of pre-mRNAs are loaded onto the nascent mRNP as it comes out of the polymerase RNA exit channel. The cotranscriptional loading of RNA processing factors includes several cleavage/polyadenylation factors that show recruitment near the 3′-end of genes as determined by ChIP assays (19–23). The ChIP assays presented in this study indicated that Pab2 is cotranscriptionally recruited to Pol II-specific genes. Given the function of Pab2 in polyadenylation control (27), we propose that the recruitment of Pab2 during transcription is important to increase the local concentration of Pab2 onto the nascent mRNP until the synthesis of a nascent poly(A) tail by the poly(A) polymerase. Pre-mRNA polyadenylation would then provide high-affinity binding sites and favor the rapid transfer of Pab2 to the growing poly(A) tail where it would function in poly(A) tail length control.
Pab2 was found to cross-link to the entire coding region of several Pol II genes. Interestingly, Pab2 showed greater enrichment at the 3′-end as compared with the 5′-end of genes (Figure 1D). The simplest explanation for the increase in association of Pab2 during transcription elongation is that Pab2 is recruited by or rapidly transferred to the nascent mRNP. This interpretation is consistent with the sensitivity of Pab2 ChIP and immunoprecipitation assays to RNase, suggesting that Pab2 is cotranscriptionally recruited via the nascent mRNP. The physical fragmentation of RNA during the sonication step is likely to be the cause for the plateau in ChIP signal detected for Pab2 beyond the middle of the ADH1, PYK1, PGK1 and MET26 genes, and reflects the size of chromatin fragments to <500 bp. Interestingly, our ChIP assays indicated that Pab2 occupancies upstream of the polyadenylation site are sensitive to RNases (Figure 4). These results could imply that the recruitment of Pab2 is mediated by direct binding to non-poly(A) sequences in the pre-mRNA prior to polyadenylation. We do not favor this interpretation, however, as studies using nuclear PABPs from various organisms indicate poor binding to nonpolyadenylated RNA as compared with poly(A) (27,56,57). Analogous to our ChIP experiments, sensitivity of ChIP signals to RNases was previously reported for the budding yeast proteins Sub2 and Gbp2 (37,58). In this case, results suggest that the TREX complex mediates the cotranscriptional recruitment of Sub2 and Gbp2 to nascent mRNPs. Similarly, we predict a model in which Pab2 is recruited to the nascent mRNP via protein interactions. A structure–function analysis of S. pombe Pab2 should provide useful insights into the mechanism that mediates the cotranscriptional recruitment of Pab2 to nascent mRNPs.
The Pab2 ChIP results described herein are in agreement with earlier electron microscopy studies that have found PABP2 in the vicinity of RNA Pol II along the entire Balbiani ring gene of Chironomus tentans (42). This previous study was limited to a single gene, however, and did not address how PAPB2 is recruited during the transcription cycle of RNA Pol II. Our study goes beyond these previous findings and provides evidence for the transcription-dependent recruitment of Pab2 to the NMT1 gene and to the coding sequences of several other yeast genes that are transcribed constitutively. Furthermore, the inclusion of an RNase step in our experiments supports the recruitment and/or the rapid transfer of Pab2 to the nascent mRNP. It remains unclear, however, whether RNA-independent interactions exist between Pab2 and the transcription machinery. Whereas the level of ADH1 mRNA was reduced after RNase-treatment as compared with a nontreated sample, segments of the ADH1 transcript could still be detected by RT-PCR following treatment with RNases (data not shown). It is therefore likely that the remaining signal detected by our immunoprecipitation assays (Figure 4) is due to the incomplete digestion of RNA.
Similar to the ChIP profile of Pab2, other cleavage/polyadenylation factors have also been shown to cross-link at promoters and coding regions in yeast and other eukaryotes (19,22,59,60). Although the functional significance for the recruitment of cleavage/polyadenylation factors during the early phase of the transcription cycle remains unclear, the multi-functional nature of some mRNA processing factors might explain this apparent discrepancy. Interestingly, cellular depletion of Drosophila PABP2 using RNAi results in nuclear accumulation of polyadenylated RNAs (61), suggesting a role for the nuclear poly(A)-binding protein in mRNA export pathways. Studies also indicate that a number of proteins important for efficient mRNA export are recruited during the transcription cycle in budding yeast (37,44,62,63) and humans (64,65). Remarkably, the ChIP profile of many mRNA export factors shows enrichment from the 5′-end to the 3′-end, similar to that of Pab2. Whereas these aforementioned evidence may support a role for the nuclear poly(A)-binding protein in mRNA export, it cannot be excluded that the accumulation of poly(A) RNA in PABP2-depleted Drosophila cells is due to defective polyadenylation. Accordingly, evidence in budding yeast indicate that mRNAs that are not properly polyadenylated are retained in the nucleus (66). Further studies are thus needed to establish whether the nuclear poly(A)-binding protein is directly involved in mRNA export.
Pab2 is not restricted to a pioneer round of translation
mRNP composition changes as it is transferred to the cytoplasm (55). Yet, the molecular details of mRNP remodeling during or after passage through the nuclear pore complex are poorly understood. Based on the different subcellular distribution of PABPC and PABP2 at steady state, cytosolic and nuclear, respectively, it has been proposed that PABPC substitutes for PABP2 after nuclear export of mRNPs. How poly(A)-bound PABP2 is replaced by PABPC during or after nuclear export has remained elusive, however. Importantly, we show here that a significant fraction of Pab2 does not get replaced by Pab1 and remains associated with translating mRNPs. This conclusion is supported by the specific cosedimentation of Pab2 with polyribosomes (Figure 7) as well as the copurification of ribosomal proteins and translation factors with Pab2 (Figure 5 and Table 2). Our studies thus provide the first evidence suggesting the association of the nuclear poly(A)-binding protein with translating mRNPs. The presence of peptides corresponding to Pab1 in the eluate of the Pab2-TAP purification (Figure 5) also suggests that both PABPs can coexist on the same poly(A) tail, in agreement with other studies (67,68).
The association of Pab2 with translating mRNPs is in contrast to previous results that suggest that mammalian PABP2 is not associated with general translation. More precisely, biochemical experiments in human cells have suggested that human PABP2 is restricted to a pioneer round of translation (31). In mammalian cells, it has been proposed that aberrant mRNAs containing nonsense codons are recognized during a pioneer round of translation that is defined by mRNAs bound by the nuclear cap-binding complex proteins, Cbp20 and Cbp80 (31,68). The detection of Pab2 in large polysomal fractions reported here (Figure 7) suggests that Pab2 is not restricted to pioneer rounds of translation, but also associates with actively translated mRNAs. It is therefore possible that the nuclear poly(A)-binding protein performs a slightly different role in mRNA translation between yeast and humans or that technical issues hampered the detection between human PABP2 and translating mRNPs. Our results do not exclude a role for Pab2 in a pioneer round of translation, however. The proteomic analysis of Pab2-associated proteins identified the fission yeast homolog of mammalian Cbp80 (Figure 5), a key constituent of the pioneer round of translation (31,68). Furthermore, two key factors involved in 5′-to-3′ decay of NMD targets, Dcp2 and Exo2, also copurified with Pab2.
The functional significance of the association between Pab2 and translating mRNPs still remains to be determined. Yet, the hypersensitivity of PAB2-null cells to different translational inhibitors (our unpublished data) is consistent with a possible role for Pab2 in translation. Accordingly, a number of predominantly nuclear proteins have recently been shown to cosediment with polysomes and modulate translation in yeast and mammals (53,69–73). Conversely, studies indicate that budding yeast Pab1 (homolog of mammalian PABPC) is a shuttling protein that is important for the proper assembly of mRNPs in the nucleus (9,10). Our findings that Pab2 is associated with translating mRNPs are in concert with the conclusions of the aforementioned studies and illustrate the integrated connections between different steps of mRNP formation in the nucleus and the fate of the mature mRNA in the cytoplasm. Future studies on Pab2 will certainly provide insight into the mechanisms that coordinate nuclear polyadenylation and translation in the cytoplasm.
FUNDING
Natural Sciences and Engineering Research Council of Canada; New Investigator Award from the Canadian Institutes of Health Research (to F.B.). Funding for open access charge: Human frontier science program organization (HFSPO).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Anita Corbett for S. cerevisiae strains and plasmids for the nuclear export assay; members of the Bachand lab for critical reading of the manuscript.
REFERENCES
- 1.Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 5.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3'-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 6.Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 9.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chekanova JA, Belostotsky DA. Evidence that poly(A) binding protein has an evolutionarily conserved function in facilitating mRNA biogenesis and export. RNA. 2003;9:1476–1490. doi: 10.1261/rna.5128903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Proudfoot N, O'Sullivan J. Polyadenylation: a tail of two complexes. Curr. Biol. 2002;12:R855–R857. doi: 10.1016/s0960-9822(02)01353-2. [DOI] [PubMed] [Google Scholar]
- 15.Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 16.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 17.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 18.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 19.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Runner VM, Podolny V, Buratowski S. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol. Cell Biol. 2008;28:1883–1891. doi: 10.1128/MCB.01714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swinburne IA, Meyer CA, Liu XS, Silver PA, Brodsky AS. Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription. Genome Res. 2006;16:912–921. doi: 10.1101/gr.5211806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 25.Kerwitz Y, Kuhn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 2003;22:3705–3714. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 27.Perreault A, Lemieux C, Bachand F. Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J. Biol. Chem. 2007;282:7552–7562. doi: 10.1074/jbc.M610512200. [DOI] [PubMed] [Google Scholar]
- 28.Cole CN, Scarcelli JJ. Transport of messenger RNA from the nucleus to the cytoplasm. Curr. Opin. Cell Biol. 2006;18:299–306. doi: 10.1016/j.ceb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Stewart M. Ratcheting mRNA out of the nucleus. Mol. Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Calado A, Kutay U, Kuhn U, Wahle E, Carmo-Fonseca M. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA. 2000;6:245–256. doi: 10.1017/s1355838200991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 32.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 33.Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- 34.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 35.Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu MC, Bachand F, McBride AE, Komili S, Casolari JM, Silver PA. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachand F, Silver PA. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004;23:2641–2650. doi: 10.1038/sj.emboj.7600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen KH, Szamecz B, Valasek L, Jivotovskaya A, Shin BS, Hinnebusch AG. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bear DG, Fomproix N, Soop T, Bjorkroth B, Masich S, Daneholt B. Nuclear poly(A)-binding protein PABPN1 is associated with RNA polymerase II during transcription and accompanies the released transcript to the nuclear pore. Exp. Cell Res. 2003;286:332–344. doi: 10.1016/s0014-4827(03)00123-x. [DOI] [PubMed] [Google Scholar]
- 43.Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol. Cell. 2007;26:145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 46.Krause S, Fakan S, Weis K, Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- 47.Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 48.Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duncan K, Umen JG, Guthrie C. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 2000;10:687–696. doi: 10.1016/s0960-9822(00)00527-3. [DOI] [PubMed] [Google Scholar]
- 51.Anderson JT, Paddy MR, Swanson MS. PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell Biol. 1993;13:6102–6113. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mangus DA, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azzam ME, Algranati ID. Mechanism of puromycin action: fate of ribosomes after release of nascent protein chains from polysomes. Proc. Natl Acad. Sci. USA. 1973;70:3866–3869. doi: 10.1073/pnas.70.12.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc. Natl Acad. Sci. USA. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benoit B, Nemeth A, Aulner N, Kuhn U, Simonelig M, Wahle E, Bourbon HM. The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic Acids Res. 1999;27:3771–3778. doi: 10.1093/nar/27.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wahle E, Lustig A, Jeno P, Maurer P. Mammalian poly(A)-binding protein II. Physical properties and binding to polynucleotides. J. Biol. Chem. 1993;268:2937–2945. [PubMed] [Google Scholar]
- 58.Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl Acad. Sci. USA. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calvo O, Manley JL. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 2005;24:1009–1020. doi: 10.1038/sj.emboj.7600575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farny NG, Hurt JA, Silver PA. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev. 2008;22:66–78. doi: 10.1101/gad.1616008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lei EP, Silver PA. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 64.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 65.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 67.Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev. Cell. 2005;9:511–522. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 68.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 71.Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 72.Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol. Cell. 2008;29:255–262. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 74.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]