Abstract
We developed a simple, direct and cost-effective approach to search for the most likely target genes of a known microRNA (miRNA) in vitro. We term this method ‘labeled miRNA pull-down (LAMP)’ assay system. Briefly, the pre-miRNA is labeled with digoxigenin (DIG), mixed with cell extracts and immunoprecipitated by anti-DIG antiserum. When the DIG-labeled miRNA and bound mRNA complex are obtained, the total cDNAs are then subcloned and sequenced, or RT–PCR-amplified, to search for the putative target genes of a known miRNA. After successfully identifying the known target genes of Caenorhabditis elegans miRNAs lin-4 and let-7 and zebrafish let-7, we applied LAMP to find the unknown target gene of zebrafish miR-1, which resulted in the identification of hand2. We then confirmed hand2 as a novel target gene of miR-1 by whole-mount in situ hybridization and luciferase reporter gene assay. We further validated this target gene by microarray analysis, and the results showed that hand2 is the top-scoring among 302 predicted putative target genes. We concluded that LAMP is an experimental approach for high-throughput identification of the target gene of known miRNAs from both C. elegans and zebrafish, yielding fewer false positive results than those produced by using only the bioinformatics approach.
INTRODUCTION
A mature miRNA is a 19–30-nt-long non-coding RNA excised from a 70-nt pre-miRNA (dsRNA hairpin) by Dicer (1–4). It is clear that miRNAs play essential roles in gene expression, development, specification and differentiation of cell fate in animals (5–8). The first miRNA found in Caenorhabditis elegans, lin-4, targets the 3′ untranslated regions (UTRs) of lin-28 and lin-14 to impede translation (9,10). Investigators have also reported miRNAs in mice, such as miR-196, which is involved in homeobox gene regulation (6), and miR-208, which is required for cardiomyocyte hypertrophy, fibrosis and βMHC expression (11). In zebrafish embryos, miRNAs are processed to silence genes during brain development and morphogenesis (7). Up to now, an estimated 5234 mature miRNAs have been found in primates, rodents, birds, fish, worms, flies, plants and viruses (see ftp://ftp.sanger.ac.uk/pub/mirbase/sequences/CURRENT /README). These miRNAs may control up to 30% of genes in animals (12–14). Thus far, however, only a few known miRNAs have been matched to their target genes (8).
The use of bioinformatics, which has been the conventional method for determining the target gene for miRNAs, matches similar sequences in a database on the basis of free-energy prediction (ΔG) and thermodynamic calculations for their binding affinities. Then, the putative genes are predicted according to the conserved region of the 3′UTR sequences (15,16). Although some target genes have been successfully predicted for a few miRNAs, a large number of potential targets are falsely predicted because of imperfect pairing between nucleotides of the miRNA and the target mRNA. Moreover, some true target sequences at the 3′UTR for a particular miRNA are extremely hard to predict because such target sequences may contain multiple elements that lack canonical seed pairing and therefore require a combinatorial binding of weak sites, such as miR214-dispatched homolog 2, as reported by Li et al. (17). Alternatively, microarray analysis has been used to compare the relative expression level of mRNA in the presence or absence of the miRNA. While this method has obtained at least 100–200 putative target genes for a known miRNA (18), it is tedious to apply, and changes in the level of mRNA often make it difficult to recognize the real target gene. Recently, Easow et al. (19) and Beitzinger et al. (20) used immunoprecipitation by an anti-HA (Ago1 fused to HA tag) monoclonal antibody, or an anti-Ago monoclonal antibody, to search for target genes of a known miRNA. However, Ago is an RNA-binding protein which therefore enables anti-HA/Ago to bind any kind of unexpected RNAs (21). Thus, non-specific binding may occur between target mRNAs and any proteins. These conditions lower the possibility of cloning the exact target gene for a known miRNA; therefore, a relatively simple, direct and cost-effective strategy to identify the target genes of miRNAs is required.
Bypassing both the bioinformatics and microarray methods, we have developed an experimental approach to search for the target gene of a known miRNA. First, we labeled pre-miRNA with digoxigenin (DIG) and then mixed it with cell extracts. The endogenous Dicer cuts this complex in vitro and generates mature miRNA. Next, this DIG-labeled miRNA is attached to its target gene(s) by the endogenous RNA-induced silencing complex (RISC). The mixtures of miRNA–target mRNA are then pulled down by anti-DIG antiserum. In order to finally identify the target genes of a given miRNA, we clone all cDNAs from total mRNAs after they are pulled down for further DNA sequencing or cloning out by reverse transcriptase polymerase chain reaction (RT–PCR). In addition, to increase the degree of certainty in our method, we also employ microarray analysis to analyze all mRNAs after they are pulled down, further validating the target genes predicted by the Labeled miRNA pull-down (LAMP) assay system we describe here.
MATERIALS AND METHODS
Experimental animals
The zebrafish AB strain and C. elegans var Bristol N2 (22) were used and maintained following the standard conditions (23). The procedures for synchronized development of C. elegans were described by Brenner (22).
Construction of pre-miRNAs
All oligonucleotide sequences used in this study are shown in Supplementary Table 1. The pre-miRNAs of C. elegans, such as pre-lin-4 (GI:434669), pre-let-7 (GI: 10799037) and the mutated pre-let-7, were produced by PCR after amplification using three primers that were designed with some overlapping sequences. Pre-lin-4 was generated by the primers Cel-Pre-lin4-1F, Cel-Pre-lin4-2F and Cel-Pre-lin4-1R. The primers were adjusted to a final concentration of 10 μM with distilled water. The mixture was heated to 95°C for 10 min, then chilled to room temperature and amplified for 35 cycles. The PCR product was ligated into the pGEMT-Easy vector (Promega). Pre-let-7 was generated under the same procedures, except that primers Cel-Pre-let7-1F, Cel-Pre-let7-2F and Cel-Pre-let7-1R were used. The mutated pre-let-7 was generated by using Cel-Pre-let7M-1F, Cel-Pre-let7M-2F and Cel-Pre-let7M-1R. The pre-miRNA of zebrafish pre-let-7 (GI:4837139) (ZF-let-7) was synthesized by using the primers ZF-Pre-let7-1F, ZF-Pre-let7-2F and ZF-Pre-let7-1R. Pre-miR-1 (GI:55859956) (ZF-miR-1) was generated by using the primers ZF-Pre-miR-1-1F, ZF-Pre-miR-1-2F, ZF-Pre-miR-1-1R and ZF-Pre-miR-1-2R.
Plasmid constructs
The 0.1-kb fragment of NotI-digested pGEMT-pre-miR-1, in which pre-miR-1 was ligated into the pGEMT-Easy vector (Promega), was ligated to NotI-digested pCMV-DsRed-1 (Clontech) or pCMLCE-(−870/787) (24) to generate pCMV-DsRed-miR-1 or pCMLC-EGFP-miR-1, respectively. The primers for detecting zebrafish hand2-3′UTR were used to amplify a 424-bp PCR product from a template of embryonic cDNA at 24 h post-fertilization (hpf). The PCR product was inserted into the pGEMT-Easy vector to produce a plasmid phand2-3′UTR. A fragment obtained from the NotI-digested phand2-3′UTR was ligated to the NotI-digested pCMV-EGFP (25) to generate pCMV-EGFP-hand2-3′UTR. Plasmid cmlc2-Luciferase-hand2-3′UTR, containing 1.2 kb of Rluc from an XhoI–XbaI-cut phRL-Null vector (Promega), was ligated into the XhoI–XbaI-cut pCMLCE-(−870/787) vector which contains a regulatory segment from −870 to +787 of zebrafish cardiac myosin light chain 2 gene (cmlc2; 21).
Cell extracts
To avoid deactivation of proteins, we collected cell extracts by the following procedures. Approximately 2000 48-hpf zebrafish embryos were collected and spun at 800 × g at room temperature for 10 min to remove water before cell extract buffer (15 mM Tris–base pH 7.5, 250 mM sucrose, 2 mM EDTA) was added. After they were evenly mixed, the mixture was spun at 800 × g for 10 min. The supernatant was saved, mixed with cell extract buffer and centrifuged again. Then, 2 mM phenylmethylsulfonylfluoride and 100 U of rRNasin (Promega) were added to the supernatant before they were broken by the ultrasonic processor (Sonics & Materials, Inc.) at grade 6 with an interval of 2 s for 30 min at 4°C. After they were centrifuged at 15 600 × g for 30 min at 4°C, the clear cell extract was collected and used immediately. The procedure of collecting the cell extract of C. elegans was the same as that used for zebrafish, except that 5000 C. elegans at stage L1 or L2–L4 were collected, washed with M9 buffer (22) and spun down at 800 × g for 10 min.
Pull-down assay of DIG-labeled pre-miRNA experiments
DIG-labeled pre-miRNA was synthesized by using the DIG RNA-labeling kit (Roche). After cell extracts were incubated with 70 μg of digoxigeninylated RNA at 4°C for 30 min, the total volume was adjusted to 1 ml with binding buffer (25 mM Tris–base pH 7.4, 60 mM KCl, 2.5 mM EDTA, 0.2% Triton X-100, 80 U of rRNasin), and the mixture was incubated at 30°C for 60 min. The sample was transferred to a tube containing 20 μl of anti-DIG agarose beads and rotated slowly overnight at 4°C. After the mixture was spun down at 15 600 × g for 30 min at 4°C, it was washed with washing buffer (20 mM Tris–base pH 7.4, 350 mM KCl, 0.02% NP-40) and spun again at for 15 min at 4°C. Washing and spinning were repeated five times before the binding buffer was added and the sample heated to 95°C for 15 min. A clear lysate was obtained after the sample was spun down at 15 600 × g for 10 min at 4°C. The purified RNA was collected when DNase I (20 U) was added and incubated at 37°C for 30 min before the phenol/chloroform extraction was performed. For the control experiment, we completely depleted Dicer from the cell lysate by using anti-Dicer antibody (Santa Cruz), following the protocol described for ExactaCruz™ (Santa Cruz). For the western blot analysis of Ago, we followed the protocol described by Ørom et al. (26), except the primary antibody against Ago (Santa Cruz) was diluted at 1:1000.
RT–PCR
RT–PCR was performed by using the total RNA extracted from the pull-down assay. RNA (1 μg) was used for first-strand cDNA synthesis with SuperScript II (Invitrogen), and reverse transcription was performed with oligo(dT)-T7 primers and Template Switch-based (TS) primers. We then mixed 20 μl of the first-strand cDNA with 130 μl of solution (3 μl of Ex Taq Polymerase, 1 μl of RNase H [5 U/µl], 3 μl of 10 mM dNTP, 15 μl of Ex Taq PCR buffer and 108 μl of DEPC-treated water) to synthesize the second strand. The mixture was incubated at 37°C for 5 min to digest mRNA, denatured at 94°C for 2 min and specific primers were annealed at 65°C for 3 min, with extension at 75°C for 30 min. The reaction was stopped by adding 7.5 μl of 1 M NaOH solution containing 2 mM EDTA and incubating at 65°C for 10 min. The double-stranded cDNA was purified using the phenol/chloroform process. PCR reactions were run with 1 μl of reverse transcription template for 35 cycles under the following primers. For amplification of C. elegans lin-41 (GI:71980712), primers Cel-lin41F and Cel-lin41R were used; for C. elegans hbl-1 (GI:4323034), primers Cel-Hbl-1F and Cel-Hbl-1R were used; for C. elegans lin-14 (GI:17568924), primers Cel-lin14F and Cel-lin14R were used; and for C. elegans lin-28 (GI:1765993), primers Cel-lin28F and Cel-lin28R were used. The primers used to amplify C. elegans eft-2 (GI:156278), which served as a positive control were Cel-eft2F and Cel-eft2R. The primers used to amplify the zebrafish lin-41 (Al794385) were ZF-lin41F and ZF-lin41R, and those used for zebrafish hand2-3′UTR (Al794385) were ZF-hand2-3′UTR-F and ZF-hand2-3′UTR-R. The primers used to amplify β-actin (GI:304429), which served as a positive control, were ZF-β-actinF and ZF-β-actinR. The unknown putative target genes were amplified by PCR using primers SP6 and TS. The resultant PCR products were ligated into the plasmid pGMET-Easy.
Maturation of DIG-labeled pre-miRNA
Cell extract, 50 μl, was mixed with 3 μg of DIG-labeled RNA and incubated at 4°C for 30 min. The total volume was adjusted to 0.5 ml with binding buffer, and the mixture was incubated at 30°C for 60 min. The RNA fragment was extracted by the phenol/chloroform method. After extraction, the products were analyzed on a 4% agarose gel and transferred onto a nitrocellulose membrane (Amersham). After RNA was ultraviolet-crosslinked to the membrane, hybridization was carried out, and signals were detected using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate reagent, as recommended by the manufacturer (Roche).
Cell culture maintenance and fluorescent signal observation
Monkey kidney COS-1 cells were used because they do not have endogenous miR-1 activity (27). COS-1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Biowest) containing 10% fetal bovine serum (Biowest), heat-inactivated by incubating for 30 min at 56°C, supplemented with 1× penicillin/streptomycin/glutamine (Biowest), and then incubated at 37°C in an atmosphere of 5% CO2 and 95% air. Fresh culture medium was provided every 2 or 3 days, and the cells were subcultured before reaching 70% confluency. About 1 × 105 cells were seeded onto each well of six-well plates for 24 h prior to transfection. COS-1 cells were transfected by constructs using the lipofectamine method (Invitrogen), according to the manufacturer's instructions. Fluorescent signals of green fluorescent protein (GFP) or red fluorescent protein (RFP) were observed in the 2-day-old COS-1 cells under fluorescence microscope (MZ FLIII, Leica).
Dual-luciferase assay
Mixtures of 15 ng of pcmlc2-Luciferase-hand2-3′UTR, 15 ng of pcmlc2-EGFP-miR-1 and 300 pg of phRG-TK (Promega) were prepared for microinjection; 2.3 nl each was microinjected into one-cell stage embryos. Forty-eight hours after injection, 20 embryos were harvested for luciferase assay by using the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was measured from three separate experiments in a Luminoskan Ascent system (Thermo Labsystems).
Microarray analysis
Microarray analysis was used to search for the targets of miR-1 by comparing the composition and level of total RNAs (3 µg) pulled down in the pool, both by the original wild-type (WT) pre-miR-1 probe and by the mutated type (MT) pre-miR-1 probe, ugcauaguaaagaaguauguau, in which three substitutive nucleotides at the 5′ end of miR-1 are underlined. The pre-miR-1 and the mutated pre-miR-1 were tagged with DIG and mixed into the cell extracts obtained from the embryos at 24–48 hpf. The miRNA/protein (miRNP)/mRNA complexes were pulled down by anti-DIG which was coated on agarose beads. The bound mRNAs were isolated and then identified by microarray analysis. The Zebrafish Oligo Microarray Kit (Agilent, Taiwan), which contains 95 000 probes covering 45 000 genes, was used, and the microarray data were analyzed by Welgene Biotech Co., Ltd. (Agilent, Taiwan) by using an Agilent Certified Service Provider Program. Because of the lack of either 28S or 18S, we added DIG-tagged cTnnT2 mRNA in the pool to monitor the amount of target mRNA in the WT and MT probe groups on the basis of the amount of DIG-tagged cTnnT2. Two independent affinity purification experiments from DIG-tagged-pre-miR-1 cell lysis and from DIG-tagged-pre-mutated-pre-miR-1 cell lysis were analyzed and ranked according to the relative enrichment in the miR-1 affinity purifications. Finally, the WT signal was normalized with the signal of MT (WT/MT) to quantify the target mRNAs. Microarray analysis was performed using a one-color strategy.
RESULTS
Normal maturation of DIG-tagged pre-miRNA
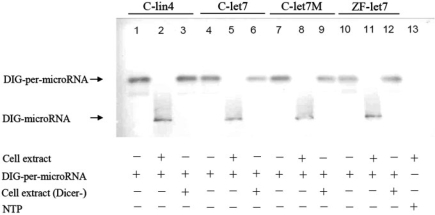
When we labeled pre-miRNAs, such as pre-lin4 and pre-let7, from C. elegans with DIG and performed northern blot analysis, we found that the mature miRNA was processed normally in the presence of cell extracts (lanes 2 and 5, Figure 1). Interestingly, the mature miRNA was processed normally for DIG-labeled Cel-pre-let7M, which contains the mutated sequences, without altering the hairpin formation, secondary structure, or Tm of Cel-pre-let7 (lane 8, Figures 1 and 2A). This is consistent with the result obtained from the zebrafish DIG-labeled pre-let7 (lane 11, Figure 1). In our method, the endogenous Dicer cuts this complex in vitro and generates mature miRNA. However, when Dicer was completely depleted from the cell extracts (Supplementary Figure 1), the miRNA was not processed (lanes 3, 6, 9 and 12, Figure 1). Taken together, we concluded that the DIG tagging does not affect the ability of Dicer to process pre-miRNA into mature miRNA.
Figure 1.
The mature microRNA (miRNA) was processed from the pre-miRNA labeled with digoxigenin (DIG). Pre-miRNAs of Cel-pre-lin4 (C-lin4), Cel-pre-let7 (C-let7), Cel-pre-let7M (C-let7M) from Caenorhabditis elegans and pre-miRNA of ZF-pre-let7 (ZF-let7) from zebrafish were labeled with DIG, and northern blot analysis was performed. The pre-miRNA was neither processed into mature miRNA in the absence of cell extracts from C. elegans (lanes 1, 4 and 7) and zebrafish (lane 10) nor in the absence of dicer (Dicer−), which was depleted by anti-dicer immunoprecipitation (lanes 3, 6, 9 and 12). However, the mature miRNA was normally processed in the presence of cell extracts (lanes 2, 5, 8 and 11), indicating that the DIG-tagged pre-miRNA does not affect the formation of mature miRNA.
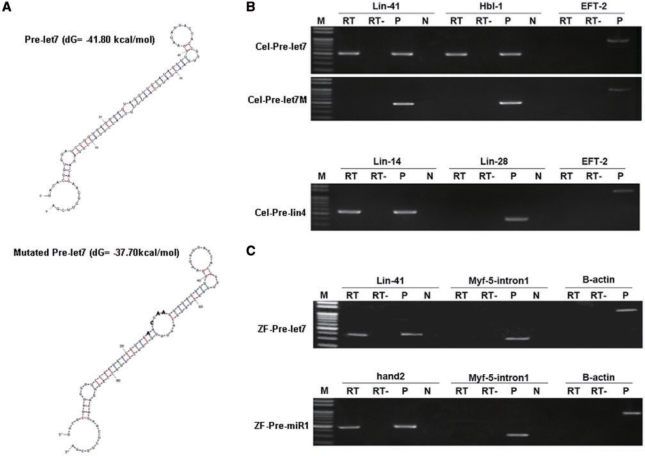
Figure 2.
Using the pull-down assay to determine the genes that are targeted by miRNA. Four miRNA-related genes were studied: two from Caenorhabditis elegans (let-7 and lin-4, Cel) and two from zebrafish (let-7 and miR-1, ZF). The target genes for Cel-pre-let7, Cel-pre-lin4 and ZF-pre-let7 are known, whereas the target gene(s) for ZF-pre-miR-1 is unknown. After pre-let7 (Cel-pre-let7, ZF-pre-let7), pre-lin4 (Cel-pre-lin4) and pre-miR-1 (ZF-pre-miR-1) were labeled with digoxigenin (DIG) and incubated with the cell extracts, RT–PCR was used to amplify the mRNA obtained from the pull-down assay. (A) The secondary structure and free energy predicted by the software program mfold (version 3.2) is shown. For the negative control, we designed a mutated sequence of pre-let7 of C. elegans (Cel-pre-let7M). The altered sequences of Cel-pre-let7M are indicated by boldface type. (B) After pull-down by DIG-Cel-pre-let7, we used RT–PCR to detect whether the known target genes, lin-41 and hb1-1, had been obtained. As expected, both lin-41 and hbl-1 were positive, but not EFT-2 (upper panel). Meanwhile, lin41, hbl-1 and EFT-2 were not positive when DIG-Cel-pre-let7M was used for pull-down and RT–PCR was used for detection (middle panel). Following a similar strategy, we detected the target genes lin-14 and lin-28 for Cel-pre-lin4 when DIG-Cel-pre-lin4 was used for pull-down (bottom panel). (C) After pull-down by DIG-ZF-pre-let7, the target gene zebrafish lin-41 was obtained, as expected, whereas neither β-actin nor myf5-intron1 (the first intron segment of the zebrafish myf5 gene) was positive, suggesting that there was no contamination of genomic DNA in the template (upper panel). Interestingly, when DIG-ZF-pre-miR-1 was used, a novel putative target gene, hand2, was obtained (bottom panel). M, molecular marker; RT, reverse transcriptase was added; RT–, reverse transcriptase was not added; P, positive control, template DNA was from C. elegans cDNA; and N, negative control, template DNA was not added.
Our method next calls for the attachment of DIG-labeled miRNA to its target gene(s) by the endogenous RISC. In order to know whether the mutated let-7 is picked up correctly by RISC, we used immunoprecipitation by following the protocol described by Ørom et al. (26) to obtain the complexes of DIG-tagged mutated let-7 from cell extracts. Then, we used western blot analysis to detect the presence of Ago, which is the most essential component within RISC. Since Ago was detected in the complex of DIG-tagged mutated let-7 (Supplementary Figure 2), we were able to confirm that the mutated let-7 is picked up correctly by RISC.
Testing whether LAMP can obtain the known target genes of mature miRNA
We next tested the ability to LAMP to identify the known target genes of two mature miRNAs from C. elegans, let-7 (Cel-pre-let7) and lin-4 (Cel-pre-lin4). When Cel-pre-let7 and Cel-pre-lin4 were labeled with DIG and incubated with the cell extracts obtained from C. elegans, we used RT–PCR to amplify the putative mRNA obtained from the pull-down assay by specific primers. For the control group, we designed a Cel-pre-let7M, which contained the mutated sequences of Cel-pre-let7 (Figure 2A). After pull-down by DIG-Cel-pre-lin4 and DIG-Cel-pre-let7, the known target genes, lin-14 and lin-28 and lin-41 and hbl-1, were obtained, respectively (Figure 2B). As expected, Cel-pre-let7M did not target the lin-41 and hbl-1 genes (Figure 2B).
Next, we examined two miRNAs from zebrafish, let-7 (ZF-pre-let7) and miR-1 (ZF-pre-miR-1). While the target gene for ZF-pre-let7 was known (28), the target gene for ZF-pre-miR-1 is unknown. Similar to the strategy used for Cel-pre-let7, the target gene zebrafish lin-41 was found after the processing of ZF-pre-let7 by LAMP (Figure 2C). Neither a housekeeping gene, such as β-actin, nor genomic DNA, such as the first intron of the zebrafish myf5 gene (23), was detected (Figure 2C). The latter was used to detect the contaminant genomic DNA.
Using LAMP to search for the unknown target genes of mature miRNA
After the DIG-ZF-pre-miR-1 was processed by LAMP, we analyzed 576 clones by the PCR method and found that there were 465 clones with an insert fragment ranging from 100 to 300 bp. There were 332 clones less than 200 bp, 133 clones between 200 and 300 bp and 11 clones more than 300 bp. We randomly selected 20 clones out of the group of 133 clones, analyzed their sequences and found that they were all meaningless clones, containing more than 10 oligo(dT) repeated sequences that were generated by random ligation of the primers used in RT–PCR. However, when we analyzed the 11 clones of more than 300 bp, we found that six of them contained the partial overlapping sequences of 3′UTR of zebrafish hand2. Furthermore, all of them contained an identical binding site for miR-1 (Supplementary Figure 3) predicted by the alignment analysis between miR-1 and hand2 3′UTR on the basis of the Miranda software results. Meanwhile, we also detected zebrafish hand2 by using hand2-specific primers (Figure 2C) in the DIG-miR-1 pull-down RNA. In summary, after using LAMP to process ZF-pre-miR-1, we analyzed 576 clones and found that there were 11 putative clones containing the target gene. Over 50% (6 of 11) of the putative clones were proven to contain the zebrafish hand2 gene, a finding which has never before been reported in zebrafish. The remaining five clones were two unknown EST clones expressing in somites (data not shown), which were co-localized with the zebrafish miR-1 signal.
Zebrafish miR-1 targets the 3′UTR of the hand2 gene
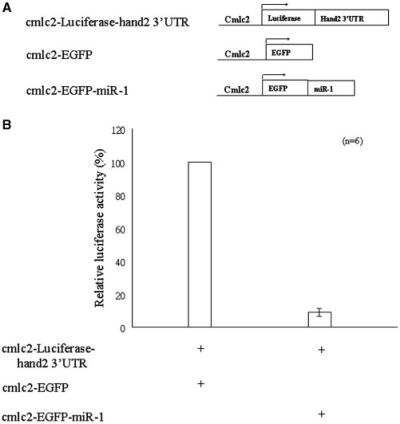
To validate whether the hand2 gene is the target gene of ZF-miR-1, we engineered three constructs: cmlc2-Luciferase-hand2-3′UTR, cmlc2-EGFP and cmlc2-EGFP-miR-1. After these plasmids were microinjected into the zygotes of zebrafish, the transgenic embryos that expressed the transgene and heart-specific GFP signal were collected, and their luciferase activity was quantified at 48 hpf. Compared to the luciferase activity driven by injection of the combined cmlc2-Luciferase-hand2-3′UTR and cmlc2-EGFP plasmids, the relative luciferase activity was dramatically reduced to 10% when embryos were injected with the combined cmlc2-Luciferase-hand2-3′UTR and cmlc2-EGFP-miR-1 plasmids (Figure 3). This finding suggests that hand2 is the target gene of ZF-miR-1.
Figure 3.
Using luciferase gene as a reporter to demonstrate that miRNA derived from zebrafish miR-1 targeted the 3′ untranslated region (UTR) of the zebrafish hand2 gene and suppressed gene expression in zebrafish embryos. (A) To validate whether the hand2 gene was the target gene of ZF-miR-1, we engineered three constructs: cmlc2-Luciferase-hand2-3′UTR, cmlc2-EGFP and cmlc2-EGFP-miR-1. (B) Compared to the luciferase activity driven by injection of the combined cmlc2-Luciferase-hand2-3′UTR and cmlc2-EGFP, the relative luciferase activity was dramatically reduced to 10% when the zebrafish embryos were injected with the combined cmlc2-Luciferase-hand2-3′UTR and cmlc2-EGFP-miR-1. The error bar indicates standard deviation (n = 6).
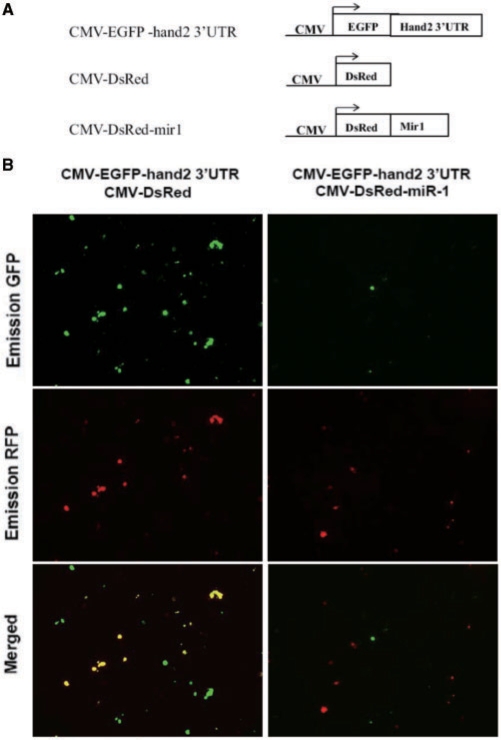
Next we investigated whether the 3′UTR of hand2 is the target site of miR-1 in vitro. Three expression plasmids, pCMV-EGFP-hand2-3′UTR, pCMV-DsRed-miR-1 and pCMV-DsRed, were constructed and transfected into COS-1 cells. After transfection, we found that both the GFP and RFP signals were observed in the COS-1 cells that were transfected with pCMV-EGFP-hand2-3′UTR and pCMV-DsRed (Figure 4, left panel). However, only the RFP signals appeared in the COS-1 cells that were transfected with pCMV-EGFP- hand2-3′UTR and pCMV-DsRed-miR-1 (Figure 4, right panel), suggesting that the miRNA of miR-1 targets the hand2 gene and results in suppressing EGFP reporter gene expression.
Figure 4.
Using fluorescent protein genes as reporters to demonstrate that miRNA derived from zebrafish miR-1 targeted the 3′UTR of the zebrafish hand2 gene and suppressed gene expression in the cell line. (A) To validate whether the hand2 gene was the target gene of ZF-miR-1, we engineered three constructs: pCMV-EGFP-hand2-3′UTR, pCMV-DsRed and pCMV-DsRed-miR-1. Then, these plasmids were transfected into COS-1 cells. (B) The expression of reporter genes was observed under fluorescence microscope at the wavelengths suitable for detection of green fluorescent protein (GFP) or red fluorescent protein (RFP).
Validation of LAMP assay system by microarray analysis
We also used microarray analysis to validate the targets of miR-1 identified from the LAMP assay system. The miRNA/protein(miRNP)/mRNA complexes isolated from DIG-tagged-pre-miR-1 (WT) and DIG-tagged-pre-mutated-miR-1 (MT) were analyzed and ranked according to the relative enrichment in the miR-1 affinity purification. After the WT signal was normalized with the signal of MT (WT/MT), we found 302 putative target genes whose WT/MT was over 1 (Supplementary Table 2). Four genes among the upper 2% of these putative target genes have already been proven to be the true target genes of miR-1 in other species (Table 1). Among them, hand2 has the highest scoring WT/MT value. Thus, it is quite reasonable to consider hand2 as the target gene of miR-1. In addition, like the DIG-tagged mutated let-7, we also used immunoprecipitation and western blot analyses to examine whether the DIG-tagged miR-1 and the DIG-tagged mutated miR-1 are picked up correctly by Ago. Results showed that Ago was detected in the complexes of both DIG-tagged miR-1 and DIG-tagged mutated miR-1 (Supplementary Figure 2). On the basis of microarray data, it is clear that the target gene cannot be accessed by DIG-tagged mutated miR-1, even though Ago can bind to it. Thus, we suggest that the correct target genes of miR-1 are not included in the complex of DIG-tagged mutated miR-1 and RISC.
Table 1.
mRNAs differentially purified by association with miR-1
| Accession number | Normalized ratio (WT/MT) | Reported in other species |
|---|---|---|
| NM_131626 (hand2) | 53 163 | * |
| NM_131397 (hsp70) | 21 741 | * |
| ENSDART00000088193 (HCN) | 17 217 | * |
| NM_001003412 (epn4) | 2381 | |
| CK873184 | 1717 | |
| ENSDART00000047113 | 1093 | |
| NM_001034186 (Nedd4a) | 995.7 | * |
| XM_001338433 | 929.5 | |
| CT638534 | 900.7 | |
| XM_692635 | 802.4 | |
| NM_173228 | 795.9 | |
| CT586204 | 731.1 | |
| NM_001007339 | 696.7 | |
| CT673410 | 673.8 | |
| NM_130970 (islet2a) | 633.2 | |
| EE707462 | 572.2 | |
| ENSDART00000041802 | 570.4 | |
| ENSDART00000054564 | 541.9 | |
| EB862259 | 541.5 | |
| CT688048 (suclg1) | 511.7 | |
| NM_001044953 | 510.6 | |
| XM_001340895 | 500.5 |
Putative target genes were selected from the microarray analysis on the basis of enrichment of ≥500-fold and located at the first upper 7% of candidates. Column 1 shows the accession number or gene name; column 2 shows that the level of mRNA expressed in the WT of pre-miR-1 is divided by that expressed in the MT of pre-miR-1 determined by using cDNA microarray hybridization with total RNA samples. DIG-tagged cTnnT2 served as a control for both WT and MT of miR-1. The last column shows that the target genes of miR-1 have been reported in other species (denoted by asterisks).
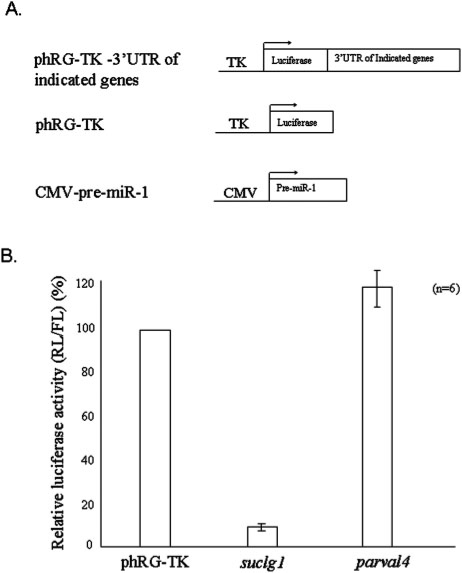
For further analysis, we also selected two other putative genes, suclg1 and parvalbumin 4, because these two genes were also expressed in the zebrafish trunk somites and were scored within the upper 7% and 25%, respectively. When we separately engineered the 3′UTR of these two cDNAs at the downstream of luciferase reporter gene and transfected into COS-1 cells with miR-1, we found that the luciferase activity was repressed in the construct containing suclg1, but not for the construct containing parvalbumin 4 (Figure 5). Therefore, based on these lines of evidence, we had now obtained confirmation for two target mRNAs, hand2 and suclg1, for miR-1 in zebrafish.
Figure 5.
Using luciferase activity assay to confirm the putative target genes for zebrafish miR-1 predicted from microarray analysis. (A) The 3′UTRs of these two cDNAs were separately engineered at the downstream of luciferase reporter gene (phRG-TK-3′UTR, as indicated) and transfected into COS-1 cells with pre-miR-1. Two other constructs—phRG-TK and CMV-pre-miR-1, served as control plasmids. (B) The luciferase activity driven by the 3′UTR from either suclg1 or parvalbumin 4 mRNA is presented in histograms. Y axis represents the relative value for Renilla reniformis luciferase to firefly luciferase after normalization with phRG-TK-transfected cells. Compared with the luciferase from the phRG-TK-transfected cells, the luciferase activity was repressed in the cells transfected with suclg1 3′UTR and pre-miR-1. The error bar indicates standard deviation (n = 6).
DISCUSSION
In this article, we present a simple, but effective, alternative by which to detect the possible target genes of known miRNAs through completely or partially complementary 3′UTR. Similar to pre-miRNA, the DIG-labeled pre-miRNA can be processed to the mature DIG-labeled miRNA by proteins, such as Dicer (Figure 1) and RISC. We take advantage of this feature and precipitate the mixture of mature DIG-labeled miRNA and its cognate target mRNAs using antiserum against DIG. As a result, the putative target gene(s) bound by the bait mature miRNA can be easily identified by subcloning or by using RT–PCR. After we analyze these cDNAs or putative clones, a target gene that interacts with the known miRNA may be found. For example, when the C. elegans lin-4 miRNA was used, the target genes lin-14 (9) and lin-28 (10) were found. When let-7 miRNA was used, the target genes hbl-1 (29) and lin-41 (30) were found as well. Moreover, when the zebrafish let-7 miRNA was used, the gene lin-41 (28) was targeted, but when zebrafish pre-miR-1 was used, we discovered a novel target gene, hand2. Therefore, we clearly demonstrated that the target genes of miRNA can be easily obtained in vitro through the endogenous proteins using our novel detection method which we have termed LAMP assay system.
We have further established that LAMP can even be used to determine different target genes for a bait miRNA at different developmental stages or from different cell types. To explain, some miRNAs, such as miR-1 and let-7, have their own gene family. Each family has similar nucleotide sequences from 1 through 8 at the 5′ end and similar levels of free energy (31). Every member of the same family has its own target gene at various developmental stages and in different tissues, such as let-7, miR-48 and miR-84 of the let-7 family of C. elegans (18). However, the target genes of miRNAs that belong to the same family, such as miR-1 and let-7, are not easily distinguished by simple computational analysis methods. Here, we demonstrate that the LAMP assay system can overcome the inherent disadvantages of the purely bioinformatics/algorithmic approach. Specifically, Cel-pre-let7M was designed to have the mutated sequence from 11 through 14 at the 5′ end of Cel-pre-let7. Cel-pre-let7 and Cel-pre-let7M had similar sequences from nucleotides 1–8 and had similar amounts of free energy. When they were used to determine the target genes by LAMP, results showed that the lin-41 and hbl-1 genes (Figure 2B) were targeted by Cel-pre-let7, but not by the mutated Cel-pre-let7. This evidence indicates that the LAMP assay system is sensitive enough to discriminate the target genes of miRNAs that belong to the same family, whereas a purely algorithmic approach would have resulted in a very puzzling and more clarification is needed. This conclusion is also strongly supported by the results following microarray analysis of ZF-pre-miR-1 and ZF-pre-miR-1M, which are DIG-tagged-pre-miR-1 (WT) and DIG-tagged-pre-mutated-pre-miR-1 (MT), respectively. Although miR-1 and miR-206 are categorized into the same family, the LAMP assay for miR-1 does not pick up the known target genes for miR-206, such as connexin 43 (32), Fstl1 and Utrn (33), because they are not included among the 302 putative targets whose WT/MT is over 1 (Supplementary Table 2). This finding strongly indicates that the LAMP assay system enables us to distinguish the target genes among miRNAs which have the same seed sequence. This advantage cannot be achieved if bioinformatics analysis is employed because many similar putative target genes should be come out among miRNAs having the same seed sequence.
In mice, it has already been reported that hand2 is one of the miR-1 targets (27). However, we noticed that the expression patterns of miR-1 and hand2 are different between zebrafish and mice; that is, mouse miR-1, but not zebrafish miR-1, displays in the heart (7,27). Thus, the results obtained from mice cannot provide the information necessary to predict whether zebrafish hand2 could be the target gene of zebrafish miR-1. Unexpectedly, however, when LAMP was used to find the target gene for ZF-pre-miR-1, we found that 6 out of 11 (50%) of putative clones contained the hand2 insert. Further study of the expression pattern of hand2 in zebrafish embryos revealed that zebrafish miR-1 is expressed in somite and pectoral fins, whereas zebrafish hand2 is expressed in heart, pharyngeal arch and pectoral fin (7,34), indicating that miR-1 and hand2 are co-expressed only in pectoral fins. Therefore, we speculate that miR-1 might enable the silencing of hand2 mRNAs in pectoral fin.
As a tool for searching target genes for known miRNAs, we believe that LAMP has general utility. We provided proof of this by using microarray analysis to compare the composition and level of mRNAs pulled down in the pool, both by the miR-1 probe (WT) and by the mutated miR-1 probe (MT). In general, the level of 28S or 18S is used to normalize the quantity of RNA used to make the comparison. However, in our study, because of the lack of either 28S or 18S, we added DIG-tagged cTnnT2 mRNA in the pool to solve this problem. Neither the WT probe nor the MT probe is able to bind cTnnt2 mRNA. Thus, it is highly unlikely that the binding capacity among anti-DIG, DIG-tagged miR-1 probe and target mRNA would encounter any interference by adding DIG-tagged cTnnT2. Based on the amount of DIG-tagged cTnnT2, we can monitor the amount of target mRNA in the WT and MT probe groups. Thus, we are able to quantify the target mRNAs in what we consider to be a feasible and reasonable manner. Specifically, by comparing the normalization ratio of WT/MT, we found 302 putative target genes, each of whose WT/MT was over 1. Four of the top five genes out of a total 302 (the upper 2%) (Supplementary Table 2) putative target genes have been proven to be the true target genes of miR-1 in other species. Since, by microarray analysis, hand2 is the top-scoring one among them, it is reasonable to suggest the high likelihood of isolating hand2 during a search for the target gene of miR-1. This is what accounts for the six clones containing the hand2 insert gene among the 11 putative clones whose insert fragment is over 300 bp when we were searching for the miR-1 target. Interestingly, we discovered another target gene, suclg1, for zebrafish miR-1 based on the priority of the WT/MT normalization ratio shown in the microarray analysis, the expression pattern shown in the whole mount in situ hybridization, and the luciferase activity shown in cell line COS-1. Taken together, we concluded that five putative genes among the upper 7% of the WT/MT normalization ratio have been validated as the target genes of miR-1 and that, therefore, the LAMP assay system is capable of determining the target genes of miR-1 by about a 20% chance (5/22) (Table 1).
Compared to the conventional bioinformatics approach, the simplicity of the LAMP assay system, as described in this study, has a number of advantages. First, it is not necessary to provide the complete and genome-wide database of 3′UTR. Second, searching for miRNA-targeting genes by using an algorithmic method is eliminated. Third, it is hard to precisely identify the target sequence for a given miRNA when a variety of prediction software programs are employed together, such as, for example, miR-101 (35). Our method also eliminates this problem. Fourth, the cDNA array chip is not necessarily required for LAMP analysis. The utility and benefit of this is particularly important when studying experimental organism whose cDNA array chips are not readily available in the commercial market. Furthermore, when combined with microarray technology, LAMP is cost-effective and has the potential to identify unknown targets of a given miRNA and is also powerful to study the dynamic changes of the target mRNAs in a biological process. Finally, when using the bioinformatics approach, a tremendous number of potential targets are predicted, while, in contrast, the LAMP assay system can rule out many improper candidates.
Compared to conventional microarray analysis, the LAMP assay system is relatively straightforward by its ability to obtain the complex of putative target mRNAs bound by DIG-labeled miRNA from cell extracts. It is well known that microarray is performed either by overexpression or by knockdown/knockout of a specific miRNA after comparing the expression level with that of wild-type embryos. In some cases, however, it should also be noted that the decrease or increase in the expression levels of genes shown on microarray may result from other mechanisms, including the influence of genes other than the target gene. In addition, there is a high probability of encountering non-specific binding by using either anti-HA monoclonal antibody (19) or anti-Ago monoclonal antibody (20). However, each of these drawbacks can be directly addressed by using our LAMP assay system since this method only labels the desired miRNA by DIG, with the correspondingly increased likelihood of cloning the exact miRNA target-mRNA.
In conclusion, the LAMP assay system is a simple and efficient method of identifying the putative target genes for a known miRNA of interest. It may also be used to find different target genes for one single miRNA from cells at different developmental stages or from different cell types. Once the miRNA of interest and its mutated miRNA, which serves as control, are labeled, they are then mixed with cell lysates extracted from the desired cells, tissues or organisms. After immunoprecipitation of the miRNA/protein (miRNP)/mRNA complexes, it is highly likely that the target mRNA will be found. The LAMP assay system could potentially detect target mRNAs at low cellular density by incorporating cell sorting by flow cytometry or LCM before the pull-down assay has been employed. Therefore, compared to either microarray or bioinformatics, the results in this study support the LAMP assay system as an overall effective means of screening candidate genes of known miRNAs. Finally, if the LAMP assay system were to be combined with microarray, for example, there is no doubt that such a combination would result in an extremely effective search tool for new miRNA target gene profiles.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council under the grant of NSC 97-2313-B-002-036-MY3, ROC.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–24. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 2.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 5.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 6.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 7.Wienholds E, Koudijs MJ, Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 8.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 10.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–641. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 11.Rooij Van E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 12.Berezikov E, Guryev V, Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 13.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 14.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 15.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li N, Flynt AS, Kim HR, Solnica-Krezel L, Patton JG. Dispatched Homolog 2 is targeted by miR-214 through a combination of three weak microRNA recognition sites. Nucleic Acids Res. 2008;13:4277–4285. doi: 10.1093/nar/gkn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev. Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;2:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 21.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YH, Lee WC, Liu CF, Tsai HJ. Molecular structure, dynamic expression, and promoter analysis of zebrafish (Danio rerio) myf-5 gene. Genesis. 2001;29:22–35. doi: 10.1002/1526-968x(200101)29:1<22::aid-gene1002>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 24.Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- 25.Wang TM, Chen YH, Liu CF, Tsai HJ. Functional analysis of the proximal promoter regions of fish rhodopsin and myf-5 genes using transgenesis. Mar. Biotechnol. 2002;4:247–255. doi: 10.1007/s10126-002-0016-y. [DOI] [PubMed] [Google Scholar]
- 26.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–47. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 28.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 30.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- 35.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.