Abstract
Phenotypic variation is critical to many aspects of biological research. Use of a captive population to address questions concerning the genetics and evolution of dental variation raises the question of how the pattern of phenotypic variation under study compares with that in a wild population of the same species. Differences in the pattern of variation within wild and captive populations may indicate different genetic and non-genetic factors, and also may have implications for how well the captive group can serve as a model for its wild type relatives.
We compared dental size measures from two Papio hamadryas populations, one captive and one wild. Lengths and widths of maxillary and mandibular second molars (M2s) were collected from 630 baboons from a captive pedigreed breeding colony housed at the Southwest National Primate Research Center in San Antonio, Texas, and 125 baboons from a wild population culled from a sisal plantation in Kibweze, Kenya.
Although the two populations consistently differed with respect to lengths and widths of the M2s, principal components analyses show that the basic pattern to variation in these molar crown traits is remarkably similar in both populations; and linear functions based on these measures cannot reliably discriminate between the two groups. This similarity in the pattern of variation among these dental crown measures in these two groups suggests that analyses to dissect their genetic architecture in captive populations is likely to be highly relevant to dental variation in wild baboons as well.
Keywords: multivariate analyses, primatology, odontology
Introduction
Captive breeding populations of animals have proven extremely valuable to many areas of scientific research. Their value is primarily attributable to the fact that the researcher can control, limit or, at least, identify and measure reliably the environmental and biological factors that might influence variation in the traits or processes of interest. Given this attribute, animals from captive breeding populations have served as research models for wild populations of their own species and, in many well-known instances, for humans as well. In either of these two cases, the validity of any captive animal model is a function of the similarity between it and the modeled species.
Captive baboons have been used extensively in anthropological research as model organisms for studies related to dental development, variation, disease, and evolution in humans and other primate species (1–3). Although the utility of captive baboons in these sorts of studies seems entirely justifiable given their phylogenetic proximity and consequent genetic, anatomic, and physiological similarities to higher primate species, it is incumbent upon the researcher to demonstrate comparability between model and modeled species with respect to each phenomenon of interest (i.e., trait, process, etc.) prior to extrapolation of results. Equally important is ascertaining that captive populations are representative of natural or wild types of their species and not a subpopulation with aberrant traits or phenotypic values (i.e., it is difficult to posit that a captive population is a model for other species if it is not adequately representative of non-captive populations of its own).
We are engaged in studies of the genetics of primate dental variation and evolution using data from captive baboons. Given the highly conserved nature of gene regulatory systems across widely diverse mammalian groups (4), the strong genetic similarity between higher primate species -- 92–95% genetic sequence homology between baboons and humans (3), the genetic similarities between phylogenetically distant taxa such as humans and mice (5), and evidence for similar patterns of skeletal morphological integration between closely related primate taxa (6–8), we began these studies not expecting significant differences in basic dental biology between wild and captive populations of the same species that have only been isolated for 1–5 generations. Yet, research of dental eruption times in baboons (9–16), for example, shows that this is not always true. Although there is considerable similarity in the timing of dental eruption and emergence across wild populations, these processes occur 1.5 years earlier in captive animals (15). Such observations motivate our view that, when possible, assessment of the degree of similarity between captive and wild populations with respect to a trait of interest is an important and prudent step in research using model organisms.
Here we report on a comparison of dental variation in a colony of captive, pedigreed, breeding baboons housed at the Southwest National Primate Research Center (SNPRC) to that seen in a wild baboon population from Kenya. Although the amount of variation in each population may not be equal, we hypothesized that this variation results from the same biological process in captive and wild populations of the same species. If true, the structure of the variance revealed through principle components analyses should be the same for both populations.
Materials
Right and left second molar metric data were collected from two populations: one held in captivity and one from a more natural habitat. Our taxonomy follows Jolly (17).
The wild baboon data are from a collection of Papio hamadryas cynocephalus (yellow baboon) skeletons housed in the Osteology Department of the National Museum of Kenya (NMK). Since 1988 more than 150 baboons have been (and continue to be) culled from 18 sites on the Dwa Sisal Estate (plantation) near Kibwezi in southeastern Kenya (2°26′S, 37°53′E). Given P. h. cynocephalus and P. h. anubis social grouping patterns, it is reasonable to assume that these animals are related and form one, or just a few large extended pedigrees similar in nature to that of the SNPRC captive colony used in our quantitative genetic analyses. The NMK data were collected from 125 permanent dentitions, consisting of 68 males, 55 females, and two first molars from juveniles whose sex was indeterminate.
The captive population data were collected from olive baboons (Papio hamadryas anubis), a few yellow baboons (Papio hamadryas cynocephalus), and their hybrids (18) housed at the Southwest National Primate Research Center (SNPRC) in San Antonio, Texas. These animals are descendant from founders that were wild caught and matings since have been controlled to inhibit inbreeding. The sample studied has a female to male sex ratio approximating 2:1, and ranges in age from 4.6 to 30 years.
While strict genetic management was (and is) employed to prevent inbreeding, all non-founder animals in this study were the result of matings that were random with respect to phenotype. Since birth or, in the case of some of the oldest of the founders, arrival at the SNPRC colony, all animals have been housed out of doors in social group cages and maintained on monkey chow diets to which they have ad libitum access. Animal care personnel and staff veterinarians provide daily maintenance as well as regular, urgent, and emergent health care to all animals throughout their stay at SNPRC in accordance with the Guide for the Care and Use of Laboratory Animals (19).
Data were collected from dental casts of 630 individuals. Dental molds were collected from anesthetized animals using a protocol described in detail elsewhere (20) and approved by the Institutional Animal Care and Use Committee in accordance with the established guidelines. Positive casts were poured with high resolution dental plaster within one week of the mold being made. Dental casts were also produced from skulls of deceased animals that are curated by Dr. J. M. Cheverud at Washington University in St. Louis. All data from the SNPRC population were collected from these plaster casts.
In living monkeys, the gumline obscures the cervix and forms the outer visual boundary for many of the teeth. Therefore, digital images were made of the molar casts following a protocol described in detail elsewhere (20) and the outline of each tooth was established as 1mm below the lowest point of the mesial and distal fovea rather than at the gumline. This was measured by sinking the tooth into a pool of titanium beads of approximately 0.15mm diameter that acted like a liquid, filling in around the molar at the designated level. Following this protocol, the outer line of the tooth was standardized and could be used for all measurements collected. Replicability of the digitizing protocol was tested for 14 molars. Measurements of the same tooth in repeated images were found to be 1.5–1.7% different.
Data Collection Methods
This study focuses on the maxillary and mandibular second molars (M2 and M2, respectively), as these are the largest of the datasets.
The NMK molars were measured using fixed-jawed dental calipers (Mitutoyo© Model NTD12–6″C). Mesiodistal lengths (md) and buccolingual widths (mesial width = mw; distal width = dw) were measured following standard definitions (13).
Data from the SNPRC baboon molars were collected from digital images of the casts (described above). Mesiodistal length was measured as the maximal length of the molar. Buccolingual distance was measured as the maximal width of the tooth oriented through the two mesial cusps, defined on the buccal and lingual sides by the contact with the titanium beads. Measurements were not taken from teeth that were worn below the landmark defined by the mesial/distal foveal depth.
Measurement errors for the caliper-metric protocols ranged between 1–5% of the average measurement. Measurement error for the data collected from the digital images ranged between 1–2% (21).
The measurements collected for these two populations are not identical, making comparisons of actual size estimates between them inappropriate. However, measurement error for all of the protocols used here are low and comparable, and the protocols were consistent within each population. Therefore, comparing the univariate distributions and multivariate structure between the two populations is not affected by the somewhat differing data collection procedures.
Analytical Methods
Initial data processing and management were done using routines implemented in the computer pedigree database program PEDSYS (22). All statistical analyses were conducted using routines implemented in the NCSS 2004 software program (23). Prior to analysis and within each sample, a linear regression procedure was used to regress out the effects of sex on each measure. All analyses described below were conducted on sex-adjusted residuals. We compared the wild Kenyan to captive pedigreed baboons on the basis of the 12 M2 crown metrics in three ways: 1.) univariate inferential statistical comparisons on a measure-by-measure basis, 2.) multivariate descriptive statistical comparisons based on latent structure of the data, and 3.) multivariate inferential statistical comparisons.
Unrelated founder sample for inferential statistics
The data from the captive baboons come from related individuals, violating the assumption of independence on which the interpretation of the results of many statistical tests rely. By contrast, we have no accurate knowledge of the patterns of kinship that may exist within the wild Kenyan sample. To address these issues, we identified a subset of 65 unrelated animals with dental data in the captive baboon population. These animals essentially are founders for the 11 pedigrees from which the captive animals in this study were obtained. We used pedigree analysis and kinship estimation routines implemented in the computer programs PEDSYS (22) and SOLAR (24) to identify these individuals. Each comparison between the wild and captive baboon populations were repeated by comparison with the captive founder sub-sample. All reports of significance, however, are based on comparisons with the captive founder sub-sample. No result from the latter comparison was accepted or reported if inconsistent with that from the former comparison with the larger pedigreed sample.
T-tests
We used Student’s two-sample t-test (with Aspin-Welch adjustment for unequal variances) to compare the means of the sex-adjusted residuals of each measure from the wild Kenyan and captive pedigreed populations. We considered test statistics with P<0.05 to be evidence of a significant difference.
Principal Components Analysis
We used the multivariate data reduction method, principal components analysis (PCA), to further aid in the comparison of the M2 metrics in the wild and captive populations. We used PCA to decompose the covariance among the sex-adjusted residuals for the 12 dental crown measures in each population, reducing the dimensionality of their M2 datasets while retaining the characteristics of each dataset that contribute most to its variance. The goal was to obtain for each population a set of low-order components: i.e., synthetic variables that are linear transformations of the original data containing the “more important” aspects of the data and account for a majority of the variance in them. We selected as the more salient components, 1.) those which incremental contribution to the variance distinguished them from the others in a scree curve and 2.) those with eigenvalues approximating or greater than 1 (25).
Prior to the principal components analyses, we eliminated an animal’s data from consideration if all three measures were not available for at least one of the four permanent M2s. Further, we eliminated from the study data from animals with too many missing measurements to support the following missing multivariate normal data estimation procedure. A regression analysis was conducted using the variable containing the missing value as the dependent variable and all variables with non-missing data from an animal as independent variables. The values of the non-missing variables from the animal containing the missing value were used in the regression equation to compute a predicted value for the missing value. This process was iterated 1000 times for each value using the imputed missing values from one run during the estimation phase of the next.
Lastly, we conducted linear discriminant analyses of the sex-adjusted residuals for these permanent M2 metric data to determine if we could obtain a set of prediction equations that would classify the individuals into their appropriate groups: i.e., wild or captive. Group membership was used as the criterion variable and the M2 crown metrics were used as the predictor variables. We used graphical and statistical analyses of the resultant discriminant function and canonical variate scores to assess whether the two groups could be separated on these metrics or whether, as expected, they overlap.
Results
Summary descriptive statistics for each sample – wild caught baboons from Kenya, captive baboons from SNPRC, and a sub-sample made up of founders for the pedigrees into which the captive baboons are organized – are presented in Table 1. Mean permanent M2 crown widths (buccolingual diameters) of the wild Kenyan baboons were consistently larger than those of the captive bred animals from SNPRC, while the contrary is true for the mesiodistal lengths. However, t-tests of the residuals (after regressing out the effects of sex) of these metrics reveal no significant differences between the wild Kenyan animals and the unrelated founders of the pedigrees to which the captive baboons belong (i.e., all P>0.21).
Table 1.
Permanent Second Molar Crown Dimensions (in mm) in Wild and Captive Baboons: Sample Descriptive Statistics.
| Wild Population | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XLM2 | XRM2 | DLM2 | DRM2 | ||||||||||
| Parameter | mdl | mw | dw | mdl | mw | dw | mdl | mw | dw | mdl | mw | dw | |
|
|
|||||||||||||
| N | 91 | 89 | 88 | 94 | 90 | 88 | 87 | 86 | 85 | 87 | 89 | 86 | |
| Mean | 12.25 | 11.47 | 10.40 | 12.22 | 11.38 | 10.38 | 12.12 | 10.09 | 9.93 | 12.22 | 10.09 | 9.86 | |
| SD | 0.84 | 0.75 | 0.69 | 0.87 | 0.79 | 0.71 | 0.82 | 0.74 | 0.74 | 0.83 | 0.70 | 0.65 | |
| Minimum | 10.1 | 9.6 | 8.8 | 10.2 | 9.5 | 8.8 | 10.1 | 8.6 | 8.4 | 9.9 | 8.6 | 8.3 | |
| Maximum | 14.2 | 13.3 | 11.9 | 14.1 | 13.2 | 11.8 | 14.0 | 12.1 | 11.9 | 14.1 | 12.0 | 11.1 | |
|
|
|||||||||||||
| Captive Population | |||||||||||||
| XLM2 | XRM2 | DLM2 | DRM2 | ||||||||||
| Parameter | mdl | mw | dw | mdl | mw | dw | mdl | mw | dw | mdl | mw | dw | |
|
|
|||||||||||||
| N | 649 | 647 | 637 | 643 | 638 | 623 | 590 | 581 | 572 | 593 | 585 | 575 | |
| Mean | 12.51 | 10.12 | 9.13 | 12.43 | 10.09 | 9.07 | 12.29 | 9.35 | 8.79 | 12.34 | 9.35 | 8.79 | |
| SD | 0.84 | 0.87 | 0.82 | 0.84 | 0.87 | 0.83 | 0.82 | 0.77 | 0.83 | 0.79 | 0.74 | 0.77 | |
| Minimum | 10.10 | 8.33 | 7.23 | 9.10 | 7.55 | 7.19 | 8.20 | 7.50 | 5.80 | 9.88 | 7.20 | 7.00 | |
| Maximum | 15.7 | 13.3 | 11.9 | 15.5 | 13.2 | 11.9 | 15.3 | 12.1 | 11.9 | 15.4 | 11.9 | 11.3 | |
|
|
|||||||||||||
| Captive Population, Founders Only | |||||||||||||
| XLM2 | XRM2 | DLM2 | DRM2 | ||||||||||
| Parameter | mdl | mw | dw | mdl | mw | dw | mdl | mw | dw | mdl | mw | dw | |
|
|
|||||||||||||
| N | 60 | 57 | 57 | 59 | 59 | 58 | 52 | 51 | 51 | 54 | 54 | 53 | |
| Mean | 12.13 | 9.60 | 8.70 | 12.09 | 9.75 | 8.76 | 11.96 | 8.94 | 8.38 | 11.99 | 9.04 | 8.45 | |
| SD | 0.57 | 0.59 | 0.58 | 0.57 | 0.52 | 0.52 | 0.54 | 0.49 | 0.54 | 0.54 | 0.51 | 0.42 | |
| Minimum | 10.9 | 8.3 | 7.2 | 11.1 | 8.6 | 7.8 | 11.0 | 7.8 | 7.2 | 10.8 | 7.7 | 7.6 | |
| Maximum | 13.4 | 11.9 | 11.2 | 13.7 | 10.9 | 8.9 | 13.2 | 10.2 | 10.0 | 13.8 | 10.1 | 9.6 | |
|
|
|||||||||||||
Principal components analysis in which we decomposed the covariance among the 12 permanent M2 measures revealed very similar results for the two. The first five components account for approximately 99.7% and 99.8% of the variance in the Kenyan and captive animals, respectively. The first through third principal components, identified as the most salient by having eigenvalues greater than or equal to 1 and by inspection of the scree plot, account for approximately 71%, 15%, and 7%–8% of the variance in these measures in each population. Examination of the component loadings from these analyses (Tables 2 and 3) amplifies the similarities between the two groups. The first principal component reflects magnitude or size; the second is a component contrasting crown length and crown width; and the third contrasts maxillary and mandibular measures. Principal components analyses of the data from the sub-sample of founders for the captive population’s pedigrees revealed the same pattern of results (not shown).
Table 2.
Principal Components Analysis of Permanent Second Molar Crown Dimensions in Wild Baboons from Kenya: Component Loadings
| Variables | Component 1 | Component 2 | Component 3 | Component 4 | Component 5 |
|---|---|---|---|---|---|
| xlm2md | −0.77943 | −0.53332 | −0.01256 | −0.08156 | −0.22932 |
| xlm2mbw | −0.86867 | 0.255708 | −0.12851 | 0.173102 | −0.06018 |
| xlm2dbw | −0.77075 | 0.192203 | −0.49384 | −0.11103 | 0.082942 |
| xrm2md | −0.80989 | −0.51882 | −0.03191 | −0.04514 | −0.22866 |
| xrm2mbw | −0.85141 | 0.282559 | −0.14545 | 0.231601 | −0.07939 |
| xrm2dbw | −0.72171 | 0.175055 | −0.51202 | −0.02565 | 0.060899 |
| dlm2md | −0.79084 | −0.527 | 0.062638 | 0.081663 | 0.229861 |
| dlm2mbw | −0.79023 | 0.273822 | 0.339985 | 0.223214 | −0.02582 |
| dlm2dbw | −0.75328 | 0.309623 | 0.258975 | −0.19408 | 0.043193 |
| drm2md | −0.78802 | −0.49016 | 0.110185 | 0.014954 | 0.232852 |
| drm2mbw | −0.82543 | 0.27725 | 0.354729 | 0.058524 | 0.00335 |
| drm2dbw | −0.75215 | 0.296817 | 0.170628 | −0.39647 | −0.00567 |
Table 3.
Principal Components Analysis of Permanent Second Molar Crown Dimensions in Captive Baboon Population: Component Loadings
| Variables | Component 1 | Component 2 | Component 3 | Component 4 | Component 5 |
|---|---|---|---|---|---|
| xlm2md | −0.76367 | −0.49165 | −0.13236 | 0.002662 | 0.046523 |
| xlm2mbw | −0.78332 | 0.301105 | −0.23389 | 0.188956 | 0.036853 |
| xlm2dbw | −0.78 | 0.317631 | −0.29681 | −0.14044 | 0.25375 |
| xrm2md | −0.78692 | −0.46715 | −0.12605 | −0.03185 | −0.01307 |
| xrm2mbw | −0.79128 | 0.37138 | −0.20619 | 0.159822 | −0.18897 |
| xrm2dbl | −0.74871 | 0.357057 | −0.25075 | −0.1883 | −0.02908 |
| dlm2md | −0.73535 | −0.54224 | −0.02214 | 0.02476 | −0.00512 |
| dlm2mbw | −0.79554 | 0.158103 | 0.292876 | 0.297911 | 0.03914 |
| dlm2dbw | −0.73571 | 0.081858 | 0.417771 | 0.024419 | 0.279188 |
| drm2md | −0.7528 | −0.51411 | −0.01046 | −0.01489 | −0.07476 |
| drm2mbw | −0.8081 | 0.208107 | 0.24715 | −0.0361 | −0.24498 |
| drm2dbw | −0.7511 | 0.172871 | 0.332795 | −0.31116 | −0.0789 |
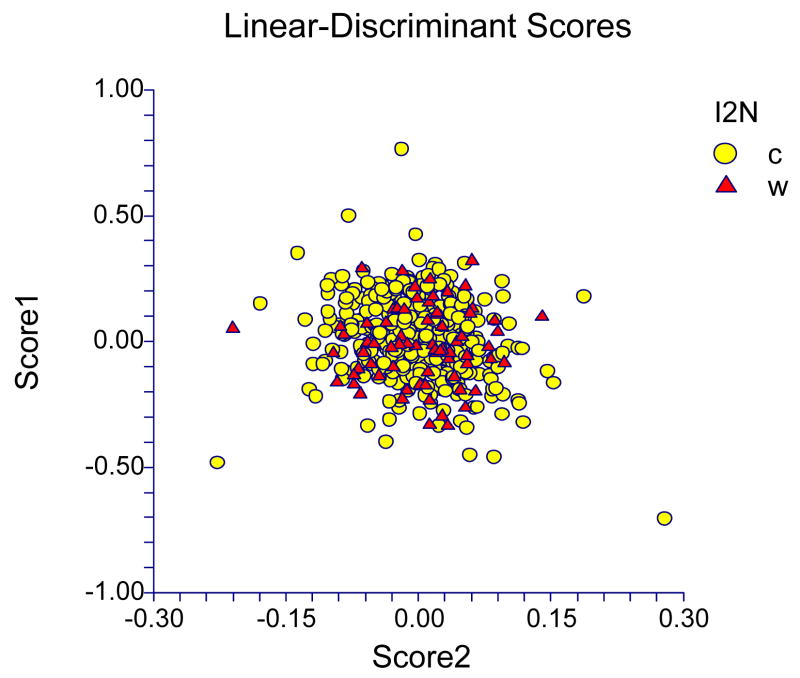
Both forward and backward stepping linear discriminant analyses failed to identify a subset of measures that could distinguish the two groups, so all M2 crown measures were used. The best linear function based on these 12 M2 crown measures from the two groups were incapable of reliably discriminating between the wild Kenyan and captive pedigreed baboons (Figure 1). Application of this function results in 44.8% of the wild baboons being classified as “captive” and 46.1% of the “captives” being classified as “wild.” The results of the associated canonical correlation analysis – i.e., a single canonical variate, canonical R=0.0667, R2=0.0045, Wilks’ Lambda=0.9955 – emphasize further the inability to discriminate between these two groups based on the 12 M2 crown measures.
Figure 1.
Plot of Linear Discriminant Function Scores from Analysis of 12 M2 Crown Metrics in Wild (W, triangle) and Captive Pedigreed Baboons (C, circle).
Discussion
The primary goal for this study was to determine whether the pattern of variation in M2 crown dimensions in captive populations of baboons was similar enough to that in wild populations of the same species to justify use of the former to study the genetics and evolution of the latter. Our results demonstrate a considerable degree of similarity between the captive and wild baboon populations in the variation of these 12 dental metrics as perhaps most evinced by the inability to construct linear discriminant functions that reliably differentiate between the two populations.
This result is perhaps not surprising given that quantitative genetic analyses have shown similarities in the genetic variance of numerous wing shape and life history phenotypes in captive and wild populations of milkweed bugs (26), and that genetic correlations yield similar patterns of craniofacial modularity across primate species (27) and genera (6, 7). Although our analyses do not yield information about the underlying genetics, a common genetic architecture could return the similarities in phenotypic variation that we find in these two baboon populations.
We interpret the results presented here as evidence that dental variation in captive baboons does reflect dental variation in wild baboons. As such, quantitative genetic analyses of molar size variation in these captive baboons are appropriate as a model for non-captive baboon biology. Further analyses are needed to determine whether or not these phenotypic similarities exist for other regions of the dentition (such as the canine complex, 28) and more complicated phenotypes such as those that describe cusp orientation (29).
Acknowledgments
We thank the National Museums of Kenya and Mr. Ogeto, Acting Head of the Department of Osteology, for permission to study the Papio hamadryas skeletons, and the Department of Osteology staff for facilitating access; J. Rogers, K.D. Carey, K. Rice and the Veterinary Staff of the Southwest Foundation for Biomedical Research and the Southwest National Primate Research Center; J. Cheverud (Washington University) for access to the SNPRC specimens that are in his care; Jeffrey Rogers, Alan Walker, and Ken Weiss for project support and development; Loren Lease for assistance with an initial analysis of these data; Laurel Buchanan, Theresa Cannistraro, Leslie Holder, Jennifer Irwin, Anne Liberatore, Mary-Louise Maas, and Danelle Pillie for assistance with data collection.
Grant sponsorship: This material is based upon work supported by the National Science Foundation under Grants 0500179 and 0130277. NIH/NCRR P51 RR013986 supports the Southwest National Primate Research Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vagtborg H. The Baboon in Biomedical Research. Proceedings of the 2nd International Symposium on the Baboon and Its Use as an Experimental Animal; San Antonio, TX. 1965; Austin, TX: The University of Texas Press; 1967. [Google Scholar]
- 2.Jolly CJ. The seed-eaters: A new model of hominid differentiation based on a baboon analogy. Man. 1970;5:5–26. [Google Scholar]
- 3.VandeBerg JL, Williams-Blangero S. Advantage and limitations of nonhuman primates as animal models in genetic research on complex diseases. J Med Primatol. 1997;26:113–119. doi: 10.1111/j.1600-0684.1997.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 4.Carroll SB. Genetics and the making of Homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]
- 5.Davideau J-L, Demri P, Hotton D, Gu T-T, MacDougall M, Sharpe PT, Forest N, Berdal A. Comparative study of MSX-2, DLX-5, and DLX-7 gene expression during early human tooth development. Pediatric Research. 1999;46:650–656. doi: 10.1203/00006450-199912000-00015. [DOI] [PubMed] [Google Scholar]
- 6.Marroig G, Cheverud JM. Size as a line of least evolutionary resistance: diet and adaptive morphological radiation in New World monkeys. Evolution. 2005;59:1128–1142. [PubMed] [Google Scholar]
- 7.Hallgrímsson B, Willmore K, Dorval C, Cooper DML. Craniofacial variability and modularity in macaques and mice. J Exp Zool (Mol Dev Evol) 2004;302B:207–225. doi: 10.1002/jez.b.21002. [DOI] [PubMed] [Google Scholar]
- 8.González-José R, Van der Molen S, González-Pérez E, Hernández M. Patterns of phenotypic covariation and correlation in modern humans as viewed from morphological integration. Am J Phys Anthropol. 2004;123:69–77. doi: 10.1002/ajpa.10302. [DOI] [PubMed] [Google Scholar]
- 9.Kuksova MI. The eruption of milk teeth in the hamadryas baboon. Sovetskaya Antropologiya. 1958;2:17–21. [Google Scholar]
- 10.Reed OM. Papio cynocephalus age determination. Am J Phys Anthropol. 1973;38:309–314. doi: 10.1002/ajpa.1330380226. [DOI] [PubMed] [Google Scholar]
- 11.Seigel MI, Sciulli PW. Eruption sequence of the deciduous dentition of Papio cynocephalus. J Med Primatol. 1973;2:247–248. doi: 10.1159/000460328. [DOI] [PubMed] [Google Scholar]
- 12.Schwendeman M, Cummins LB, Moore GT, McMahan CA. Age estimation in baboons (Papio cynocephalus) using dental characteristics. Lab Anim Sci. 1980;30:860–864. [PubMed] [Google Scholar]
- 13.Swindler DR. Primate Dentition. New York: Cambridge University Press; 2002. [Google Scholar]
- 14.Swindler DR, Meekins D. Dental development of the permanent mandibular teeth in the baboon, Papio cynocephalus. Am J Hum Biol. 1991;3:571–580. doi: 10.1002/ajhb.1310030606. [DOI] [PubMed] [Google Scholar]
- 15.Phillips-Conroy JE, Jolly CJ. Dental eruption schedules of wild and captive baboons. Am J Primatol. 1988;15:17–29. doi: 10.1002/ajp.1350150104. [DOI] [PubMed] [Google Scholar]
- 16.Kahumbu P, Eley RM. Teeth emergence in wild olive baboons in Kenya and formulation of a dental schedule for aging wild baboon populations. Am J Primatol. 1991;23:1–9. doi: 10.1002/ajp.1350230102. [DOI] [PubMed] [Google Scholar]
- 17.Jolly CJ. Species, subspecies, and baboon systematics. In: Kimbel WH, Martin LB, editors. Species, Species Concepts, and Primate Evolution. New York: Plenum Press; 1993. pp. 67–107. [Google Scholar]
- 18.Williams-Blangero S, VandeBerg JL, Blangero J, Konigsberg L, Dyke B. Genetic differentiation between baboon subspecies: relevance for biomedical research. Am J Primatol. 1990;20:67–81. doi: 10.1002/ajp.1350200202. [DOI] [PubMed] [Google Scholar]
- 19.National Research Council. Guide for the Care and Use of Laboratory Animals. Washington, DC: National Academy Press; 1996. [Google Scholar]
- 20.Hlusko LJ, Weiss KM, Mahaney MC. Statistical genetic comparison of two techniques for assessing molar crown size in pedigreed baboons. Am J Phys Anthropol. 2002;117:182–189. doi: 10.1002/ajpa.10022. [DOI] [PubMed] [Google Scholar]
- 21.Hlusko LJ. Identifying the genetic mechanisms of dental variation in cercopithecoid primates. Penn State University; 2000. PhD thesis. [Google Scholar]
- 22.Dyke B. PEDSYS: A pedigree data management system Version 2.0. San Antonio, TX: Southwest Foundation for Biomedical Research; 1996. [Google Scholar]
- 23.Hintze J. Number Crunching Statistical Systems. Kaysville; Utah: 2001. NCSS and PASS. http://www.NCSS.com. [Google Scholar]
- 24.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kachigan SK. Multivariate Statistical Analysis: A Conceptual Introduction. 2. Radius Press: New York; 1991. [Google Scholar]
- 26.Rodríguez-Clark KM. Effect of captivity on genetic variance for five traits in the large milkweed bug (Oncopeltus fasciatus) Heredity. 2004;93:51–61. doi: 10.1038/sj.hdy.6800479. [DOI] [PubMed] [Google Scholar]
- 27.Cheverud JM. Quantitative genetic analysis of cranial morphology in the cotton-top (Saguinus Oedipus) and saddle-back (S. fuscicollis) tamarins. J Evol Biol. 1996;9:5–42. [Google Scholar]
- 28.Kieser JA, Groeneveld HT. Patterns of metric variability in the dentition of Papio usinus. Am J Primatol. 1988;14:141–151. doi: 10.1002/ajp.1350140205. [DOI] [PubMed] [Google Scholar]
- 29.Hlusko LJ, Maas ML, Mahaney MC. Statistical genetics of molar cusp patterning in pedigreed baboons: implications for primate dental development and evolution. J Exp Zoolog B Mol Dev Evol. 2004;302:268–83. doi: 10.1002/jez.b.21. [DOI] [PubMed] [Google Scholar]