Abstract
Biofilm formation by pathogenic bacteria plays a key role in their pathogenesis. Previously, the pstA gene was shown to be involved in the virulence of Mycobacterium avium subspecies paratuberculosis (M. ap), the causative agent of Johne's disease in cattle and a potential risk factor for Crohn's disease. Scanning electron microscopy and colonization levels of the M. ap mutant indicated that the pstA gene significantly contributes to the ability of M. ap to form biofilms. Digital measurements taken during electron microscopy identified a unique morphology for the ΔpstA mutant, which consisted of significantly shorter bacilli than the wild type. Analysis of the lipid profiles of the mycobacterial strains identified a novel lipopeptide that was present in the cell wall extracts of wild-type M. ap, but missing from the ΔpstA mutant. Interestingly, the calf infection model suggested that pstA contributes to intestinal invasion of M. ap. Furthermore, immunoblot analysis of peptides encoded by pstA identified a specific and significant level of immunogenicity. Taken together, our analysis revealed a novel cell wall component that could contribute to biofilm formation and to the virulence and immunogenicity of M. ap. Molecular tools to better control M. ap infections could be developed utilizing the presented findings.
Keywords: M. paratuberculosis, pathogenesis, virulence, biofilm formation, pstA, immunogenicity
1. Introduction
Mycobacterium avium subsp. paratuberculosis (M. ap) is the causative agent of Johne's disease (paratuberculosis), a disease that infects ruminants world-wide, most notably dairy cattle (17). It is also possible that exposure to M. ap could play a role in the development of Crohn's disease in humans (4). Experts estimate that Johne's disease results in a $220 million loss per year in the USA alone (22). Currently, there is no effective control strategy for Johne's disease and infection with M. ap is hard to diagnose and monitor because of the chronic nature of the disease. Additionally, it is very difficult to remove M. ap from the environment, which threatens any effective control strategy. In recent studies, M. ap was shown to be ubiquitous in animal environments (11,23), especially among wildlife animals (6). One reason M. ap may be difficult to eliminate from the environment and to treat with antibiotics is the possibility that the M. ap bacilli may form biofilm-like structures. Biofilm formation has been described before in both M. smegmatis (24,25), M. tuberculosis (20), and M. avium subspecies avium (M. avium) (34). In M. smegmatis and M. avium, biofilm formation is affected by glycopeptidolipids (GPLs) that are a major component of mycobacterial cell wall (3). The biosynthesis of GPLs in M. avium is initiated by an operon containing two large genes encoding for non-ribosomal peptide synthetases (pstA and pstB) that incorporate four amino acids (Phe, Thr, Ala and alaninol) into the core lipopeptide molecules. In this report, we examined the contribution of the pstA gene in forming cell wall lipids and the impact of the disruption of the pstA gene product on biofilm formation and virulence of M. ap.
Biofilm formation is considered a virulence phenotype in both gram-negative (e.g. Vibrio cholera) (13) and gram-positive bacteria (e.g. Staphylococcus aureus) (5). The main hypothesis behind this characterization is the ability of bacterial biofilms to induce a persistent source of infection and to resist antibiotics (8). Recently, M. avium, a bacterium closely related to M. ap, was found to form biofilms especially in water pipe systems of large cities (9,18). This phenotype can exacerbate the problems associated with M. avium infection in AIDS patients (29). A similar situation could also exist in animal pastures where infected and naïve animals come in close contact. Clinically-infected cows can shed 106-108 CFU/gm of fecal material that can easily contaminate animal surroundings for a long period of time. The ability of M. ap to form a biofilm could increase the survival of this pathogen under stress conditions and could increase the infection rate among cattle herds. Understanding the genetic basis of biofilm formation in M. ap will greatly enhance our knowledge of the pathogenesis of M. ap.
The pstA gene (∼12 kb) participates in GPL biosynthesis and biofilm formation in M. avium as a non-ribosomal peptide synthetase (nrp) (34). In some mycobacteria, the function of PstA is a result of two different genes, pstA and pstB (also referred to as nrp and mps) (10). A transposon insertional mutant in the pstA coding sequence of M. avium failed to form a biofilm in a water recirculation system (10,34). Through screening a transposon mutant library of M. ap, an insertion mutant of the pstA gene was shown to be attenuated in a mouse model of paratuberculosis with significant reduction in tissue colonization of the mutant (26). In this report, our analysis indicated that M. ap is able to form biofilms while its isogenic mutant, ΔpstA, is deficient in this phenotype, indicating the likely involvement of the pstA gene in biofilm formation. On the cellular level, electron microscopy analysis displayed a significant reduction in extracellular matrix of biofilms formed by the ΔpstA mutant with significantly shorter bacilli than both wild type and complemented strains. Further lipidomic analysis of the M. ap strains identified a unique lipopeptide in the wild type and pstA complemented strains that was absent from the ΔpstA mutant. Moreover, studies in cattle indicated the involvement of pstA in intestinal invasion and immunogenicity of M. ap.
2. Results
2.1. Phylogenetic analysis of the pstA gene
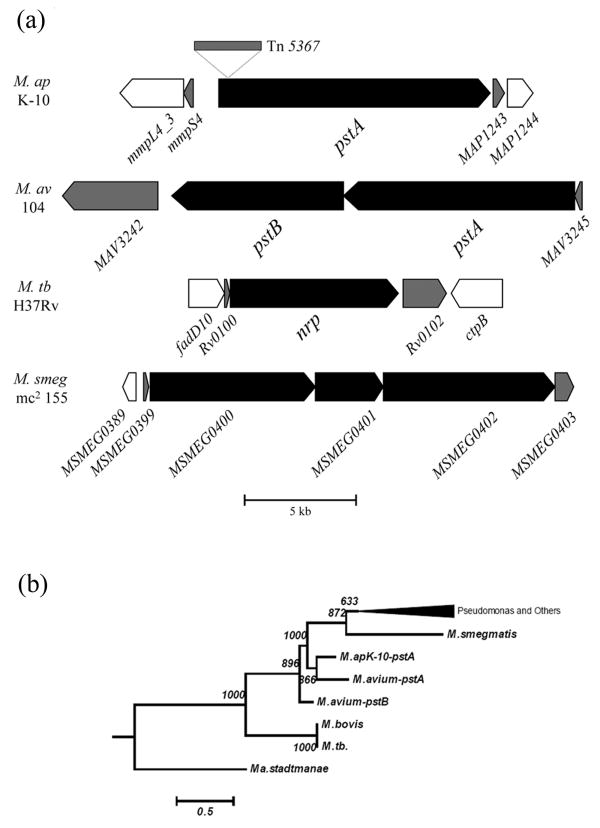
A large-scale screening strategy of a bank of M. ap transposon mutants identified a mutant with an insertion in the pstA gene to be attenuated in a murine model of paratuberculosis (26). Sequence analysis indicated that the Tn5367 transposon was inserted at base 465 (3.8 %) from the predicted start codon of the pstA gene (Fig. 1a). Earlier reports indicated the involvement of orthologues of the pstA gene in biofilm formation in M. avium (pstB) (34) and M. smegmatis (mps) (25). BLASTp analysis of the pstA sequences identified 14 orthologues with significant E-scores (<10-5 and > 25% overlap) (GenBank release 147, April 2005). Alignment of the orthologues identified one clade where sequences from both M. ap and M. avium were present closest to other mycobacterial species, indicating that pstA is conserved during mycobacterial evolution (Fig. 1b) and could play an important role in the pathobiology of M. ap.
Fig. 1.
Sequence analysis of pstA genes in different mycobacteria. (a) Schematic organization of the pstA gene with location of the Tn5367 insertion sequence. Orthologus of the pstA gene were also shown in M. avium, M. tuberculosis H37Rv (M. tb) and M. smegmatis (mc2155). A scale bar is also shown. (b) Phylogenetic tree based on the sequences of the pstA gene and its orthologues. A bar representing phylogenetic distances and values for 1000 boot-strapping is shown.
2.2. Biofilm formation by M. ap
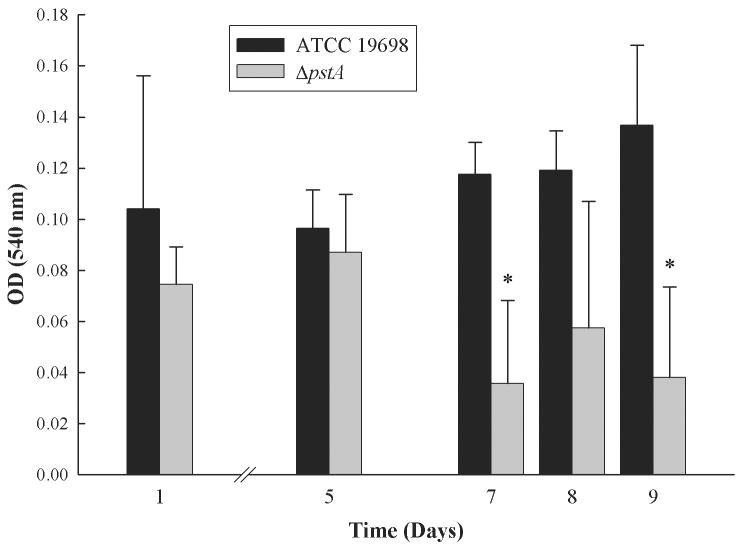
A recent analysis of the M. avium ΔpstB mutant suggested a role for this gene in biofilm formation (34). Sequence analysis indicated that the pstB coding sequence is not present in M. ap; however, the pstA gene in M. ap and the pstB gene in M. avium are closely related and hence, may perform a similar function. To examine the contribution of the pstA gene to biofilm formation in M. ap, we adopted a standard method on PVC surfaces that was previously used to quantify biofilm formation in M. avium (34). In this assay, the extent of biofilm structure is measured by the level of bacterial adherence to the PVC surfaces with crystal violet, a bacterial staining dye. Repeated measurements (at least 3 times) of biofilm formation of both the wild type and ΔpstA mutant showed a significant (P < 0.05) reduction in the ability of the ΔpstA mutant to form a biofilm compared to its parent strain of M. ap ATCC 19698, especially at 7 and 9 days post inoculation (Fig. 2).
Fig. 2.
Biofilm formation in M. ap and the ΔpstA mutant. Bacterial cultures of the wild type or ΔpstA mutant were grown on PVC 96-well plates. The plates were washed and subsequently stained with crystal violet. Optical density readings of the wild type and ΔpstA mutant were measured after 1, 5, 7, 8 and 9 days of growing, showing significantly reduced (P < 0.05) biofilm formation of the ΔpstA mutant at 7 and 9 days samples (denoted by asterisks). A representative of triplicate experiments is shown here.
To quantify the contribution of the pstA gene to biofilm formation in M. ap, we estimated the number of bacterial colonies participating in forming biofilms in the wild type, its isogenic pstA mutant and the complemented strain of M. ap ΔpstA∷pstA. Cultures of all strains were allowed to grow in falcon tubes in the presence of a PVC tube strip. Following 10 days of incubation, a larger biofilm structure was formed on the sides of PVC strips of M. ap wild type compared to the ΔpstA mutant. Further counting of the bacterial cells forming biofilms compared to the planktonic cells indicated that 51.6 % of ATCC 19698 participated in biofilm formation while only 31.4 % of the ΔpstA participated in biofilm formation. On the other hand, 54.1 % of cells of the complemented strain, ΔpstA∷pstA, participated in biofilm formation, an indication of the ability of the complemented strain to restore biofilm formation in M. ap. Taken together, binding to PVC (plates and strips) indicated the ability of M. ap to form a biofilm was significantly reduced in absence of pstA.
2.3. Visualization of M. ap biofilms
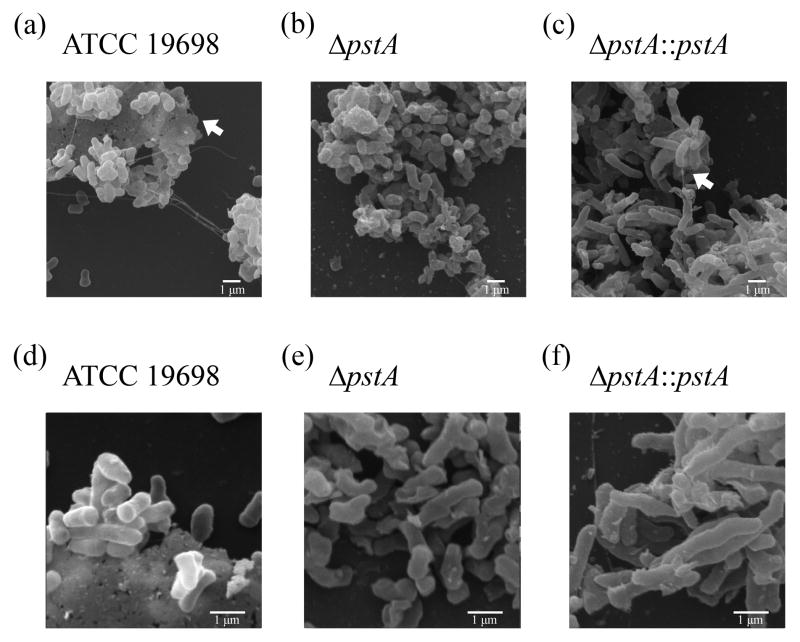
To further characterize biofilm formation on the cellular level, we examined mycobacterial bacilli with different genetic backgrounds following attachment to glass coverslips at 5, 10 and 24 hrs post incubation. Cultures of the wild type, mutant and complemented strains of M. ap were analyzed using scanning electron microscopy (SEM) following incubation in presence of HBSS (34) or Middlebrook 7H9 broth that is usually used to grow M. ap cultures. As expected, all characteristics of biofilm communities (presence of aggregates with extracellular matrix) were clearly evident in all cultures of M. ap ATCC 19698, ΔpstA∷pstA and to a lesser extent in the ΔpstA mutant (Fig. 3). A clear difference observed in the ΔpstA mutant is the lack of extensive extracellular matrix materials that were present in both M. ap ATCC 19698 and the ΔpstA∷pstA strains. Similar biofilm characteristics were observed in samples analyzed from HBSS (Fig. 3) or 7H9 broth cultures (data not shown), suggesting the ability of M. ap to form biofilms, regardless of the culture medium. Additionally, biofilm structures were clearly developed as early as 5 hrs post-incubation in all examined strains as well as at 10 and 24 hrs post-incubation. Notably, all formed structures contained extracellular matrix, a clear distinction from bacterial aggregates.
Fig. 3.
Visualization of M. ap biofilms. Scanning electron microscopy of biofilms formed by wild-type M. ap ATCC 19698, ΔpstA mutant and complemented strain, ΔpstA∷pstA. Samples were analyzed at 5 hrs post inoculation of M. ap grown on glass coverslips in presence of HBSS buffer at 10,000 X (a, b, c) or 20,000 X (d, e, f). Arrows point to extracellular matrix in different images. All analyses were repeated twice from independent cultures. Representative images are shown here.
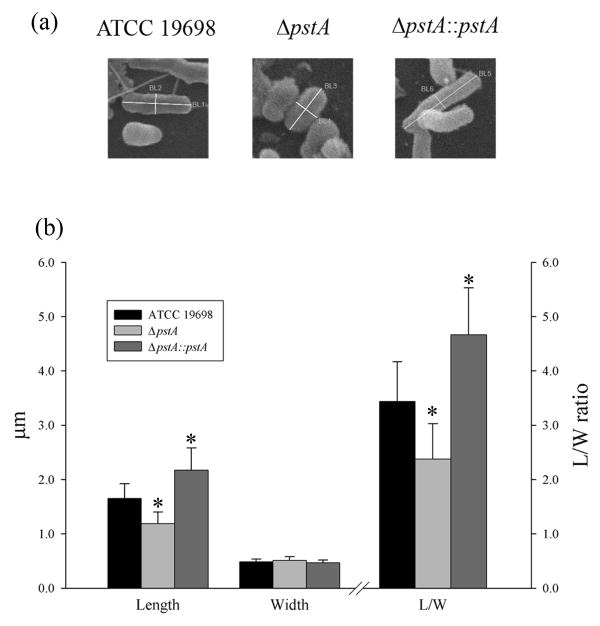
Further analysis of SEM images revealed an interesting phenotype characteristic for the ΔpstA mutant that was shorter in length compared to the other strains. To examine this phenotype further, we measured the dimensions of individual, 90 mycobacterial bacilli collected from different sample preparations of all strains. Interestingly, bacilli of the complemented strain, ΔpstA∷pstA were significantly longer (average length 2.1 μm) than the ΔpstA mutant (1.2 μm) or the wild type (1.7 μm) strains (Fig. 4). However, all bacilli showed similar width (∼0.5 μm), suggesting a role for the pstA-encoded protein in bacilli elongation, a fact that was supported by the analysis of length/width (L/W) ratio. The L/W ratio maintained the same relationship in length between strains, suggesting that cell length but not width is the reason for the observed phenotype. Overall, biofilm visualization confirmed the ability of M. ap to form a biofilm and that biofilm formation and perhaps cell size appears to be significantly affected by the disruption of the pstA gene.
Fig. 4.
Analysis of bacilli dimensions in M. ap strains with variable pstA gene content. (a) Examples of bacilli used for measuring both length and width among examined strains. (b) A histogram showing the average of bacilli length and width from different strains of M. ap. Both the length and width of 30 individual bacilli from each of the M. ap ATCC 19698, ΔpstA and ΔpstA∷pstA cultures were measured. Also, the lengths to width (L/W) ratios were calculated. Bars represent averages with ±SD for each analyzed sample. Significant differences with p< 0.05 are denoted by stars. The ratio units are displayed on the right hand of the graph.
2.4. The lipid profile of the ΔpstA mutant
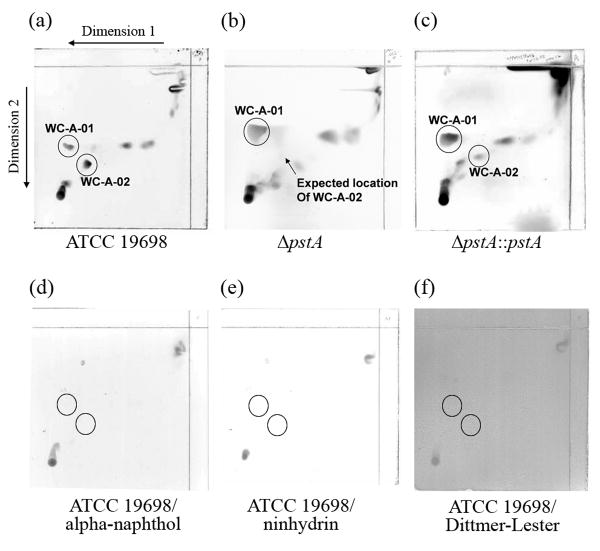
The biosynthesis of GPLs in M. avium is initiated by an operon encoding for non-ribosomal peptide synthetases (pstA and pstB) (3). It is possible that disruption of the pstA gene in M. ap could disrupt the synthesis of this or a similar lipopeptide. To test this hypothesis, the whole lipid profile of the ΔpstA mutant was compared to that of the wild-type M. ap ATCC 19698. Using two-dimensional thin-layer chromatography (2D-TLC) in the five standard solvent systems (A through E) spanning the whole polarity, one difference between the two lipid profiles was identified within the polar system A. This lipid, termed WC-A-02, was found only in the wild-type ATCC 19698 but not the transposon mutant, ΔpstA (Fig. 5a and b). No other differences were identified within the lipid composition of the parental strain and the transposon mutant strain. Moreover, when the lipid profile of the complemented strain (ΔpstA∷pstA) was examined, the presence of the WC-A-02 lipid was partially restored (Fig. 5c).
Fig. 5.
Lipidomic analysis of M. ap strains used in this study. Thin layer chromatography (TLC) of lipid extracts from the wild-type ATCC 19698 (a), the ΔpstA mutant (b) and the pstA complemented strain (ΔpstA∷pstA) (c). TLC plates stained with alpha-naphthol (d), ninhydrin (e) or Dittmer- Lester (f) of M. ap ATCC 19698 samples are also shown,. Key lipopeptides in different TLC runs are encircled.
Additional structural analyses of the nature of the unique WC-A-02 lipid by a differential staining protocol employed before (7). The generated staining profile of WC-A-03 suggested the presence of a lipopeptide without a sugar moiety (Fig. 5d) or a free primary amino group (Fig. 5e). Furthermore, this lipid is not a phospholipids (Fig. 5f). Since no primary amino group could be detected a peptide should be N-terminal linked to another moiety such as a fatty acyl chain. Thus, the most likely structure of WC-A-02 should be a lipopeptide. In general, the lipidomic analysis indicated that the pstA gene is involved in the generation of a cell wall lipid with the strong indication that it is involved in the biosynthesis of a lipotripeptide since the non-ribosomal peptide synthetase contains three amino acid binding modules.
2.5. Virulence of the M. ap ΔpstA mutant in cattle
Previously, we showed that a pstA insertional mutant of M. ap is not able to efficiently colonize the intestines and livers of infected mice (26). Because of the sampling strategy used in that study, the early stages of tissue colonization were not analyzed. To examine the role of pstA in M. ap virulence in cattle, we examined the ability of the M. ap ΔpstA mutant to transverse the intestinal epithelial cells using a recently described bovine model for Johne's Disease (33). Both the wild type and its isogenic mutant, ΔpstA, were surgically inoculated directly to the ileum of a paratuberculosis-free calf. Infection was allowed to proceed for 2 hrs before samples from mesenteric lymph nodes and liver were harvested for colony counts. Typically, the competitive index of virulence (CIV) of virulent strains should be 1. In the case of the ΔpstA mutant, the CIV was < 0.4 compared to the wild-type M. ap ATCC 19698 (Fig. 6) indicating that the ΔpstA had a reduced ability to transverse the intestine to the liver and mesenteric lymph nodes. The low CIV suggested that the pstA gene product can play a key role in intestinal invasion, particularly during early stages of infection.
Fig. 6.
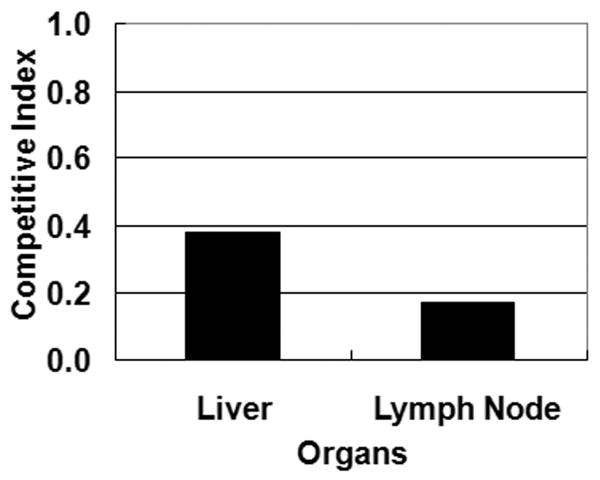
Contribution of pstA to the virulence of M. ap in cattle. Competitive index of the calf intestinal invasion assay for the liver and lymph node samples collected at 2 hrs following intra-intestinal inoculation of an equal mix of the wild-type ATCC 19698 and ΔpstA mutant.
2.6. Immunogenicity of PstA
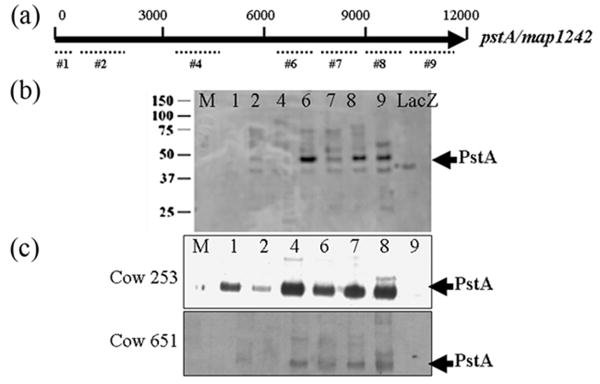
The large pstA coding sequence was divided up into 9 segments for cloning and expression in E. coli. Seven of the 9 segments were successfully cloned, expressed and purified (Fig. 7a). All 7 purified proteins were detected by immunoblot analysis using monoclonal antibodies directed against the protein tag, MBP (data not shown). However, when identical immunoblots were probed with sera from rabbits exposed to M. ap, strong signals were detected only for PstA-6, PstA-8 and PstA-9 peptides, which encompass the C-terminal half of this large protein (Fig. 7b). These data suggest that these regions of the C-terminus of PstA might be surface exposed and hence, the most immunogenic. To examine the immunogenicity of PstA peptides in naturally infected animals, the PstA peptides were analyzed by immunoblot using sera from a total of 6 Johne's disease infected cows and 7 disease-free animals. Consistently, sera from infected animals (N=6) gave more bands with PstA peptides than when rabbit sera were used, including the peptides numbered 6, 7 and 8 (Fig. 7c). Interestingly, more PstA fragments with higher intensities were detected with a serum sample from a cow suffering from sub-clinical infection (cow 253) compared to clinically-infected cows of Johne's disease. As expected, sera from control cows (N=7) did not yield any reactivity with PstA peptides suggesting the specificity of the PstA (data not shown).
Fig. 7.
Immunogenicity of PstA using bovine serum samples and immunoblot analysis. (a) A simplified structural map of pstA gene displaying regions used for peptide selection and expression. (b) An example of immunoblot of peptides #1 to 9 that were probed with a serum sample collected from a rabbit exposed to M. ap. (c) Examples of immunoblots probed with serum samples collected from a Johne's disease positive cows (ID#253, top and ID#651, bottom). Lane assignments for (b) and (c): M-Protein size markers; 1- PstA-1; 2- PstA-2; 4- PstA-4; 6- PstA-6; 7-PstA-7; 8- PstA-8; 9- PstA-9 and LacZ negative control. Kilodalton sizes are indicated to the left of the figures while arrows to the right point to the correct bands representing the PstA fragments.
3. Discussion
PstA is the second largest gene in the M. ap genome with 12 kb (15) containing three modules, suggesting that this non-ribosomal peptide synthetase encodes for the biosynthesis of a lipotripeptide, which makes it different from the pstA gene in M. avium (2 modules) or in M. smegmatis (4 modules). Importantly, the identification of a transposon mutant (26) and construction of the complemented mutant for this very large gene has enabled a thorough examination of the role PstA plays in biofilm formation and pathogenicity. Biofilm formation is an important phenotype for pathogens that could be transmitted through environmental sources. Under sub-optimal manure processing for dairy herds, M. ap was shown to persist for long periods of time (12). It is possible that M. ap bacilli contaminating animal environments (11,23) persist in biofilms or biofilm-like structures. Experiments described here represent our attempt to characterize the ability of M. ap to form a biofilm and begin to dissect the genetic factors involved in biofilm formation of this important pathogen. Our analysis indicated that M. ap bacilli are able to form biofilms with a significant role played by the pstA gene product. Similar to M. avium (10), biofilm formation in M. ap was not completely dependent on the activity of PstA, a conclusion that was supported by the presence of a small level of biofilm structures in the ΔpstA mutant when biofilms were visualized by electron microscopy. Interestingly, SEM revealed a significant difference in bacilli length, depending on the presence of a fully active pstA gene, suggesting a role for pstA in elongation of mycobacterial cells. However, whether cell elongation/division has a causal relationship with biofilm formation that is mediated by pstA needs further studies Interestingly, the pstA gene was over-expressed in the ΔpstA∷pstA complemented strain and their bacilli were the longest among other bacilli. It is possible that pstA plays several roles important for the biology of M. ap bacilli growth and biofilm formation.
In M. smegmatis, the presence of active groEL1 gene and both short-chain fatty acids and the available iron play a role in biofilm maturation (19,21). In Fig. 2, biofilm formation of the ΔpstA mutant was significantly less than that of the wild type on day 7 and 9, but not in earlier time points. This result suggests pstA may contribute to biofilm formation in M. ap during the maturation stage, similar to the role played by groEL1 in M. smegmatis. The lipidome analysis of the wild-type M. ap and its isogenic, ΔpstA mutant identified a particular cell wall lipid (WC-A-02) that was missing in the lipidome of the ΔpstA mutant. The employed lipid extraction protocol suggested that the WC-A-02 could be a component of cell wall of M. ap, similar to the lipopeptide component of glycopeptidolipids (GPLs) present in M. avium (28). The identified lipid spot does not represent a glyco- or glycopeptidolipids (negative with α-naphthol staining) nor a phospholipids (negative Lester-Dittmer staining). Furthermore, it does not contain amino acids with NH2 groups nor does it have a free N-terminus. Since the fatty acyl chain within the GPLs of M. avium is a long β-hydroxy fatty acyl attached to a phenylalanine, this might be the case for M. ap as well, especially, after comparative genomics reveals a high similarity between the pstA of M. avium and M. ap. It is possible that the synthetase encoded by the pstA gene participates in the pathway that leads to the formation of this particular lipotripeptide.
We also employed a standard protocol for colony counting to quantify the contribution of the pstA gene to biofilm formation on solid surfaces, a key phenotype that cannot be quantified by SEM analysis. Our colony counting protocol showed that inactivation of the pstA gene yielded a mutant that was still able to form biofilm but to a lesser extent compared to the wild-type stain, suggesting the presence of other genes that could participate in the M. ap biofilms. In M. avium, several genes were shown to contribute to biofilm formation (34). It is possible to identify the whole set of genes important for biofilm formation by screening the M. ap mutant library that was screened before for virulence genes (26). In this screen, the pstA gene was shown to be involved in virulence. Whether biofilm-related genes other than pstA are also involved in M. ap virulence remains to be investigated.
An important goal in this study was to analyze the role played by the pstA gene on the virulence of M. ap, especially during early stages of Johne's disease. We demonstrated that the gene was expressed during infection, as segments of the protein were detected by sera from Johne's disease cows indicating its immunogenicity. Previously, GPLs of M. avium were shown to induce proinflammatory responses (2). Furthermore, the calf model of infection suggested a role for pstA in early stages of M. ap invasion across intestinal barriers. It is possible that the early translocation event tested in the calf model is dependent on the ability of the pstA gene product to interact with intestinal cells which can lead to rapid spreading from the site of primary infection (intestine) to other organs such as spleen and liver. A role for interacting with host cells were suggested before for GPLs in M. avium (28) and could explain the inefficiency of intestinal invasion by the ΔpstA mutant observed in the calf model of invasion employed here. However, the virulence of ΔpstA mutant should be tested on more calves in the future.
In summary, our results and others analyzing M. smegmatis (25) and M. avium (34) strains indicate that the pstA gene could be involved in biofilm formation in both virulent and avirulent strains of mycobacteria. More experiments are needed to focus on the role played by pstA and host cells in M. ap pathogenesis and to further decipher the molecular interactions activate between host cells and mycobacterial cell wall components, in general. Such analysis can also improve our understanding of the pathogenesis of the inflammatory bowl diseases including Johne's and Crohn's diseases. Peptides derived from pstA sequence could be further developed to a diagnostic tool to differentiate infected from healthy animals. In general, findings presented here will facilitate the analysis of other mycobacterial species and can provide a model for the analysis of other biofilm-forming pathogens.
4. Materials and Methods
4.1. Bacteria and plasmids
Mycobacterium avium ss. paratuberculosis (M. ap) strains used in this study include the type strain ATCC 16968, its isogenic mutant ΔpstA and the complemented strain ΔpstA∷pstA. All strains used in this study were grown in 7H9 broth (Difco) supplemented with Mycobactin J, 10 % ADC and 0.05 % Tween 80 (32). The ΔpstA mutant was generated using transposon mutagenesis of ATCC 19698, as described previously (26). The ΔpstA mutant was grown in Middlebrook media supplemented with kanamycin (30 μg/mL). The complemented strain (ΔpstA∷pstA) was grown in media supplemented with kanamycin (30 μg/mL) in addition to hygromycin (100 μg/mL). To construct the complementation vector (pVV16-MAP1242), the sequence of the MAP1242 (pstA) was amplified in two overlapping fragments of the whole ORF (∼12 Kb) by using the primers (pstA1-4) listed in Table 1. The fragments were amplified with High Fidelity Taq/Tgo Polymerase (Roche) in a MyCycler (BioRad) thermal cycler with 35 cycles of 95 °C for 1 min, 67 °C for 1 min and 72 °C for 8 min. PCR products were purified from a gel and the ends were digested with NdeI, BamHI, and HpaI restriction enzymes before the ligation with T4 ligase (Promega) into the pVV16, an E. coli-mycobacteria shuttle vector. Positive transformants were selected on LB agar plates containing kanamycin after transforming into E. coli DH5α. The plasmid pVV16-MAP1242 was transformed into the ΔpstA mutant strain by electroporation with 1000 Ω, 25 μF, and 1.5 kV using standard protocols (30). Transformants were selected on 7H11 agar plates supplemented with 10 % OADC, mycobactin J (2 μg/mL), kanamycin (50 μg/mL) and hygromycin (100 μg/mL).
Table 1.
Primers used in this study.
| Primer | Purpose | Sequence |
|---|---|---|
| pstA-1 | pstA cloning | GGGGCATATGGGTGGAGACTTGGGG |
| pstA-2 | pstA cloning | CACCGGATCCAGCCCCTGCACCTCGATCTCG |
| pstA-3 | pstA cloning | GGACGGATCCGGTGATCGTATTCAACTACCT |
| pstA-4 | pstA cloning | CGGCGTTAACACCTATCACCAGAGC |
| pstA-P1F | Peptide production | ATCCTCTAGAGTGGGTGGAGACTTGGGGGAG |
| pstA-P1R | Peptide production | GATGGCAAGCTTTTAGGACAGCTC |
| pstA-P2F | Peptide production | ATCCTCTAGACTGGACCCGTCCGTCGCCAACCGG |
| pstA-P2R | Peptide production | GCGCAAGCTTAGTCCTCCGGCGACGACGCCAC |
| pstA-P3F | Peptide production | ATCCTCTAGACTGCTGGTCGCGGAGCAGGTCAG |
| pstA-P3R | Peptide production | GCGCAAGCTTCAGGCGATGTTGATGTCCTCGAT |
| pstA-P4F | Peptide production | ATCCTCTAGAGCGCAGCATCACAGCGGTCAGGA |
| pstA-P4R | Peptide production | GCGCAAGCTTCAGGTGACGAAGCCGCGGTACGG |
| pstA-P5F | Peptide production | ATCCTCTAGACTGGCGGCCCGCGACGTGCCGGC |
| pstA-P5R | Peptide production | GCGCAAGCTTACAGCGCCGCCAGCGCCGACGG |
| pstA-P6F | Peptide production | ATCCTCTAGACTGCACGCGGCGTTGGTGATCGG |
| pstA-P6R | Peptide production | GCGCAAGCTTCACAACGTGTAATCGACATACTG |
| pstA-P7F | Peptide production | ATCCTCTAGACAGCGCGCCCAGTTCGGTGACCT |
| pstA-P7R | Peptide production | GCGCAAGCTTCACACTGAGAAGTCGAAGGCCAG |
| pstA-P8F | Peptide production | ATCCTCTAGAGAGATCTTCGGCGCGCTCCTGAA |
| pstA-P8R | Peptide production | GCGCAAGCTTCAGGTCCGATCGTCCAGGCCGAG |
| pstA-P9F | Peptide production | ATCCTCTAGACTCAAGCCGTACAGCTAC |
| pstA-P91R | Peptide production | GCGCAAGCTTCAGAGCAAGCCCAGCAACTGCAG |
4.2. Biofilm formation assays
Cultures of M. ap ATCC 19698 and its isogenic ΔpstA mutant grown to log phase (OD600 = 1.0) were centrifuged and resuspended to OD600 = 1.0 using Hanks' Balanced Salt Solution (HBSS) (BioWhittaker/Cambrex). Aliquots of 200 μl of bacterial resuspension were dispensed into a 96-well, polyvinyl chloride (PVC) plate (Falcon) in triplicate. Plates were incubated at 37 °C without agitation. To quantify biofilm formation, 15μl of 1 % crystal violet was added to each well and incubated for 15 min. Plates were then washed 3 times to remove bacteria that had not adhered to the PVC plate. Ethanol (95 %) was added to each well and the absorbance of wells was recorded at a 540 nm wavelength using a Thermodynamic plate reader (Fisher Scientific). The net absorbency for each sample is recorded after subtracting out the absorbency of sterile culture media used as a control. All assays were performed in triplicates and repeated 3 times starting from new M. ap cultures.
To quantify the percentage of biofilm formation in M. ap strains, cultures grown to OD600=1.0 were transferred to 50 ml Falcon tubes in which a 5 x 2 cm2-strip of PVC tube was placed inside and incubated at 37 °C with slow shaking (50 rpm). At 10 days post inoculation, PVC strips were washed and M. ap cells adhered to PVC tubes were scrapped thoroughly, until no film was visible, and rinsed with HBSS for colony counting on Middlebrook 7H10 agar plates. Aliquots were also collected from planktonic cultures (cultures in Falcon tubes not attached to the PVC strips) of each examined strain. The number of colony forming units (CFU) retrieved from the PVC tube was divided by the CFU from the 50-ml tube to represent the percentage of cells forming biofilms (biofilm percentage).
4.3. Scanning electron microscopy
Bacterial cultures, when OD600 reached 1.0, were sonicated at 50 Watts for 2 min or until well-dispersed in a water-filled cup horn sonicator (Fisher Scientific), washed once with HBSS and resuspended in HBSS or Middlebrook 7H9 broth. On a 24-well cell culture plate, 500 μl of the bacterial suspensions were added onto an autoclaved 12-mm circular coverslip placed in the wells. The plate was sealed with parafilm to prevent liquid evaporation. After 24 hr of incubation at 37 °C, each coverslip was washed once with 1 ml HBSS and fixed by incubating in 500 μl of 2.0 % glutaraldehyde in a 0.1 M phosphate buffer, pH 7.4, overnight at 4 °C. The samples were dehydrated through an alcohol series with 30 %, 50 %, 70 %, 80 %, 90 %, 95 % and 100 % ethanol followed by drying with molecular sieve for 5 min. Dehydrated specimens were dried by the critical point procedure using 100 % sieve-dried ethanol (Samdri 780-A, Tousimis) as the intermediate fluid and liquid CO2, as the transitional fluid. All samples were then splatter coated with gold-palladium to a thickness of approximately 20 nm and observed with a Hitachi S-570 scanning electron microscope at 10kV accelerating voltage (31).
4.4. Lipid extraction and thin-layer chromatography (TLC)
Bacterial cells from M. ap cultures were harvested after 8 weeks of growth by scraping from Middlebrook 7H11 agar plates with supplements as described above. Cells were lyophilized prior to lipid extraction and total cell lipids were extracted twice with chloroform/methanol (2:1) (30 ml per gram of lyophilized cells) at 55 °C for 3 hrs. Crude lipid extracts were dried down under nitrogen and subjected to a Folch wash (chloroform/methanol/water 4:2:1 ml). The organic phase was transferred to a new tube and dried under nitrogen. Whole cell lipids were resuspended in chloroform/methanol (2:1) at a concentration of 10 mg ml-1. Two-dimensional thin layer chromatography was performed on 10 x 10 cm aluminum-backed silica gels 60 F254. Samples of a 200 μg/each of total cell lipids were separated in 5 systems with different polarities (A→E) using standard TLC protocols (7). For example, system A was composed of chloroform/ methanol/water 100:14:0.8 for dimension 1 and chloroform/acetone/methanol/ water 50:60:2.5:3 for dimension 2. Separated lipids were visualized by spraying with 10 % CuSO4 in 8 % H3PO4 followed by heating until spots appeared. Differential staining for structural analyses of the lipid WC-A-02 were performed with α-naphthol (sugar moieties), ninhydrin (free primary amino groups), and Dittmer-Lester (phospholipids) followed by heating for α-naphthol and ninhydrin as described before (7).
4.5. Calf intestinal invasion assay
Both the wild-type M. ap ATCC 19698 and its isogenic mutant, ΔpstA mutant were grown to OD600=1.0. Before animal inoculation, cultures were centrifuged, resuspended in PBS and equal volumes of cultures were mixed. A male Holstein calf, 14 days of age, was purchased from a dairy herd that has been Johne's disease-free for the last 5 years and that is maintained by the School of Veterinary Medicine, University of Wisconsin-Madison. Sera and fecal samples collected from the calf and the dam were examined and demonstrated to be negative for evidence of M. ap infection by means of an IDEXX ELISA test (16) before inclusion in the study. All animal waste and disposable utensils used throughout the experiment were autoclaved before disposal. A total of 10 ml of the culture mixture was inoculated into 10 cm of occluded ileum of the anaesthetized calf as described before (33). Following infection (1-2 hrs), samples were taken from mesenteric lymph nodes, small intestine, liver and spleen. Duplicate tissue homogenates were processed for bacterial counting using Middlebrook 7H10 agar in the presence or absence of kanamycin supplement. The identity of colonies retrieved from animals was verified using PCR. The percentage of wild type and mutant counts was applied in the following formula to calculate the competitive index of virulence (33).
4.6. Production of PstA peptides and antibodies
The 12 kb coding sequence of pstA was too large to clone the entire open reading frame (ORF) into a protein expression vector. Therefore, the coding sequence was divided into 9 segments for cloning and protein production. A maltose binding protein (MBP) fusion for each of these PstA segments was constructed using the pMAL-c2 vector (New England Biolabs, Beverly, MA, USA) as described before (27). The upstream and downstream primers for each segment are listed in Table 1. Because only segment 6 of PstA appeared to show the strongest reactivity, that segment was selected for production of polyclonal antibody to serve as a positive control for subsequent immunoblot analysis. To generate fragment 6 antibodies, 2 New Zealand white rabbits were immunized with the PstA-6 protein. Pre-immunization sera were collected one day before initial injections of antigen. Immunizations began using 200 μg of purified protein injected intradermally (ID) into each of two rabbits. Twenty-eight days later a boost of 100 μg peptide was administered subcutaneously (SC). A second boost was given on day 42 comprising 50 μg peptide ID and 50 μg SC. A final boost with 50 μg peptide delivered ID and SC was given at day 70. All immunizations were performed using Freund's incomplete (Sigma-Aldrich, St. Louis, MO, USA) as the adjuvant.
4.7. Immunoblot analysis
Polyacrylamide gel electrophoresis was performed using 12% (w/v) polyacrylamide gels. Electrophoretic transfer of proteins onto pure nitrocellulose (Schleicher and Schuell, Keene, NH, USA) was accomplished with the Bio-Rad Trans Blot Cell (Bio-Rad Laboratories, Richmond, CA, USA) with sodium phosphate buffer (25 mM, pH 7.8) at 0.8 A for 90 min. After transfer, filters were blocked with phosphate-buffered saline (PBS; 150 mM NaCl, 10 mM NaPO4, pH 7.4) plus 2% bovine serum albumin (BSA) and 0.1% Tween 20, referred to hereafter as PBS-BSA. Rabbit antisera were diluted 1:1000 in PBS-BSA and all cattle sera were diluted 1:300. All cattle serum samples (N=13) were obtained from the National Animal and Disease Center's (NADC) herd where their Johne's disease status is well-documented. Also, the status of the source animals for sera was blinded until the end of the experiment. The primary sera were incubated on the blot at room temperature for 2 h. After three washes in PBS plus 0.1 % Tween-20, blots were incubated for 1.5 h in goat anti-bovine-peroxidase (ThermoScientific-Pierce) diluted 1:20,000 in PBS-BSA. The blots were again washed three times as described above and developed for chemiluminesence using Supersignal detection reagents (ThermoScientific-Pierce).
4.8. Sequence and statistical analyses
The sequence of the pstA gene in M. ap and its orthologues in other bacteria were downloaded from the GenBank database and prepared for BLASTp analysis (1). Only sequences with E-value < 10-5 within at least 25 % of the overall sequence were included in constructing the phylogenetic tree of orthologues sequences. Alignments of sequences were generated by the CLUSTALW program implemented in the MEGA 3.1 Software (14). Phylogenetic trees were generated from the aligned sequences using Neighbor-Joining algorithm followed by bootstrapping (1000 X).
For statistical analysis, Student's t-test implemented in Microsoft Excel program was used to evaluate differences in the bacterial colonization and the ability to form a biofilm between M. ap strains used in this study. A level of P-value <0.05 is considered statistically significance among paired samples.
Acknowledgments
We would like to acknowledge Bassam Abomoelak, Sarah K. Ward and Kathy L. Talaat for reading the manuscript. Research reported here is supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (AGRICCREE 2003-02230) and the Animal Formula Fund (WIS01093) as well as Johne's Disease Integrated Program (2004-35605-14243) for AMT and R01 AI-51283 for JMI. Portions of this study were also supported by the USDA-Agricultural Research Service. Specimen preparation and SEM imaging was performed at the University of Wisconsin Laboratory for Biological and Biomaterials Specimen Preparation, Imaging, and Characterization (BBPIC) with assistance by Joseph A. Heintz.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bhatnagar S, Schorey JS. Exosomes Released from Infected Macrophages Contain Mycobacterium avium Glycopeptidolipids and Are Proinflammatory. J Biol Chem. 2007;282:25779–25789. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cangelosi GA, Do JS, Freeman R, Bennett JG, Semret M, Behr MA. The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrobial Agents & Chemotherapy. 2006;50:461–468. doi: 10.1128/AAC.50.2.461-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng J, Bull TJ, Dalton P, Cen S, Finlayson C, Hermon-Taylor J. Mycobacterium avium subspecies paratuberculosis in the inflamed gut tissues of patients with Crohn's disease in China and its potential relationship to the consumption of cow's milk: A preliminary study. World Journal of Microbiology & Biotechnology. 2005;21:1175–1179. [Google Scholar]
- 5.Cucarella C, Tormo MA, Ubeda C, Trotonda MP, Monzon M, Peris C, Amorena B, Lasa I, Penades JR. Role of Biofilm-Associated Protein Bap in the Pathogenesis of Bovine Staphylococcus aureus. Infect Immun. 2004;72:2177–2185. doi: 10.1128/IAI.72.4.2177-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson WR, Manning EJB, Nettles VF. Culture and serologic survey for Mycobacterium avium subsp paratuberculosis infection among Southeastern white-tailed deer (Odocoileus virginianus) J Wildl Dis. 2004;40:301–306. doi: 10.7589/0090-3558-40.2.301. [DOI] [PubMed] [Google Scholar]
- 7.Eckstein TM, Chandrasekaran S, Mahapatra S, McNeil MR, Chatterjee D, Rithner CD, Ryan PW, Belisle JT, Inamine JM. A major cell wall lipopeptide of Mycobacterium avium subspecies paratuberculosis. Journal of Biological Chemistry. 2006;281:5209–5215. doi: 10.1074/jbc.M512465200. [DOI] [PubMed] [Google Scholar]
- 8.Falkinham JO. Growth in catheter biofilms and antibiotic resistance of Mycobacterium avium. J Med Microbiol. 2007;56:250–254. doi: 10.1099/jmm.0.46935-0. [DOI] [PubMed] [Google Scholar]
- 9.Falkinham JO, III, Norton CD, LeChevallier MW. Factors Influencing Numbers of Mycobacterium avium, Mycobacterium intracellulare, and Other Mycobacteria in Drinking Water Distribution Systems. Appl Environ Microbiol. 2001;67:1225–1231. doi: 10.1128/AEM.67.3.1225-1231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman R, Geier H, Weigel KM, Do J, Ford TE, Cangelosi GA. Roles for Cell Wall Glycopeptidolipid in Surface Adherence and Planktonic Dispersal of Mycobacterium avium. Appl Environ Microbiol. 2006;72:7554–7558. doi: 10.1128/AEM.01633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenstein RJ, Collins MT. Emerging pathogens: is Mycobacterium avium subspecies paratuberculosis zoonotic? Lancet. 2004;364:396–397. doi: 10.1016/S0140-6736(04)16781-0. [DOI] [PubMed] [Google Scholar]
- 12.Grewal SK, Rajeev S, Sreevatsan S, Michel FC. Persistence of Mycobacterium avium subsp paratuberculosis and other zoonotic pathogens during simulated composting, manure packing, and liquid storage of dairy manure. Applied & Environmental Microbiology. 2006;72:565–574. doi: 10.1128/AEM.72.1.565-574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13:7–10. doi: 10.1016/j.tim.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci USA. 2005;102:12344–12349. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning EJB, Kucera TE, Gates NB, Woods LM, Fallon-McKnight M. Testing for Mycobacterium avium subsp Paratuberculosis infection in asymptomatic free-ranging tule elk from an infected herd. J Wildl Dis. 2003;39:323–328. doi: 10.7589/0090-3558-39.2.323. [DOI] [PubMed] [Google Scholar]
- 17.Nordlund KV, Goodger WJ, Pelletier J, Collins MT. Associations between subclinical paratuberculosis and milk production, milk components, and somatic cell counts in dairy herds. Journal of the American Veterinary Medical Association. 1996;208:1872–1876. [PubMed] [Google Scholar]
- 18.Norton CD, LeChevallier MW, Falkinham JO. Survival of Mycobacterium avium in a model distribution system. Water Res. 2004;38:1457–1466. doi: 10.1016/j.watres.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Ojha AK, Anand M, Bhatt A, Kremer L, Jacobs WR, Hatfull GF. GroEL1: A Dedicated Chaperone Involved in Mycolic Acid Biosynthesis during Biofilm Formation in Mycobacteria. Cell. 2005;123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR, Jr, Hatfull GF. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008;69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ojha AK, Hatfull GF. The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol Microbiol. 2007;66:468–483. doi: 10.1111/j.1365-2958.2007.05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ott SL, Wells SJ, Wagner BA. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev Vet Med. 1999;40:179–192. doi: 10.1016/s0167-5877(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 23.Pickup RW, Rhodes G, Bull TJ, Arnott S, Sidi-Boumedine K, Hurley M, Hermon-Taylor J. Mycobacterium avium subsp. paratuberculosis in Lake Catchments, in River Water Abstracted for Domestic Use, and in Effluent from Domestic Sewage Treatment Works: Diverse Opportunities for Environmental Cycling and Human Exposure. Applied and Environmental Microbiology. 2006;72:4067–4077. doi: 10.1128/AEM.02490-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Recht J, Kolter R. Glycopeptidolipid Acetylation Affects Sliding Motility and Biofilm Formation in Mycobacterium smegmatis. J Bacteriol. 2001;183:5718–5724. doi: 10.1128/JB.183.19.5718-5724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recht J, Martinez A, Torello S, Kolter R. Genetic Analysis of Sliding Motility in Mycobacterium smegmatis. J Bacteriol. 2000;182:4348–4351. doi: 10.1128/jb.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin SJ, Wu CW, Steinberg H, Talaat AM. Identification of Novel Virulence Determinants in Mycobacterium paratuberculosis by Screening a Library of Insertional Mutants. Infec Immun. 2006;7:3825–3833. doi: 10.1128/IAI.01742-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talaat AM, Ward SK, Wu CW, Rondon E, Tavano C, Bannantine JP, Lyons R, Johnston SA. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: Insights from genome-wide transcriptional profiling. J Bacteriol. 2007;189:4265–4274. doi: 10.1128/JB.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vergne I, Prats M, Tocanne JF, Laneelle G. Mycobacterial glycopeptidolipid interactions with membranes: a monolayer study. FEBS Lett. 1995;375:254–258. doi: 10.1016/0014-5793(95)01219-5. [DOI] [PubMed] [Google Scholar]
- 29.von Reyn CF, Arbeit RD, Tosteson ANA, Ristola MA, Barber TW, Waddell R, Sox CH, Brindle RJ, Gilks CF, Ranki A, Bartholomew C, Edwards J, Falkinham JO, O'Connor GT, Jacobs NJ, Maslow J, Lahdevirta J, Buhler S, Ruohonen R, Lumio J, Vuento R, Prabhakar P, Magnusson M. The international epidemiology of disseminated Mycobacterium avium complex infection in AIDS. AIDS. 1996;10:1025–1032. doi: 10.1097/00002030-199610090-00014. [DOI] [PubMed] [Google Scholar]
- 30.Wards BJ, Collins DM. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol Lett. 1996;145:101–105. doi: 10.1111/j.1574-6968.1996.tb08563.x. [DOI] [PubMed] [Google Scholar]
- 31.Woo SR, Heintz JA, Albrecht R, Barletta RlG, Czuprynski CJ. Life and death in bovine monocytes: The fate of Mycobacterium avium subsp. paratuberculosis Microb Pathog. 2007;43:106–113. doi: 10.1016/j.micpath.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Wu CW, Glasner J, Collins MT, Naser S, Talaat AM. Whole Genome Plasticity among Mycobacterium avium subspecies: Insights from Comparative Genomic Hybridizations. J Bacteriol. 2006;188:711–723. doi: 10.1128/JB.188.2.711-723.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu CW, Livesey M, Schmoller SK, Manning EJB, Steinberg H, Davis WC, Hamilton MJ, Talaat AM. Invasion and Persistence of Mycobacterium paratuberculosis During Early Stages of Johne's Disease in Calves. Infec Immun. 2007;75:2110–2119. doi: 10.1128/IAI.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamazaki Y, Danelishvili L, Wu M, MacNab M, Bermudez LE. Mycobacterium avium genes associated with the ability to form a biofilm. Appl Environ Microbiol. 2006;72:819–825. doi: 10.1128/AEM.72.1.819-825.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]