Abstract
Respiratory infections caused by nontypeable Haemophilus influenzae (NTHi) are a major medical problem. Evidence suggests that the ability to form biofilms on mucosal surfaces may play a role in NTHi pathogenesis. However, the factors that contribute to NTHi biofilm cohesion remain largely unknown. In this study we investigated the biofilm growth and detachment phenotypes of eight NTHi clinical strains in vitro. We found that the majority of strains produced biofilms within 6 hours when cultured statically in tubes. Biofilm formation was inhibited when culture medium was supplemented with proteinase K or DNase I. Both enzymes also caused significant detachment of pre-formed NTHi biofilms. These findings indicate that both proteinaceous adhesins and extracellular DNA contribute to NTHi biofilm cohesion. Treatment of NTHi biofilms cultured in centrifugal filter devices with DNase I, but not with proteinase K, caused a significant decrease in fluid convection through the biofilms. These results suggest that extracellular DNA is the major volumetric component of the NTHi biofilm matrix. Mechanical or enzymatic disruption of NTHi biofilms cultured in microtiter plates significantly increased their sensitivity to killing by SDS, cetylpyridinium chloride, chlorhexidine gluconate, povidone iodine and sodium hypochlorite. These findings indicate that biocide resistance in NTHi biofilms is mediated to a large part by the cohesive and protective properties of the biofilm matrix. Understanding the mechanisms of biofilm cohesion and biocide resistance in NTHi biofilms may lead to new methods for treating NTHi-associated infections.
Keywords: Biocide, Biofilm, Extracellular DNA, Nontypeable Haemophilus influenzae, Resistance
1. Introduction
Haemophilus influenzae is a gram-negative bacterium that comprises part of the normal nasopharyngeal flora of most humans [1]. Most strains lack capsular polysaccharides and are referred to as nontypeable H. influenzae or NTHi [2]. Some NTHi strains cause otitis media, which is a major medical problem that can lead to childhood hearing loss [3]. NTHi also causes bronchitis, sinusitis and other chronic respiratory infections [3].
Biofilms are communities of bacteria, encased in a self-synthesized extracellular matrix, growing attached to a biotic or abiotic surface [4]. Evidence suggests that the ability to form biofilms may contribute to NTHi pathogenesis [1]. Matrix-encased biofilm-like structures have been observed on middle ear tissue of experimentally infected chinchillas [5], and on middle-ear mucosa biopsy specimens obtained from children undergoing tympanostomy tube placement [6]. Structures consistent with a biofilm have also been observed on cultured human airway epithelial cells [7] and on abiotic surfaces in vitro [8]. Like most biofilms, NTHi biofilms cultured in vitro exhibit increased resistance to killing by antibiotics compared to the resistance exhibited by planktonic cells [9-11]. Biofilm formation may account for the recalcitrance of many chronic NTHi infections to antibiotic therapy [1].
Numerous studies have identified NTHi cellular components, such as pili, surface proteins and lipooligosaccharide, that contribute to mucosal surface attachment and colonization [12,13]. Few studies, however, have identified cellular components that mediate biofilm-specific processes such as intercellular adhesion and biocide resistance. NTHi strains carrying mutations in the pilin genes hifA [14] or pilA [15] exhibit reduced biofilm formation in vitro. Adhesive pili have been shown to mediate biofilm formation in numerous other bacteria [16,17]. However, not all NTHi strains produce pili [18]. Sialylated lipooligosaccharides may also play a cohesive role in NTHi biofilms [19,20]. Several studies have shown that DNA is present in the matrix of NTHi biofilms recovered from the middle ear of the chinchilla [21-23], although no cohesive role for matrix DNA was identified. To date, there is no evidence that the NTHi biofilm matrix contains polysaccharide adhesins, which are common matrix components in most other bacterial biofilms [24].
The purpose of the present study was to identify extracellular components that mediate NTHi biofilm cohesion, and to further characterize NTHi biofilm resistance. We tested the ability of various matrix-degrading enzymes, detergents, antiseptics and disinfectants to inhibit, detach or kill biofilms produced by eight NTHi clinical strains in vitro. Our findings demonstrate that both proteinaceous adhesins and extracellular DNA mediate biofilm cohesion and biocide resistance in NTHi biofilms.
2. Results
2.1 Biofilm formation by NTHi strains
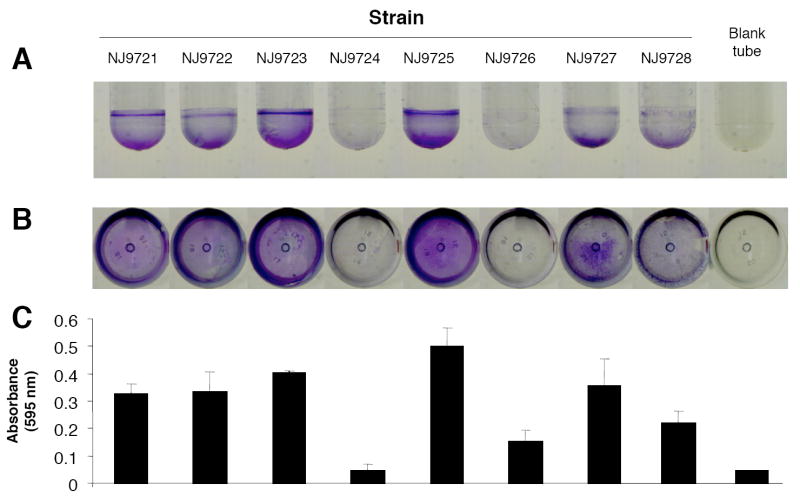
Eight NTHi clinical strains were tested for their ability to form biofilms in polystyrene culture tubes (Fig. 1). After staining the tubes with crystal violet, which stains bacterial cells and biofilm matrix components but not polystyrene [25], a visible biofilm was evident on the surface of the tube for most strains (Fig. 1A and B). The amount of biofilm formation varied among the eight strains, but was highly reproducible within each strain. For example, strains NJ9724 and NJ9726 consistently formed weaker biofilms than did the other six strains. Biofilm formation was evident at both the air-liquid interface and at the bottom of the tube. Biofilms were tenaciously attached to the surface of the tube and were resistant to detachment by vortex agitation. The biofilm phenotypes were stable after three passages on agar. Quantitation of crystal violet binding revealed that most strains produced a detectable amount of stainable, surface-associated biomass (Fig. 1C).
Fig. 1.
Test tube biofilm assay. NTHi strains were cultured for 24 h in 17-mm × 100-mm polystyrene tubes. Tubes were rinsed with water and stained with crystal violet. Panels (A) and (B) show photographs of stained tubes taken from the side and bottom, respectively. Also shown is a control tube that was inoculated with sterile BHI broth, cultured and stained with crystal violet using the same procedure. In panel (C), tubes were destained with 33% acetic acid and the absorbance of the dye solution (at 595 nm) was measured in a spectrophotometer. Values in panel (C) show mean absorbance and range for duplicate tubes.
Quantitation of biofilm formation by CFU enumeration revealed that strong biofilm-forming strains such as NJ9725 produced 107-108 biofilm cells per tube after 24 h, which constituted approximately 20% of the total CFU per tube. After 48 h, the amount of crystal violet staining remained the same, but the total CFU decreased to <102 per tube.
2.2 Inhibition of NTHi biofilm formation by DNase I and proteinase K
DNase I and proteinase K, when added to the culture medium at the time of inoculation, significantly inhibited biofilm formation by NTHi strain NJ9725 as determined by crystal violet staining (Fig. 2A). Both enzymes also inhibited biofilm formation by the other five NTHi strains that exhibited a strong biofilm phenotype (data not shown). When assayed by CFU enumeration, the presence of DNase I in the growth medium resulted in significantly less surface-associated biofilm cells (Fig. 2B). The total CFU/tube values were approximately the same for cultures grown with or without DNase I or proteinase K, indicating that neither enzyme severely affected cell growth or viability. These findings confirm that both proteinaceous adhesins and extracellular DNA contribute to NTHi biofilm cohesion.
Fig. 2.
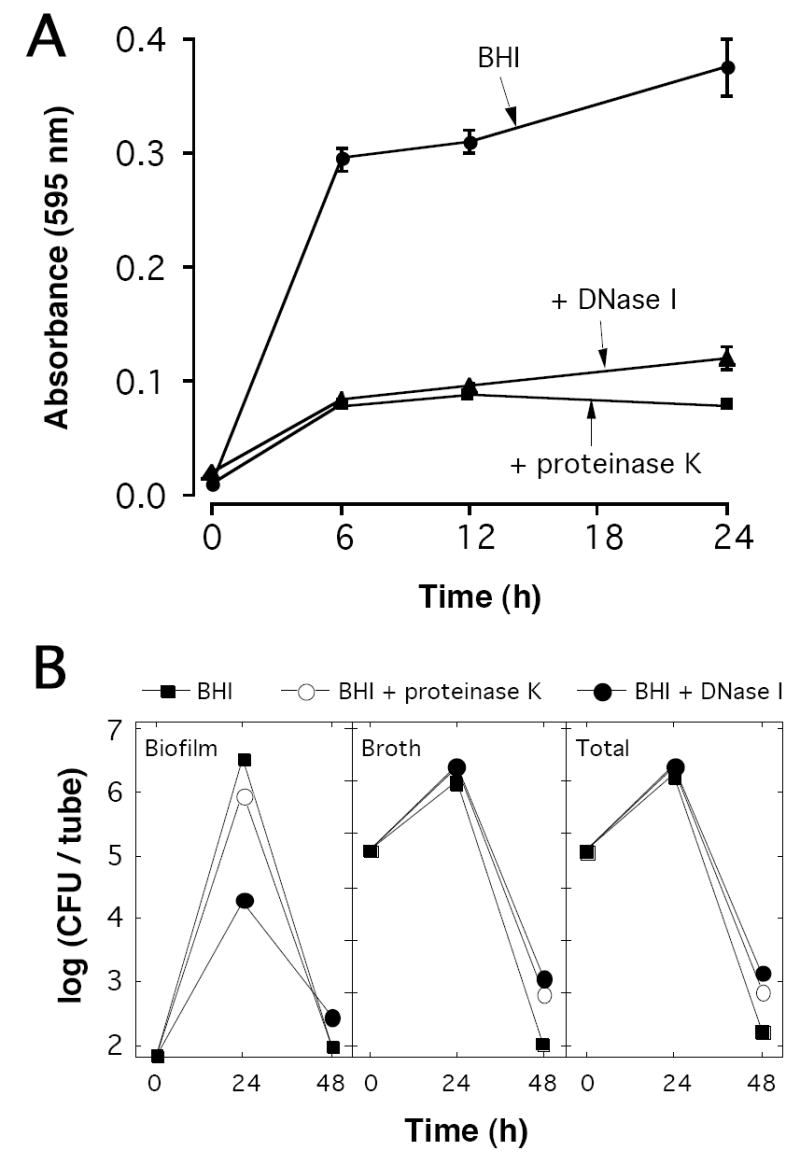
DNase I and proteinase K inhibit NTHi biofilm formation. (A) NTHi strain NJ9725 was cultured in polystyrene tubes in BHI broth or BHI broth supplemented with 1 mg mL-1 of DNase I or proteinase K. After increasing amounts of time, tubes were washed with water and biofilms were quantitated by crystal violet staining. Values show mean absorbance and range for duplicate tubes. (B) Quantitation of NTHi biofilm formation by CFU enumeration. NJ9725 biofilms were cultured in tubes and the surface-associated (biofilm) and planktonic (broth) CFU/tube values were measured separately after 24 and 48 h. The graph on the right shows the total (biofilm + broth) CFU/tube values.
2.3 Detachment of NTHi biofilms by DNase I and proteinase K
To further confirm that proteinaceous adhesins and DNA mediate NTHi biofilm cohesion, we treated 24-h-old biofilms for 1 h with 1 mg mL-1 proteinase K or DNase I (Fig. 3). Proteinase K caused significant detachment of biofilm biomass in all strains (Fig. 3A). A time-course study indicated that detachment of NJ9725 biofilms by proteinase K was very rapid, with most of the biomass detaching within 15 sec (Fig. 3B). In contrast, DNase I caused only partial detachment of biofilms produced by most NTHi strains after 1 h of treatment (Fig. 3C). These findings suggest that proteinaceous adhesins are a major mediator of intercellular cohesion in mature NTHi biofilms. NTHi biofilms were not detached by treatment with the carbohydrate-modifying agent sodium metaperiodate (data not shown), which has been shown to detach biofilms whose cohesion depends on polysaccharide adhesins [26,27].
Fig. 3.
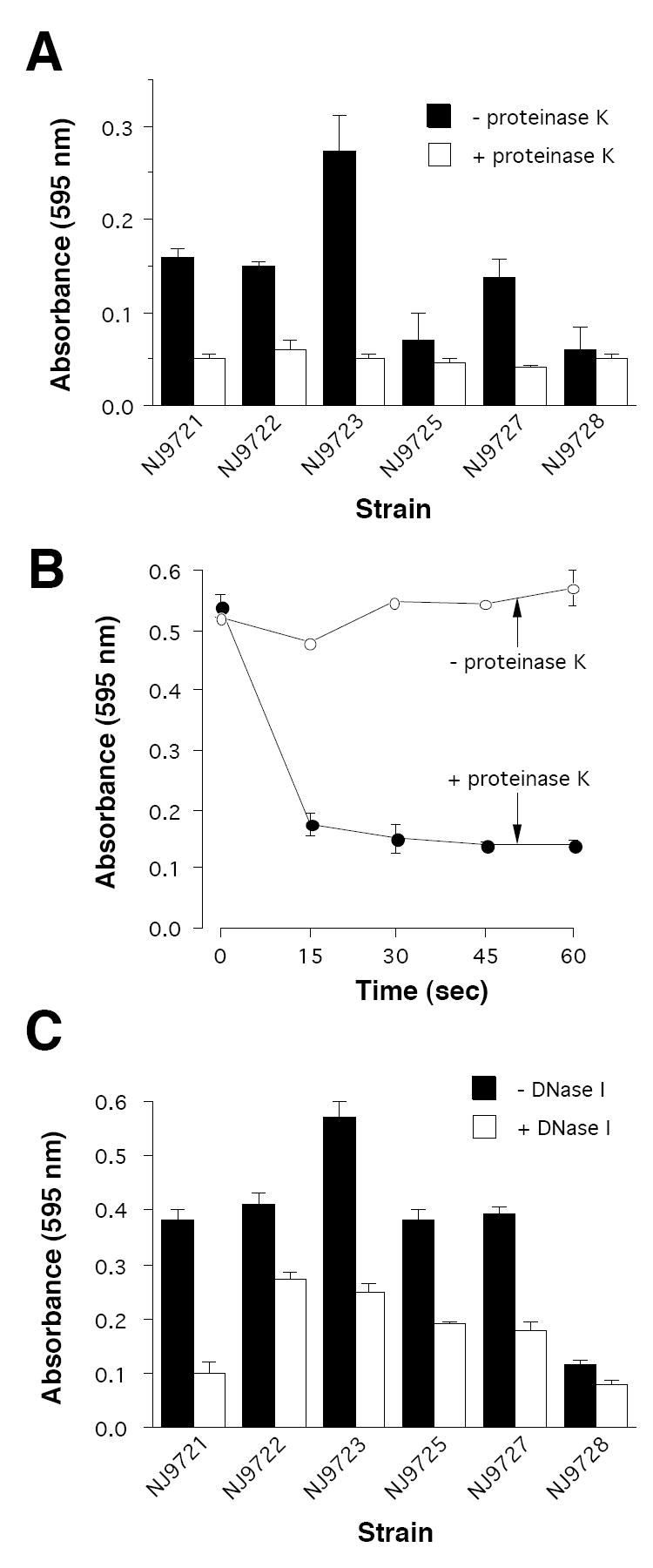
Detachment of 24-h-old NTHi biofilms by DNase I and proteinase K. All enzymes were tested in enzyme buffer at a concentration of 1 mg mL-1. (A) Detachment of biofilms in 1 h by proteinase K. (B) Time course for proteinase K-induced detachment of NJ9725 biofilms. (C) Detachment of biofilms in 1 h by DNase I. In all panels, biofilms were quantitated by crystal violet staining. Values show mean absorbance and range for duplicate tubes.
2.4 Extracellular DNA facilitates fluid convection in NTHi biofilms
To determine whether the presence of proteins and DNA in the NTHi biofilm matrix significantly alters the physical properties of the biofilms, NJ9725 biofilms were cultured for 24 h in centrifugal filter devices in the presence or absence of proteinase K and DNase I, and the devices were then subjected to low-speed centrifugation for increasing amounts of time (Fig. 4). The volume of broth that flowed through the biofilm was <50% of the volume that flowed through control filter devices inoculated with sterile BHI broth, indicating that NTHi biofilms inhibit bulk fluid convection. Fluid convection through NJ9725 biofilms cultured in the presence of proteinase K was not significantly different from fluid convection through biofilms cultured in BHI broth alone (Fig. 4). However, fluid convection through biofilms cultured in the presence of DNase I was significantly less than fluid convection through biofilms cultured in BHI broth alone. These differences were significant at all time points tested. An identical decrease in fluid convection was observed when biofilms cultured in BHI broth alone were perfused with DNase I in enzyme buffer during the centrifugation step (data not shown). The fact that a decrease in biofilm porosity was observed when biofilms were treated with Dnase I, but not with proteinase K, suggests that DNA constitutes the major volumetric component of the NTHi biofilm matrix.
Fig. 4.
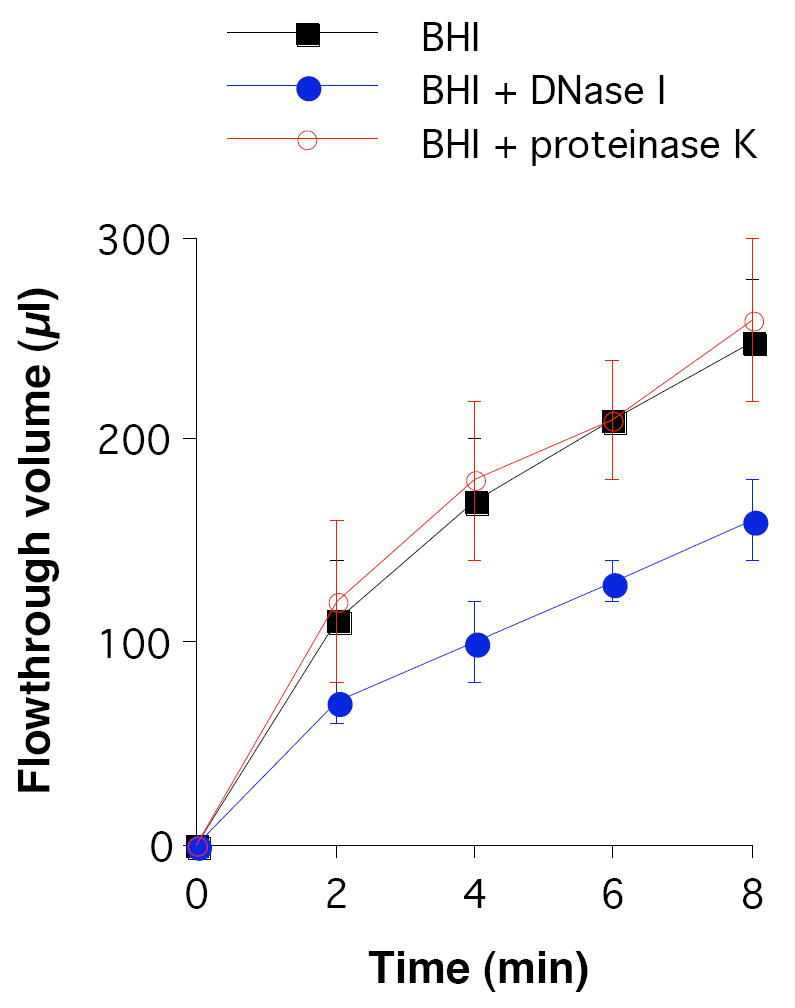
Fluid convection through 24-h-old NTHi NJ9725 biofilms cultured in centrifugal filter devices. Biofilms were cultured in BHI broth or BHI broth supplemented with 1 mg mL-1 DNase I or proteinase K. The devices were subjected to low-speed centrifugation and the flowthrough volume was measured after increasing amounts of time. Data represent mean and SD for three different experiments, each performed in duplicate devices.
2.5 NTHi biofilm resistance
Previous studies showed that NTHi biofilms cultured in vitro exhibit increased resistance to killing by antibiotics compared to the resistance exhibited by NTHi planktonic cells [9-11]. We sought to determine whether NTHi biofilms also exhibit resistance to killing by various detergents, antiseptics and disinfectants (Fig. 5). NJ9725 biofilms cultured in microtiter plates were rinsed with water and then treated for 5 min with 0.015% SDS, 0.004% CPC, 0.03% chlorhexidine gluconate, 0.004% povidone iodine or 0.00012% sodium hypochlorite. All agents were tested at concentrations sufficient to kill 99% of NJ9725 planktonic cells in 5 min. After treatment, biofilms were rinsed with BHI broth, scraped from the surface of the well, diluted and plated on agar. Treatment of NJ9725 biofilms with SDS, CPC, chlorhexidine gluconate and povidone iodine resulted in a significant decrease in CFU/well values (0.9-1.8 log units). Sodium hypochlorite treatment caused a 0.2 log unit decrease in CFU/well values, which was not statistically significant. In contrast, NJ9725 biofilm cells that were scraped from the surface of the well and dispersed by vortex agitation exhibited a 5-6 log unit decrease in CFU/well values when treated for 5 min with the same concentrations of agents, which was significantly greater than the decrease in biofilm CFU/well values (Fig. 5). These findings suggest that resistance to biocide killing may be mediated by the inherent protective nature of the biofilm matrix and its ability to retard penetration of agents into the biofilm.
Fig. 5.
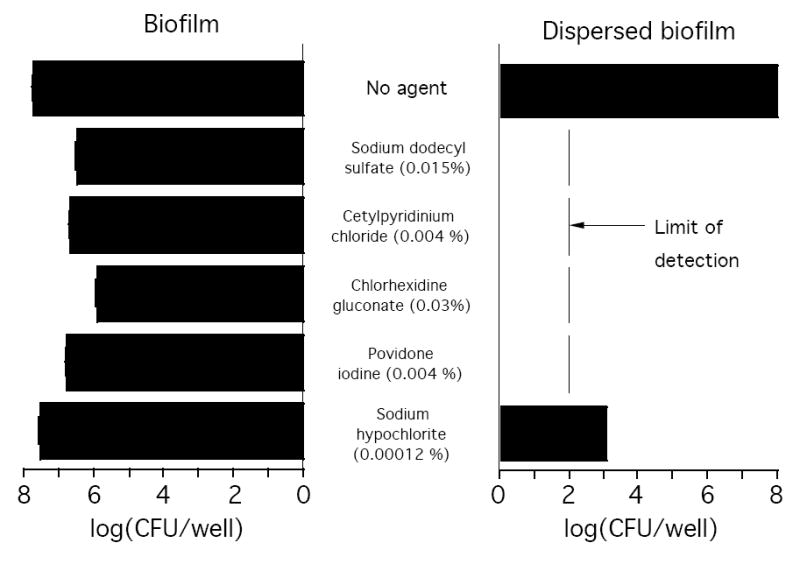
NTHi biofilms exhibit increased resistance to killing by detergents, antiseptics and disinfectants. NTHI NJ9725 biofilms cultured for 24 h in 12-well microtiter plates (left panel), or biofilm cells detached from the surface and dispersed by vortex agitation (right panel), were treated with the indicated concentration of agent for 5 min. Surviving CFUs were enumerated by dilution plating. Values show mean CFU/well values and range for duplicate wells.
We also measured the ability of DNase I and proteinase K to sensitize NTHi biofilms to killing by 0.004% CPC (Fig. 6). In one experiment, NJ9725 biofilms cultured in microplate wells were rinsed with water and then treated for 10 min with DNase I buffer alone (mock pre-treatment) or DNase I buffer containing 1 mg mL-1 DNase I. Biofilms were then rinsed with water and their sensitivity to killing by CPC was measured (Fig. 6A). DNase I pre-treatment alone, without CPC treatment, did not cause a significant decrease in biofilm CFU/well values under these conditions. However, biofilms pre-treated with DNase I were significantly more sensitive to CPC killing than were mock pre-treated biofilms (2.7 log unit reduction versus 1.0 log unit reduction, respectively). In another experiment, the ability of CPC to kill cells detached from NJ9725 biofilms by DNase I or proteinase K was measured (Fig. 6B). Enzyme-detached cells were efficiently killed by CPC. These findings demonstrate that enzymatic degradation of the biofilm matrix sensitizes NTHi biofilm cells to biocide killing.
Fig. 6.
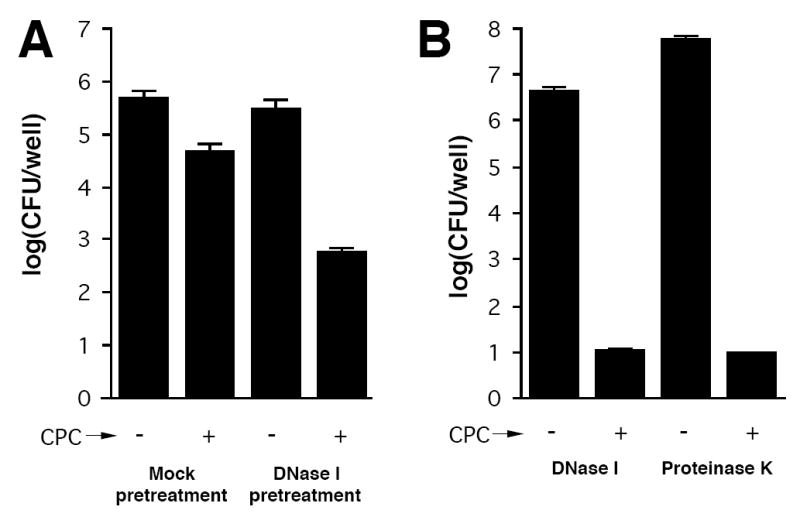
DNase I and proteinase K sensitize NTHi NJ9725 biofilms to killing by CPC. (A) Biofilms cultured for 24 in microtiter plates were rinsed with water and treated for 10 min with DNase I buffer (mock pre-treatment) or DNase I buffer containing 1 mg mL-1 of DNase I. Biofilms were then rinsed with water, treated for 5 min with 0.004% CPC and rinsed with water to remove the CPC. Surviving CFUs were enumerated by dilution plating. (B) Biofilm cells detached by 1 mg mL-1 DNase I or proteinase K in enzyme buffer were treated for 5 min with 0.004% CPC and surviving CFUs were enumerated by dilution plating.
3. Discussion
Biofilm formation has been invoked to explain the chronic nature of some cases of otitis media and the failure to culture NTHI from middle ear effusions [28,29]. Experimental evidence for NTHi biofilm formation includes the visualization of biofilm-like structures on middle ear tissue of human subjects and experimentally infected chinchillas [5,6], the apparent involvement of pili in biofilm cohesion in vitro [14,15], and significant biofilm-mediated antibiotic resistance in vitro [9-11]. However, no evidence for a biofilm-specific programmed switch in gene expression, or for the production of a biofilm-associated glycocalyx or extracellular polysaccharide, has been reported. These finding have led some authors to question whether NTHi strains form true biofilms [30]. The results of the present study demonstrate that NTHi biofilms exhibit phenotypes characteristic of many other bacterial biofilms including proteinaceous intercellular adhesins, an adhesive DNA matrix, and increased biocide resistance. Our findings support the hypothesis that NTHi strains form biofilms.
Our results suggest that proteinaceous adhesins mediate a significant amount of intercellular adhesion in NTHi biofilms. This hypothesis is based on the observation that proteinase K significantly inhibited biofilm formation by biofilm-forming NTHi strains, and that proteinase K treatment rapidly detached pre-formed NTHi biofilms from surfaces. Pre-formed NTHi biofilms were also rapidly detached by the anionic detergent SDS at concentrations above its critical micelle concentration (unpublished data), which further supports the hypothesis that proteinaceous adhesins mediate biofilm cohesion [31]. Further studies are needed to determine whether these proteinaceous intercellular adhesins are adhesive pili or other surface adhesins.
Previous studies have shown that DNA constitutes a matrix adhesin in many bacterial biofilms [32-35]. Our findings indicate that DNA also functions as a matrix adhesin in NTHi biofilms. This hypothesis is based on the observation that DNase I inhibited NTHi biofilm formation and detached pre-formed NTHi biofilms. In addition, treatment of NTHi biofilms with DNase I significantly impeded fluid convection through the biofilm, suggesting that extracellular DNA constitutes the major volumetric component of the NTHi biofilm matrix. These findings are consistent with the observation that double-stranded DNA is present in the NTHi biofilm matrix [22]. The genome of H. influenzae contains a homologue of cidA, a murein hydrolase that regulates DNA release in Staphylococcus aureus biofilms [36-37]. These observations raise the possibility that the NTHi biofilm matrix contains genomic DNA released from lysed cells.
Previous studies have shown that NTHi biofilm cells exhibit increased resistance to killing by antibiotics compared to the resistance exhibited by planktonic cells [9-11]. Our findings demonstrate that NTHi biofilms also exhibit increased resistance to killing by various biocides including detergents, antiseptics and disinfectants. The fact that mechanical or enzymatic disruption of NTHi biofilms rendered them sensitive to biocide killing suggests that the biofilm matrix may act as a barrier that prevents penetration of biocidal agents into the biofilm. This hypothesis is further supported by the fact that DNase I sensitized NTHi biofilms to CPC killing even though the enzyme did not cause a significant decrease in biofilm CFUs. Biofilm matrix polymers may act as a diffusion barrier which prevents access of biocides to the biofilm cells, or by forming physical complexes with biocides, thereby sequestering the agents within the matrix [38]. NTHi biofilm resistance may also result from the slow growth rate of biofilm cells, the presence of “persister” cells within the biofilm or from other unknown phenotypic differences [39,40].
Understanding the mechanisms of NTHi biofilm cohesion and biocide resistance could lead to the development of biofilm-specific agents for the management and treatment of NTHi-associated infections. Agents that target and destabilize the biofilm matrix may provide a useful biofilm-control strategy [41]. One such agent may be DNase I, which inhibits NTHi biofilm formation, detaches pre-formed NTHi biofilms, and sensitizes pre-formed NTHi biofilms to biocide killing. It has already been proposed that Pulmozyme®, a recombinant human DNase I used to treat cystic fibrosis patients, may be useful for treating respiratory infections caused by Streptococcus pneumoniae [42]. The ability of DNase I to sensitize biofilms to biocide killing may further increase its effectiveness against NTHi-associated infections [43].
4. Materials and methods
4.1 Bacterial strains
Eight NTHi strains, designated NJ9721-NJ9728, were isolated from sputum samples of eight different patients at University Hospital, Newark, New Jersey. Strains were collected in accordance with the Human Subjects Protection Program of the University of Medicine and Dentistry of New Jersey. The disease status of the patients was not disclosed. Strains were identified as H. influenzae using the RapID NH kit (Remel). The identity of the strains was confirmed by partial 16S rRNA sequence analysis as previously described [44]. The 16S rRNA sequences were compared to those of 330 different H. influenzae isolates described by Sacchi et al. [45]. The sequence of strain NJ9724 was identical to that of 16S type 5, the sequence of strain NJ9726 was identical to that of 16S type 7, and the sequences of strains NJ9722 and NJ9728 were identical to that of 16S type 59, all nontypeable strains. The sequences of strains NJ9721, NJ9723, NJ9725 and NJ9727 did not match those of any of the 330 H. influenzae isolates previously described but were phylogenetically most closely related 16S rRNA sequences of other nontypeable strains. All strains were confirmed to be nontypeable by using a multiplex PCR assay for identification of nontypeable and serotype b H. influenzae as described by Billal et al. [46]. H. influenzae strain Eagan [47] was used as a serotype b positive control for the multiplex PCR assay.
4.2 Media and growth conditions
Stock cultures of all strains were stored at -80°C in 10% glycerol. Bacteria were passaged weekly on chocolate agar and stored at 4°C. Strains were re-cultured from stock cultures after 2-3 passages. Bacteria were grown in Brain Heart Infusion (BHI) broth or on BHI agar plates (1.5% agar). In all experiments, BHI broth and agar were supplemented with 8 g l-1 glucose, 10 mg l-1 NAD, and 10 mg l-1 hemin. Unless otherwise indicated, all cultures were incubated statically at 37°C in 10% CO2.
4.3 Test tube biofilm assay
A loopful of cells from a BHI agar plate was transferred to a polypropylene microcentrifuge tube containing 100 μl of BHI broth. The cells were dispersed with a disposable pellet pestle, vortexed for 30 sec, and an additional 1 mL of BHI broth was added to the cells. The inoculum was then passed through a 5-μm pore-size syringe filter to remove large clumps of cells as previously described [48]. Cells were diluted to 105-106 CFU mL-1 in BHI broth. Aliquots of cells (1 mL each) were transferred to 17-mm × 100-mm polystyrene test tubes (Falcon no. 352051) and incubated for 24 h. For CFU enumeration, biofilms were rinsed twice with water and then treated with 100 μg mL-1 of proteinase K for 5 min, which efficiently detached the biofilm cells resulting in a uniformly turbid cell suspension (see below). Cells were serially diluted in BHI broth and plated on agar. For crystal violet staining, biofilms were washed three times with water, stained for 1 min with 200 μl of Gram’s crystal violet (Fisher no. 23255960) and then rinsed with water and dried. The amount of biofilm biomass was quantitated by destaining the biofilms for 10 min with 1 mL of 33% acetic acid (vol/vol) and then measuring the absorbance of the crystal violet solution in a BioRad microtiter plate reader set to 595 nm.
4.4 Biofilm detachment assay
Biofilms were cultured for 24 h in polypropylene tubes as described above. Biofilms were rinsed three times with water and then treated with 1 mL of 50 mM Tris-HCl (pH 8.0) containing 1 mg mL-1 proteinase K (Sigma); 100 mM NaCl, 1 mM CaCl2 containing 1 mg mL-1 bovine DNase I (Sigma); 0.02-1.2% SDS in water; or 0.002-1.0% cetylpyridinium chloride (CPC) in water. Control tubes were treated with 1 mL of the appropriate buffer or water alone. Tubes were incubated for the indicated amount of time, after which biofilms were quantitated by CFU enumeration or crystal violet staining as described above.
4.5 Biofilm inhibition assay
Biofilms were cultured in polypropylene tubes as described above, except that BHI broth was supplemented directly with 1 mg mL-1 bovine DNase I or proteinase K. Control biofilms were cultured in BHI broth alone. After 6-48 h, biofilms were quantitated by CFU enumeration or crystal violet staining as described above.
4.6 Fluid convection assay
Measurement of fluid convection through NTHi biofilms was carried using a centrifugal filter device assay as previously described [49]. Briefly, filtered inocula were prepared in BHI broth, or in BHI broth supplemented with DNase I or proteinase K, as described above. Aliquots of inocula (600-μl each) were transferred to the sample reservoirs of Microcon centrifugal filter devices (No. YM-100; 100,000 MW cut-off; Millipore) and the devices were incubated for 24 h. The devices were then centrifuged for 2 min at 4,300 × g in a swinging bucket rotor, and the flowthrough volume was measured. The 2-min centrifugation step was repeated three additional times, and the cumulative flowthrough volume was re-measured after each centrifugation step.
4.7 Biofilm killing assays
Filtered inocula were prepared in BHI broth as described above. Aliquots of inocula (2-mL each) were transferred to the wells of a 12-well tissue-culture-treated microtiter plate (Falcon no. 353043). After incubation for 24 h, biofilms were rinsed with water and then treated with 2 mL of 0.015% SDS, 0.004% CPC, 0.03% chlorhexidine gluconate, 0.004% povidone iodine, or 0.00012% sodium hypochlorite. These concentrations were chosen because they are slightly higher than the concentrations needed to kill 99% of NTHi planktonic cells in 5 min as determined in control experiments. None of the agents caused significant detachment of the biofilms at the concentration tested. Control biofilms were treated with 2 mL of water. After 5 min, agents were removed and the biofilms were rinsed three times with water. Biofilm cells were scraped into 1 mL of fresh BHI broth using a cell scraper, transferred to a tube, and mixed by vortex agitation for 15 sec. Serial decimal dilutions of cells were plated on agar for CFU enumeration.
To measure biocide killing of mechanically-disrupted biofilms, biofilms were rinsed with water, scraped into 2 mL of water using a cell scraper, transferred to a tube, and mixed by vortex agitation for 15 sec. Aliquots of cells (100 μl each) were transferred to the wells of a 96-well, round bottom microtiter plate (Falcon no. 353227). A total of 100 μl of agent (at twice the test concentration) was added to each well and mixed. Control wells received 100 μl of water. After 5 min, serial decimal dilutions in BHI broth were made directly into adjacent wells, and the dilutions were plated on agar for CFU enumeration. Control experiments indicated that NTHi cells were not lysed by water during the brief resuspension and incubation steps.
To measure biocide killing of enzyme-treated biofilms, biofilms were rinsed three times with water and treated for 10 min with 2 mL of DNase I or proteinase K in enzyme buffer as described above. Control wells were treated with the appropriate enzyme buffer alone. Cells detached by the enzymes were transferred to a tube, mixed by vortex agitation for 15 sec, treated for 5 min with 0.004% CPC in 96-well microtiter plates, and then quantitated by dilution plating as described above. For DNase I-treated biofilms, which still retained a significant amount of surface-associated biofilm after enzyme treatment, biofilms were rinsed three times with water, treated with 0.004% CPC for 5 min, and rinsed three more times with water. Biofilm cells were then scraped from the surface and enumerated by dilution plating as described above.
4.8 Statistics and reproducibility of results
All biofilm growth, inhibition, detachment and killing assays were performed in duplicate tubes, which usually exhibited < 10% variation in A595 or CFU values. Fluid convection assays were performed in duplicate centrifugal filter devices. All assays were performed on at least three separate occasions with nearly identical results. The significance of differences between means was calculated using a Student’s t-test. A P value of < 0.05 was considered significant.
4.9 Nucleotide sequence accession numbers
The 16S rRNA sequences of NTHi strains NJ9721-NJ9728 were deposited in the GenBank database under accession numbers EU597468-EU597475, respectively.
Acknowledgments
We thank Darshini Shah (University Hospital, Newark, NJ) for help in procuring H. influenzae strains, and Joseph W. St. Geme, III, (Duke University Medical Center) for providing genomic DNA from H. influenzae strain Eagan. Supported by Public Health Service award DE15124 (to J.B.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Erwin AL, Smith AL. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol. 2007;15:355–362. doi: 10.1016/j.tim.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Foxwell AR, Kyd JM, Cripps AW. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol Mol Biol Rev. 1998;62:294–308. doi: 10.1128/mmbr.62.2.294-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy TF. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr Opin Infect Dis. 2003;16:129–134. doi: 10.1097/00001432-200304000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;73:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun. 2005;73:3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. J Am Med Assoc. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starner TD, Zhang N, Kim G, Apicella MA, McCray PB., Jr Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am J Respir Crit Care Med. 2006;174:213–220. doi: 10.1164/rccm.200509-1459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallaher TK, Wu S, Webster P, Aguilera R. Identification of biofilm proteins in non-typeable Haemophilus influenzae. BMC Microbiol. 2006;6:65. doi: 10.1186/1471-2180-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slinger R, Chan F, Ferris W, Yeung SW, St Denis M, Gaboury I, Aaron SD. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn Microbiol Infect Dis. 2006;56:247–253. doi: 10.1016/j.diagmicrobio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Starner TD, Shrout JD, Parsek MR, Appelbaum PC, Kim G. Subinhibitory concentrations of azithromycin decrease nontypeable Haemophilus influenzae biofilm formation and diminish established biofilms. Antimicrob Agents Chemother. 2008;52:137–145. doi: 10.1128/AAC.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starner TD, Zhang N, Kim G, Apicella MA, McCray PB., Jr Haemophilus influenzae forms biofilms on airway epithelia. Am J Respir Crit Care Med. 2006;174:213–220. doi: 10.1164/rccm.200509-1459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilsdorf JR, McCrea KW, Marrs CF. Role of pili in Haemophilus influenzae adherence and colonization. Infect Immun. 1997;65:2997–3002. doi: 10.1128/iai.65.8.2997-3002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St Geme JW., III Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell Microbiol. 2002;4:191–200. doi: 10.1046/j.1462-5822.2002.00180.x. [DOI] [PubMed] [Google Scholar]
- 14.Murphy TF, Kirkham C. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2002;2:7. doi: 10.1186/1471-2180-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurcisek JA, Bookwalter JE, Baker BD, Fernandez S, Novotny LA, Munson RS, Jr, Bakaletz LO. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol Microbiol. 2007;65:1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- 16.Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH, Kaplan JB. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2001;40:542–554. doi: 10.1046/j.1365-2958.2001.02422.x. [DOI] [PubMed] [Google Scholar]
- 17.Nika JR, Latimer JL, Ward CK, Blick RJ, Wagner NJ, Cope LD, Mahairas GG, Munson RS, Jr, Hansen EJ. Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect Immun. 2002;70:2965–2975. doi: 10.1128/IAI.70.6.2965-2975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mhlanga-Mutangadura T, Morlin G, Smith AL, Eisenstark A, Golomb M. Evolution of the major pilus gene cluster of Haemophilus influenzae. J Bacteriol. 1998;180:4693–4703. doi: 10.1128/jb.180.17.4693-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greiner LL, Watanabe W, Phillips NJ, Shao J, Morgan A, Zaleski A, Gibson BW, Apicella MA. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glucans. Infect Immun. 2004;72:4249–4260. doi: 10.1128/IAI.72.7.4249-4260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swords WE, Moore ML, Godzicki L, Bukofzer G, Mitten MJ, VonCannon J. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect Immun. 2004;72:106–113. doi: 10.1128/IAI.72.1.106-113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong W, Pang B, West-Barnette S, Swords WE. Phosphorylcholine expression by nontypeable Haemophilus influenzae correlates with maturation of biofilm communities in vitro and in vivo. J Bacteriol. 2007;189:8300–8307. doi: 10.1128/JB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leroy M, Cabral H, Figueira M, Bouchet V, Huot H, Ram S, Pelton SI, Goldstein R. Multiple consecutive lavage samplings reveal greater burden of disease and provide direct access to the nontypeable Haemophilus influenzae biofilm in experimental otitis media. Infect Immun. 2007;75:4158–4172. doi: 10.1128/IAI.00318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutherland IW. The biofilm matrix – an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 25.O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan JB, Velliyagounder K, Chandran R, Rohde H, Mack D, Knobloch JKM, Ramasubbu N. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte M, Knobloch JK, Heilmann C, Herrmann M, Mack D. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 28.Post JC. Direct evidence of bacterial biofilms in otitis media. Laryngoscope. 2001;111:2083–2094. doi: 10.1097/00005537-200112000-00001. [DOI] [PubMed] [Google Scholar]
- 29.Rayner MG, Zhang Y, Gorry MC, Chen Y, Post JC, Ehrlich GD. Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. J Am Med Assoc. 1998;279:296–299. doi: 10.1001/jama.279.4.296. [DOI] [PubMed] [Google Scholar]
- 30.Moxon ER, Sweetman WA, Deadman ME, Ferguson DJP, Hood DW. Haemophilus influenzae biofilms: hypothesis or fact? Trends Microbiol. 2008;16:95–100. doi: 10.1016/j.tim.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Izano EA, Wang H, Ragunath C, Ramasubbu N, Kaplan JB. Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J Dent Res. 2007;86:618–622. doi: 10.1177/154405910708600707. [DOI] [PubMed] [Google Scholar]
- 32.Izano EA, Sadovskaya I, Wang H, Vinogradov E, Ragunath C, Ramasubbu N, Jabbouri S, Perry MB, Kaplan JB. Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb Pathogen. 2008;44:52–60. doi: 10.1016/j.micpath.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen FC, Tao L, Scheie AA. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol. 2005;187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 35.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 36.Bayles KW. The biological role of death and lysis in biofilm development. Nature Rev Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 37.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Nat Acad Sci USA. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mah T-F, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 39.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Lewis K. Persister cells, dormancy and infectious disease. Nature Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 41.Xavier JB, Picioreanu C, Rani SA, van Loosdrecht MCM, Stewart PS. Biofilm-control strategies based on enzymic disruption of the extracellular polymeric substance matrix - a modelling study. Microbiology. 2005;151:3817–3832. doi: 10.1099/mic.0.28165-0. [DOI] [PubMed] [Google Scholar]
- 42.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, Johnson C, Hu FZ, Stoodley P, Ehrlich GD, Post JC. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macfadyen C, Gamble C, Garner P, Macharia I, Mackenzie I, Mugwe P, Oburra H, Otwombe K, Taylor S, Williamson P. Topical quinolone vs. antiseptic for treating chronic suppurative otitis media: a randomized controlled trial. Trop Med Int Health. 2005;10:190–197. doi: 10.1111/j.1365-3156.2004.01368.x. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan JB, Schreiner HC, Furgang D, Fine DH. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J Clin Microbiol. 2002;67:5427–5433. doi: 10.1128/JCM.40.4.1181-1187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sacchi CT, Alber D, Dull P, Mothershed EA, Whitney AM, Barnett GA, Popovic T, Mayer LW. High level of sequence diversity in the 16S rRNA genes of Haemophilus influenzae isolates is useful for molecular subtyping. J Clin Microbiol. 2005;43:3734–3742. doi: 10.1128/JCM.43.8.3734-3742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Billal DS, Hotomi M, Suzumoto M, Kobayashi I, Fujihara K, Yamanaka N. Rapid identification of nontypeable and serotype b Haemophilus influenzae from nasopharyngeal secretions by the multiplex PCR. Int J Ped Otorhinolaryngol. 2007;71:269–274. doi: 10.1016/j.ijporl.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Moxon ER, Murphy PA. Haemophilus influenzae bacteremia and meningitis resulting from survival of a single organism. Proc Nat Acad Sci USA. 1978;75:1534–1536. doi: 10.1073/pnas.75.3.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaplan JB, Fine DH. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl Environ Microbiol. 2002;68:4943–4950. doi: 10.1128/AEM.68.10.4943-4950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganeshnarayan K, Shah SM, Libera MR, Santostefano A, Kaplan JB. Poly-N-acetylglucosamine matrix polysaccharide impedes fluid convection and transport of the cationic surfactant cetylpyridinium chloride through bacterial biofilms. Appl Environ Microbiol. 2009 doi: 10.1128/AEM.01900-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]