Abstract
This report describes the adaptation of the biotin ligase BirA - biotin acceptor sequence (BAS) labeling system to biotinylate specific human immunodeficiency virus 1 (HIV-1) proteins in vivo. Two HIV-1 clones were constructed, with the BAS introduced into the matrix region of gag or the integrase region of pol. Specific biotinylation of target proteins in virions was observed when molecular clones were co-expressed with BirA. Both BAS-containing viruses propagated in SupT1 T-cells although replication of the integrase clone was delayed. Further studies demonstrated the integrase insertion yielded an approximate 40% reduction in single-round infectivity as assessed on MAGI-5 indicator cells, as well as in the in vitro integration activity of preintegration complexes extracted from acutely-infected C8166-45 T-cells. Biotinylation of the integrase BAS tag furthermore rendered this virus non-infectious. The matrix viral clone by contrast displayed wild-type behavior under all conditions tested. These results therefore establish a system whereby biotinylated matrix protein in the context of replication-competent virus could be used to label and capture viral protein complexes in vivo.
Keywords: human immunodeficiency virus, insertion mutagenesis, biotinylation
1. Introduction
Peptide labeling of proteins is a useful tool for investigating protein structure and function, and identifying protein-protein interactions in vivo. Human immunodeficiency virus 1 (HIV-1), the etiologic agent of acquired immunodeficiency syndrome, belongs to the genus Lentivirus in the Retroviridae family. A large focus of HIV-1 research is devoted to the identification and characterization of cellular proteins that interact with viral proteins as a means to identify new targets for therapeutic interventions. Many protein-based strategies have been developed for this purpose. Two commonly used purification strategies are single- and tandem-affinity purification (TAP). Single tag protocols are less labor intensive and more efficiently capture target proteins compared to TAP, but exhibit high levels of background. TAP is more specific, but the recovery rate of bait proteins from lysates is quite poor (~5%) and therefore TAP can miss low abundant proteins. Furthermore, TAP requires two insertions in the same protein which typically increases the chances of protein misfolding and/or inactivation.
The strength of the streptavidin (SA)-biotin bond, the strongest known non-covalent bond, is several orders greater than that of antibody-antigen or other affinity purification systems. Biotinylated proteins can be efficiently purified in a single-step under high-stringency conditions. In the last two decades the biotin-avidin interaction has been developed as a strategy to isolate protein complexes (Chen et al., 2005; de Boer et al., 2003; Furuyama and Henikoff, 2006; Penalva and Keene, 2004). Co-expression of the E. coli biotin ligase BirA with a minimal biotin acceptor sequence (BAS) catalyzes the covalent attachment of biotin to a central lysine residue (Beckett et al., 1999; Schatz, 1993). The minimum identified BAS length is 13 amino acids, comparable to several affinity tags (Schatz, 1993).
This study demonstrates the adaptation of the BAS-BirA system to label and capture HIV-1 integrase (IN) and matrix (MA) protein complexes. MA, encoded as the N-terminal portion of the Gag polyprotein, plays a role in both the efferent and afferent stages of the viral life cycle. During virus assembly, MA regulates the interaction between Gag and the plasma membrane (Zhou et al., 1994). After cell entry and reverse transcription, a small portion of virion-derived MA remains associated with the preintegration complex (PIC; Bukrinsky et al., 1993; Lin and Engelman, 2003; Miller et al., 1997). IN, encoded as the C-terminal end of the Gag-Pol precursor protein, catalyzes the insertion of the reverse transcript into a cell chromosome. We show that virion-associated MA and IN proteins were biotinylated both specifically and efficiently when proviral DNAs bearing the BAS tags were co-transfected with a BirA expression plasmid, or when the proviruses were transfected into 293T cells stably expressing BirA. Characterization of the BAS-containing molecular clones demonstrated that the infectivity of the MA-BAS virus with and without the added biotin modification was similar to wild-type HIV-1. The IN-BAS virus, by contrast, replicated poorly due to a loss of IN activity, which was furthermore abolished by biotinylation.
2. Materials and Methods
2.1. Plasmids
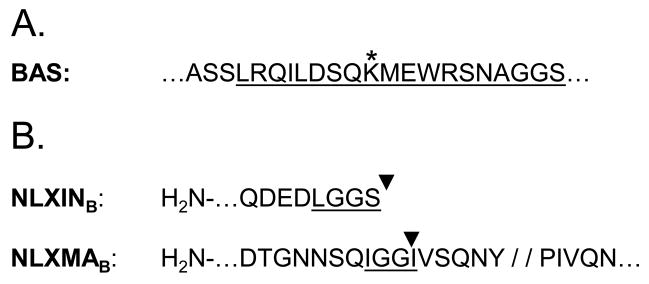
The BirA expression plasmid, pc6BirA, was constructed by transferring the birA ORF and intronic sequences from pEV-BirA (a kind gift of John Strouboulis) into pcDNA6/V5-His (Invitrogen, Carlsbad, CA). A series of plasmids were constructed to transfer a previously described ~22 residue BAS (Fig. 1A; de Boer et al., 2003) into the C-terminal regions of MA and IN in the HIV-1NLX molecular clone (Brown et al., 1999). Plasmid pUCWTpolBam-Spe was made by modifying the 3′ end of the pol gene in pUCWTpol (Limon et al., 2002) by PCR-directed mutagenesis to CTAGGGGGATCCAAGACTAGT, which contained CTA in place of the TAA stop codon (italics) and downstream BamHI and SpeI sites (underlined). Plasmid pUCWTpol-SR2 was constructed by ligating a BamHI-SpeI BAS linker, made by annealing AE2217 (5′-PO4-GATCCGCCAGCAGCCTGCGGCAGATCCTGGACAGTCAGAAGATGGAGTGGCGGAGCAACGCCGGGGGGAGCTAA-3′) and AE2218 (5′-PO4-CTAGTTAGCTCCCCCCGGCGTTGCTCCGCCACTCCATCTTCTGACTGTCCAGGATCTGCCGCAGGCTGCTGGCG-3′), with BamHI/SpeI-digested pUCWTpolBam-Spe. The IN-BAS proviral clone pNLXINB (Fig. 1B) was made by swapping the AgeI-PflMI fragment from pUCWTpol-SR2 for the corresponding fragment in pNL43/XmaI (Brown et al., 1999).
Figure 1.
Creation of BAS-containing IN and MA molecular clones. (A) The canonical BAS, as described in de Boer et al. (2003). The underlined portion defines the common inserted sequence, as indicated by inverted black triangles in panel B. The asterisk denotes the biotin-acceptor lysine residue within the BAS. (B) BAS placement. The BAS in NLXINB is followed by a stop codon after the C-terminal Ser residue in panel A. NLXMAB harbors the BAS insertion 5 residues N-terminal of the MA-capsid protease cleavage site (indicated by double slash) to permit correct Gag polyprotein processing (Muller et al., 2004). Underlines denote heterologous BAS-abutting linker amino acid residues.
For MA, plasmid pTZ18UPL/BssH2-SpeI was made by ligating the 796 bp BssHII-SpeI fragment from pNL43/XmaI with BssHII/SpeI-digested pTZ18U/PL (Dorfman et al., 1993). PCR-directed mutagenesis was used to build pAB1, containing unique AgeI and BsrGI sites in the 3′ and 5′ regions of the MA and CA gag reading frames, respectively. A 123 bp fragment was generated from pUCWTpol-SR2 template using PCR with primers AE2224 (5′-GCTGACACCGGTAACAACAGCCAGATAGGAGGCCTGCGGCAGATCCTGGACAGC -3′; Age I site underlined) and AE2225 (5′-GGTTCTGTACAATAGGGTAATTTTGGCTGACTATGCTCCCCCCGGCGTTGCTCCGCC -3′; BsrGI site underlined). AgeI/BsrGI-digested DNA was then ligated to AgeI/BsrGI-digested pAB1. Plasmid pNLXMAB was made by swapping the resulting BssHII-SpeI fragment for the corresponding fragment in pNL43/XmaI (Fig. 1B).
2.2. Cells
2.2.1. Cell Culture
293T and MAGI-5 (Pirounaki et al., 2000) cells were propagated in Dulbecco’s modified eagle media (DMEM) supplemented with 10% Fetalclone III (Hyclone, Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin. The MAGI-5 cell media was supplemented with 0.1 mg/ml G-418, 25 μg/ml puromycin, and 0.1 mg/ml Hygromycin B. C8166-45 and SupT1 T-cells were maintained in RPMI 1640 media supplemented with 10% Fetalclone III, 100 U/ml penicillin, and 100 μg/ml streptomycin.
2.2.2. 293T.BirA cells
5×105 293T cells were seeded into a single well of a six-well dish, grown overnight, then transfected with 2 μg pc6BirA using TransIT-LT1 as directed by the manufacturer (Mirus Bio, Madison WI). 48 h post-transfection, the cells were passed into a 10 cm2 dish and propagated in complete DMEM media containing 20 μg/ml Blasticidin S (Invivogen, San Diego, CA). Cells maintained for two weeks were then cloned by limiting dilution into 96-well plates, and grown until colonies were visible. Wells containing single colonies were expanded and examined for BirA expression by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using an anti-BirA antibody (Genway Biotech, San Diego, CA), HRP-conjugated anti-chicken IgG secondary antibody (Pierce Biotechnology, Rockford, IL), followed by visualization by chemiluminescence (Pierce Biotechnology). All Western blot images were acquired by flatbed scanning of developed films and, if necessary to improve quality, sharpened and adjusted for brightness/contrast using Adobe Photoshop.
2.3. Virus production
Viruses were produced by transient transfection of 293T or 293T.BirA cells. Cells were seeded into 10 cm2 dishes at 60% confluency. Cells were transfected with 20 μg of viral molecular clone DNA using TransIT-LT1. For pseudotyping with vesicular stomatitis virus glycoprotein G (VSVg), 15 μg of viral molecular clone was premixed with 5 μg of VSVg expression vector pMD2.G (Addgene Plasmid Repository). Media was collected every 24 h for 72 h, clarified by centrifugation at 4000 ×g for 5 min., and concentrated using Centricon-100 concentrator units as directed by the manufacturer (Millipore, Billerica MA).
2.4. Detection of biotinylated proteins
Viruses produced by transient transfection as described above were harvested at 24 h, clarified, and concentrated by ultracentrifugation through 20% sucrose (w/v in phosphate-buffered saline (PBS)). Afterwards, the media and sucrose were aspirated and virus pellets resuspended directly in 0.25 ml 1x SDS-PAGE sample buffer. For cell lysates, the 293T cells were washed with PBS, harvested, and lysed with 0.5 ml M-PER solution (Pierce Biotechnology). Biotinylated proteins were detected by SDS-PAGE and Western blotting using SA-HRP conjugate (GE Healthcare, Piscataway, NJ) and chemiluminescence (Pierce Biotechnology), followed by exposure to film. Viral proteins were similarly detected using antigen specific antibodies followed by species specific HRP-conjugated secondary antibodies (GE Healthcare). For SA-capture experiments, samples were resuspended in 0.5 (virions) or 1 ml (cells) RIPB buffer (50 mM Tris (pH 7.5)/150 mM NaCl/1% NP-40/0.5% sodium deoxycholate/0.1% SDS) and clarified by centrifugation. 50 μl was removed for a “Pre” sample, then the lysates were incubated with 25 μl SA-sepharose (GE Healthcare) overnight at 4°C. The beads were pelleted by centrifugation and a “post” sample was removed. The beads were washed in triplicate with 1 ml RIPB buffer, bound proteins were released by boiling in 100 μl 1x sample buffer and detected by SDS-PAGE and Western blot. Quantitative imaging was performed using a FluorChem FC2 imaging system (Alpha Innotech Corp., San Leandro, CA)
2.5. Virus replication and reverse transcriptase (RT) assays
SupT1 cells were inoculated overnight with equivalent levels of viruses (as determined by RT assay), washed, and propagated for 22 days. Cells were passed every two days by 1:2 dilution in fresh media. 300 μl of supernatant was sampled at the days indicated, clarified by centrifugation (12,000 ×g, 5 min), and stored at −20°C until the end of the time-course. RT activity was measured by [32P]TTP incorporation (Goff et al., 1981; Willey et al., 1988). RT reactions were reduced to 40 μl for compatibility with a high-throughput microtiter plate system (Millipore, Billerica, MA). Samples were measured in triplicate, and data is presented with standard deviation. The results of the replication studies are representative of at least two independent experiments. MAGI assays were performed as previously described (Belshan et al., 2006). Results represent data from nine independent infections.
2.6. In vitro integration assays
PIC-containing lysates were produced using VSVg-pseudotyped HIV essentially as described (Engelman, 2009), except 1×108 C8166-45 T-cells were infected by spinoculation (O’Doherty et al., 2000) in the presence of 8 μg/ml polybrene, and the cells were lysed with buffer k (20 mM HEPES, pH 7.4/150 mM KCl/5 mM MgCl2/1 mM dithiothreitol) containing 0.1% TritionX-100 (Farnet and Haseltine, 1990). Integration assays were developed using nested real-time PCR (Lu et al., 2005) of reaction products with the following modifications. First, linker sequence (5′-ATGCCACGTAAGCGAAACTGC -3′) was added to the 5′ end of the HIV-specific primer in the first PCR. The linker primer was used in place of one of the HIV primers in the subsequent real-time PCR reaction to reduce background HIV amplification, mimicking the nested PCR design developed by Brussel and Sonigo (2003) for analyzing chromosomal integration during infection. Secondly, the vector pTZ19R, which is commercially available (Fermentas, Glen Burnie, MD), was used instead of pTZ18U/PL as the DNA target during in vitro integration. In vitro integration values were normalized to the levels of viral cDNA present in cell extracts (Lu et al., 2005). Real-time PCR reactions were performed with an iQ5 system and iQ SYBR Green Supermix (Bio-rad laboratories, Hercules, CA).
3. Results
3.1. Specific biotinylation of HIV-1 proteins in vivo
The BAS-BirA labeling system has been used to purify proteins in a wide variety of systems including ribonucleoprotein complexes (Penalva and Keene, 2004), nucleosomes (Furuyama and Henikoff, 2006), cell surface proteins (Chen et al., 2005), and transcription factors in BirA transgenic mice (de Boer et al., 2003). To adapt this system to label HIV-1 proteins in replication competent viruses the BAS was inserted into the C-terminal portion of the MA or IN protein within the gag and pol reading frames, respectively, of the NLX molecular clone of HIV-1 (details shown in Fig. 1).
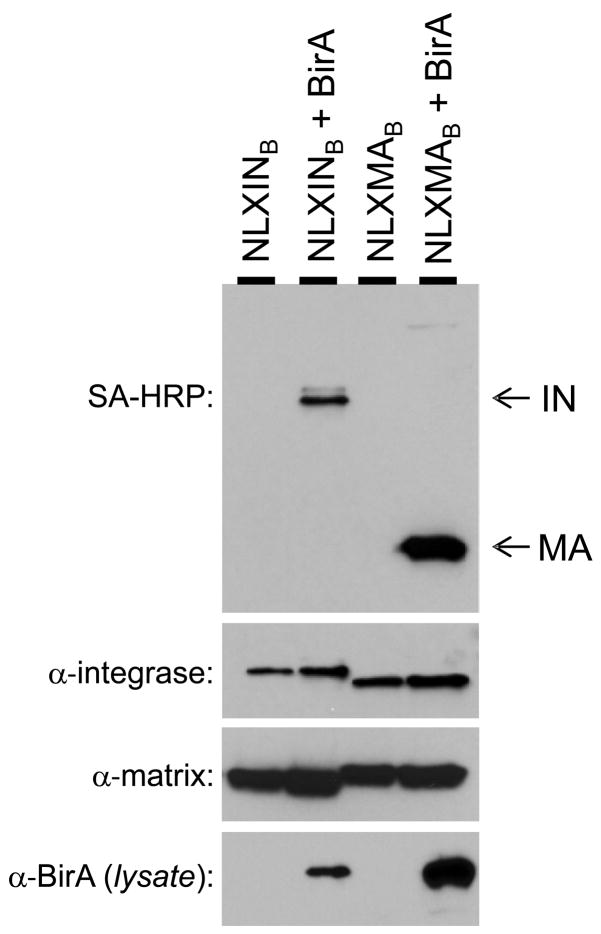
To demonstrate specific biotinylation of HIV proteins in vivo, 293T cells were transfected with the molecular clones with and without the pc6BirA expression vector. At 24 h post-transfection, cell-free viral supernatants and cells were harvested, and virus particles were concentrated by ultracentrifugation through 20% sucrose cushions prior to lysis. Biotinylated proteins were detected by Western blot using SA-HRP. As shown in the top panel of Figure 2, IN and MA were biotinylated when either tagged molecular clone was co-transfected with the BirA expression plasmid (bottom panel). A small amount of a ~55 kDa biotinylated protein, corresponding to the size of full-length Gag, was observed in virus pellets derived from cells co-transfected with pNLXMAB and pc6BirA DNA. Biotinylation of MA and IN was not detected in virions produced in the absence of BirA, despite similar levels of expression as determined by Western blotting with IN- and MA-specific antibodies (middle panels). As expected, the BAS-containing MA and IN proteins were slightly larger in size than their wild-type counterparts.
Figure 2.
Specific biotinylation of IN and MA in HIV-1 virions. 293T cells were transfected with NLX molecular clones containing the BAS in IN (pNLXINB) or MA (pNLXMAB), alone or with the BirA expression plasmid as indicated. Virion lysates (top three panels) were probed by Western blotting using the indicated reagents. The bottom panel indicates the cellular level of BirA expression.
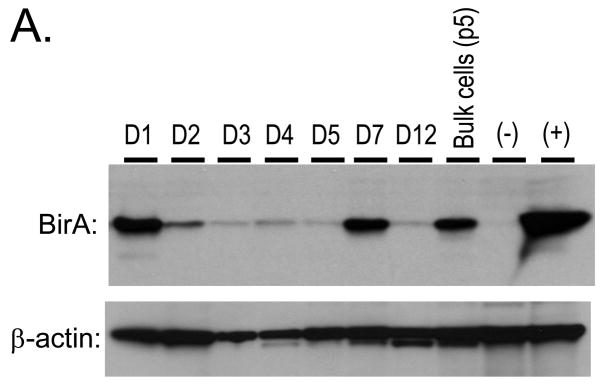
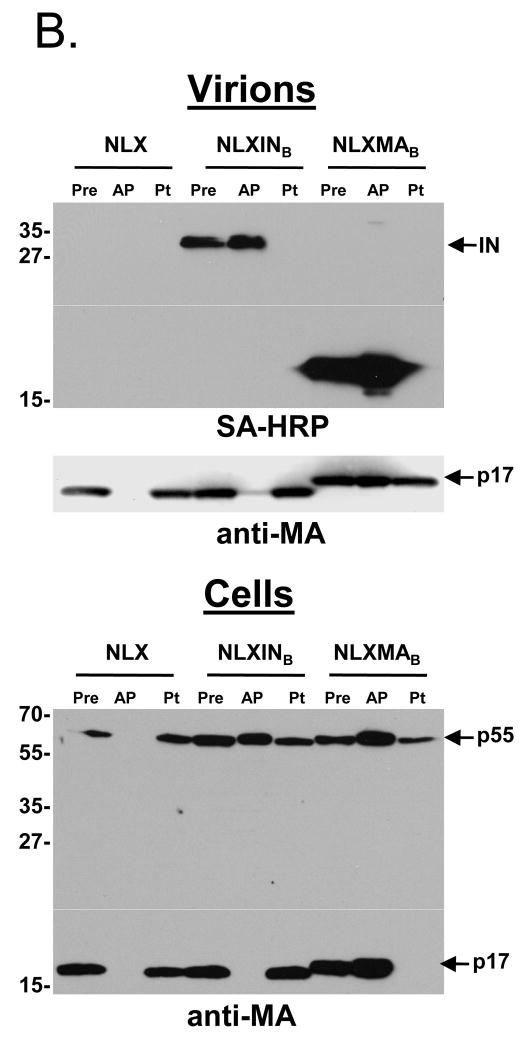
3.2. 293T.BirA cells and affinity capture of HIV-1 proteins
To increase the efficiency of target protein biotinylation, a 293T cell line stably expressing BirA was constructed. Cells transfected with pc6BirA were selected with 20 μg/ml blasticidin S antibiotic for two weeks. Cells were diluted in 96-well plates to obtain single cell clones. Numerous cell lines were expanded and tested for expression of BirA by Western blotting (Fig. 3A) with anti-BirA antibody. The cell lines varied in their expression of BirA. The BirA-expressing cell line D7 displayed better growth characteristics than D1 and was designated the primary 293T.BirA cell line. The cells were used to examine biotinylation and affinity purification of both MA and IN by transient transfection (Fig. 3B). Cells were transfected with pNLX, pNLXMAB, or pNLXINB, and resulting viruses purified by ultracentrifugation and lysed in immunoprecipitation buffer. A pre sample was taken prior to overnight incubation with SA-sepharose beads. The next day, post (Pt) samples were recovered from supernatants after bead pelleting, the beads were washed extensively, and affinity purified (AP) bead-bound proteins were released by boiling. Proteins from both virion and cell lysates were detected by SDS-PAGE and Western blot with SA-HRP or anti-MA antibody. Both IN-BAS and MA-BAS, but not wild-type HIV-1NLX proteins, were affinity purified from virion samples (top panel, AP lanes), demonstrating specific capture of biotinylated proteins. Consistent with their molar ratios, a larger amount of MA was captured compared to IN. A small amount of the p55 Gag polyprotein could be detected in the NLXMAB AP sample upon over-exposure of blots (not shown). Interestingly, a small amount of MA was captured via IN biotinylation, suggesting that MA and IN can associate in virion lysates and may also do so in intact virions. Biotinylated proteins were importantly not observed in the post capture samples (Pt lanes), indicating quantitative binding of all detectable biotinylated proteins. Similar levels of MA were detected in all pre samples, indicating that both BAS-tagged clones were expressed and released from cells at levels similar to wild-type HIV-1NLX (middle panel). Moreover, significant levels of Gag or Gag-Pol polyproteins were not detected in virion samples (not shown), indicating that neither insertion adversely effected polyprotein processing. The detection of MA in the post NLXMAB sample (middle panel, right lane) suggested only a portion of the BAS-tagged protein was biotinylated. Comparison of the post and pre samples of triplicate experiments by quantitative Western blot imaging indicated that 30% to 60% of total MA-BAS was biotinylated.
Figure 3.
Construction and characterization of 293T cell lines stably expressing BirA, and SA-capture of biotinylated IN and MA. (A) 293T cells were transfected with pc6BirA and selected using Blasticidin as described in Materials and Methods. Clonal 293T.BirA cell lines were obtained by limiting dilution, expanded, and assayed for BirA expression by Western blotting. (B) SA capture of biotinylated IN and MA proteins from purified virions and cell lysates. Virus was produced by transfection of 293T.BirA cells as described in the legend of Fig. 2. Virus and cell lysates were resuspended in RIPB buffer, a pre-sample removed (Pre), and viral proteins captured with SA-agarose beads (AP). After the beads were pelleted, a post sample was removed (Pt), then the beads were washed extensively with RIPB buffer. Bound proteins were released by boiling in sample buffer, and samples separated by SDS-PAGE were detected using SA-HRP or anti-MA antibody as denoted beneath each blot. Approximate MW sizes are shown to the left of the blots, and viral proteins are indicated on the right.
A high level of background was observed in affinity purified cell lysates blotted with SA-HRP. Though this revealed biotinylated IN and MA, Gag and Gag-Pol specific bands were not discernable (data not shown). MA and p55 Gag were readily detected in affinity captured cell samples using anti-MA antibody (bottom panel). A surprisingly large level of p55, but not MA, was recovered via biotinylated IN (NLXINB AP lane). Although Gag-Pol was not discernable due to the relatively high levels of non-specific cell binding proteins, we speculate that p55 Gag recovery was due to its association with Gag-Pol during IN-BAS pull-down. Accordingly, p55 Gag was not recovered from cells expressing wild-type HIV-1NLX. The p55 Pre levels were similar among samples, indicating the BAS-tagged viruses expressed Gag at levels that were comparable to wild-type HIV-1NLX.
3.3. Replication profiles of NLXINB and NLXMAB
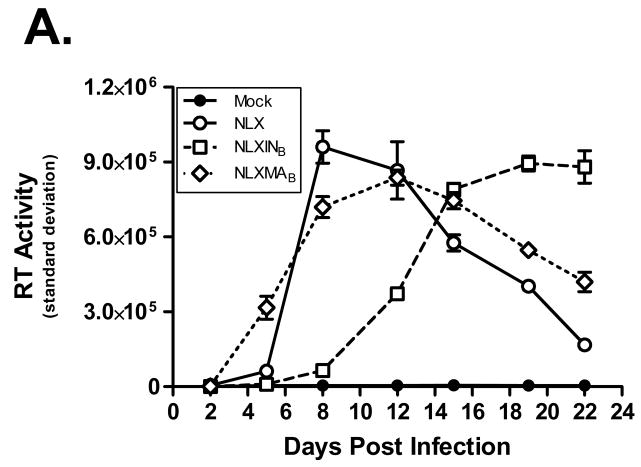
Next the replication capacity of the BAS-containing molecular clones was assessed. Viruses were produced by transient transfection of 293T cells, concentrated by ultracentrifugation through a sucrose cushion, and normalized by RT assay. SupT1 cells were inoculated overnight, washed, and propagated for 22 days to assess virus replication kinetics (Fig. 4A). Samples were measured in triplicate, and the standard deviation is shown in the graph. Both NLXMAB and NLXINB were replication competent. NLXMAB replicated with kinetics similar to wild-type virus, whereas the replication of NLXINB was delayed in comparison to either NLX or NLXMAB.
Figure 4.
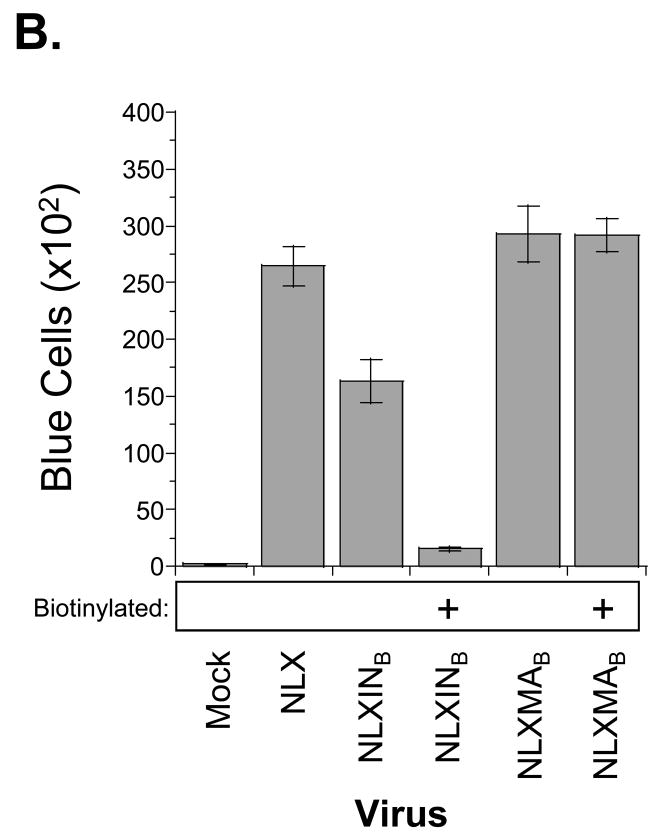
Replication profiles of BAS-containing viruses. (A) Replication kinetics in SupT1 cells; note the replicating virus is not biotinylated due to the lack of BirA expression in these cells. Results are representative of at least two independent experiments. (B) MAGI-5 cell titer of biotinylated viruses. Virus infectivity was determined after 2 d post-infection by fixing and staining the cells for β-galactosidase expression. Error bars denote the standard deviation from 9 independent infections.
To determine the fitness of the biotinylated IN and MA viruses, their infectivities were measured in single-round assays using MAGI-5 indicator cells as described previously (Belshan et al., 2006; Pirounaki et al., 2000). Normalized virus amounts, produced with or without co-transfection with pc6BirA, were inoculated overnight onto MAGI-5 cells. The cells were washed and incubated an additional 24 h. Following fixation and staining for β-galactosidase expression, the number of positive cells was counted to measure titer (Fig. 4B). Similar to its replication in SupT1 cells, NLXMAB infected MAGI-5 cells at a level comparable to wild-type NLX, and the biotinylation of MA did not affect the efficiency of infection. Consistent with its delayed replication in SupT1 cells, NLXINB displayed an ~40% reduction in MAGI-5 cell titer as compared to wild-type HIV-1NLX. This reduction was exacerbated by the addition of the biotin moiety, suggesting that biotinylation of the C-terminus of IN inactivates the virus.
3.4. The loss of NLXINB infectivity is due to loss of IN function
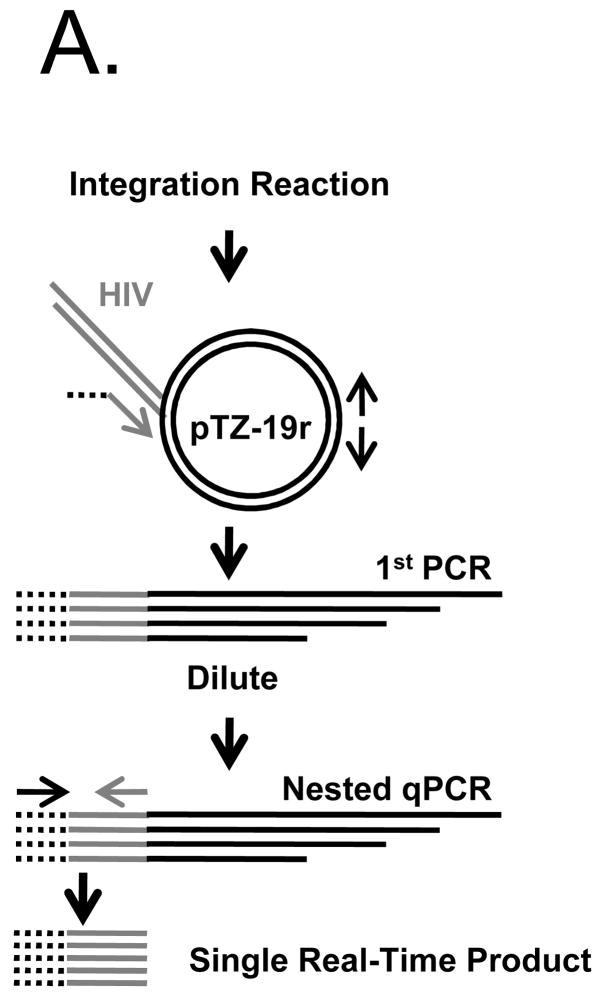
As the BAS was tacked onto the IN protein, it was critical to determine the level of IN function. For this, the in vitro activity of PICs produced by each virus was assayed using a sensitive real-time integration assay (Lu et al., 2005). The nested PCR method used to quantify the level of in vitro integration is outlined in Fig. 5A. The initial PCR step contains one HIV-specific primer, and two back-to-back target DNA primers to amplify integration events. This reaction is diluted 1000-fold, and integration events are quantified with a nested PCR using HIV-specific primers. We performed the assay as described by Lu et al., but added a linker to the 5′ end of the HIV-specific primer in the first PCR (Fig. 5A, dotted lines). The linker primer was used in place of one of the HIV primers in the qPCR to reduce background HIV amplification. For these experiments, a standard curve was generated by serial dilution of reaction products formed by wild-type HIV-1NLX PICs. Integration product formation was in turn normalized to the levels of reverse transcription that occurred under the different infection conditions (Lu et al., 2005).
Figure 5.
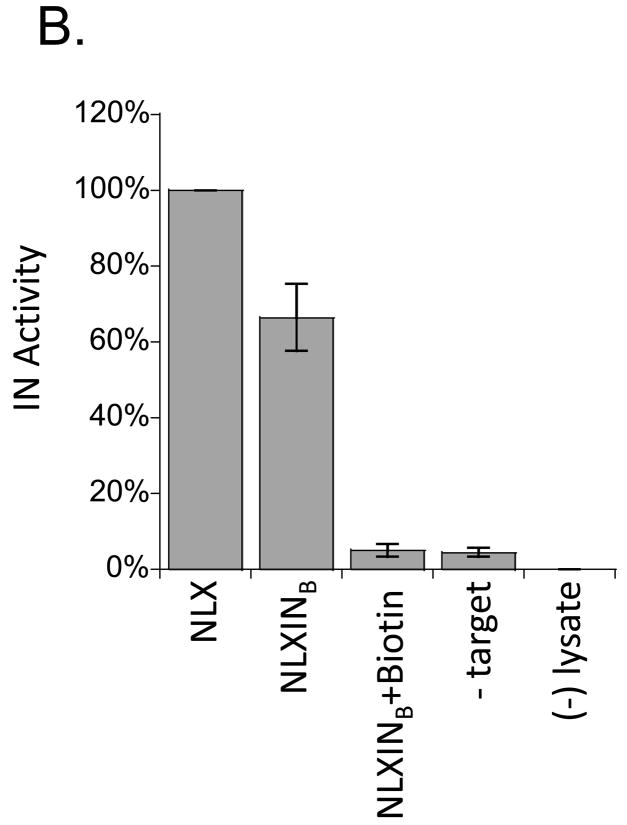
NLXINB yields defective PICs. (A) Schematic representation of nested PCR assay used to measure integration activity. The linker primer is represented by a black dashed line, the HIV sequence as a gray line, and target plasmid and primers as solid black lines. (B) PIC activity of NLXINB produced in the absence and presence of IN biotinylation. Samples were prepared in parallel, and results normalized for levels of cDNA synthesis are provided as percentage of activity compared to wild-type HIV-1NLX. No target (-Target) and uninfected lysate ((−) lysate) controls are shown. Data is representative of two independent experiments.
Addition of the BAS to the C-terminus of IN resulted in an approximate 40% reduction in IN activity, in complete agreement with the effect on MAGI-5 cell titer (compare Fig. 5B to Fig. 4B). Biotinylation further reduced IN activity to background levels. The specificity of in vitro integration was confirmed for each experiment using no target DNA and uninfected lysate controls. Combined, these results suggest that the addition of the BAS to the C-terminus of IN reduces viral titer through a direct effect on IN catalytic function. Biotinylation of this BAS furthermore abolishes PIC function and NLXINB infectivity.
4. Discussion
This report describes the successful adaptation of the BirA-BAS protein labeling system to tag HIV-1 proteins in vivo. We constructed two biotinylatable viruses, with the long-term goal of identifying physiologically-relevant protein complexes. We demonstrated site-specific biotinylation of the C-termini of both MA and IN. Biotinylation of BAS-containing proteins were not observed when viruses were produced in the absence of BirA. Quantitative image analysis indicated that 30% to 60% of total MA-BAS was biotinylated, but all detectable biotinylated protein was recovered by affinity purification. Both NLXINB and NLXMAB produced levels of Gag similar to wild-type HIV-1NLX. Moreover, we detected no defects in Gag processing in either tagged clone. Together these data indicate that both NLXINB and NLXMAB express and process Gag polyprotein at levels comparable to wild-type HIV-1NLX. Finally, BirA was stably expressed in 293T cells, which enabled labeling of viral proteins by single plasmid transfection. Efforts are currently underway to construct HIV-1-permissive BirA-expressing cell lines to facilitate non-invasive labeling of replicating HIV-1.
The MA insertion was well tolerated, and the resulting virus replicated similar to wild-type. The virus with the insertion at the C-terminus of Gag-Pol likewise functioned normally during the efferent stage of the life cycle (Fig. 2 and 3). The kinetics of viral growth through multiple rounds of replication was noticeably delayed from the wild-type, suggesting that NLXINB likely suffered a defect(s) during the afferent phase. Indeed, detailed analyses revealed a roughly 40% drop in single-round titer attributable to a loss of IN catalytic function. Biotinylation of this BAS, though tolerated during HIV-1 assembly and release, furthermore rendered the ensuing PIC functionally inactive. Due to these somewhat discouraging results, the BAS was inserted at two additional locations within the IN reading frame: after amino acid Glu-212, which lies at the boundary between the catalytic core domain and C-terminal domain (CTD), and after Gly-247 within the CTD. These insertion sites were based on results from Puglia et al. (2006), who demonstrated that recombinant IN proteins containing ~19 amino acid insertions at these positions displayed near wild-type levels of in vitro integration activities. However, neither of these clones yielded infectious virus (data not shown). Thus, the current study failed to identify a biotin-acceptor position in IN that could be used to isolate functionally-relevant complexes at the afferent arm of the HIV-1 life cycle, for example during PIC formation and nuclear import.
The BAS-BirA system offers a novel method to tag replicating HIV. The BirA-mediated biotinylation of target proteins is advantageous to most conventional protein tag systems because of its specificity, the small target sequence, covalent attachment of the biotin tag, and the affinity of the SA-biotin interaction (Kd= 10−15 M). The strength of this interaction enables the capture of complexes in low abundance in cells without excessive over-expression of bait protein. Target protein complexes can be isolated using a single-step method with low background. This system has been used to purify proteins in a wide variety of systems both in vitro and in vivo (Fernandez-Suarez et al., 2008), including ribonucleoprotein complexes (Penalva and Keene, 2004), nucleosomes (Furuyama and Henikoff, 2006), cell surface proteins (Chen et al., 2005), and transcription factors in BirA transgenic mice (de Boer et al., 2003). These studies suggest this system will be a useful method to identify the components of viral-protein complexes. Though this study failed to identify a useful site for IN biotinylation, a fraction of virion-derived MA protein remains PIC-associated, indicating that the fully functional NLXMAB virus may serve well to investigate the composition of protein complexes involved in both the efferent and afferent steps of the virus life cycle. A future goal is to use NLXMAB to investigate this question, as well as to identify other BAS-containing derivatives for the analysis of physiologically relevant HIV-1 replication intermediates.
Acknowledgments
The authors wish to thank John Strouboulis for the pEV-BirA expression plasmid, Lee Ratner for the anti-MA antibody, and Duane Grandgenett for the anti-IN antibody. M.B. is supported by the Nebraska Center for Virology (NIH P20 RR01635) and the Creighton Health Futures Foundation; A.E. is supported by NIH grant AI39394.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beckett D, Kovaleva E, Schatz P. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshan M, Mahnke LA, Ratner L. Conserved amino acids of the human immunodeficiency virus type 2 Vpx nuclear localization signal are critical for nuclear targeting of the viral preintegration complex in non-dividing cells. Virology. 2006;346:118–126. doi: 10.1016/j.virol.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Brown HE, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussel A, Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol. 2003;77:10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman T, Luban J, Goff SP, Haseltine WA, Gottlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A. Isolation and analysis of HIV-1 preintegration complexes. In: Prasad VR, Kalpana GV, editors. HIV Protocols. 2. The Humana Press; Totowa, New Jersey: 2009. pp. 135–149. [DOI] [PubMed] [Google Scholar]
- Farnet CM, Haseltine WA. Integration of Human Immunodeficiency Virus Type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Chen TS, Ting AY. Protein-protein interaction detection in vitro and in cells by proximity biotinylation. J Am Chem Soc. 2008;130:9251–9253. doi: 10.1021/ja801445p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S. Biotin-tag affinity purification of a centromeric nucleosome assembly complex. Cell Cycle. 2006;5:1269–1274. doi: 10.4161/cc.5.12.2889. [DOI] [PubMed] [Google Scholar]
- Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon A, Devroe E, Lu R, Ghory HZ, Silver PA, Engelman A. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J Virol. 2002;76:10598–10607. doi: 10.1128/JVI.76.21.10598-10607.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J Virol. 2003;77:5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Vandegraaff N, Cherepanov P, Engelman A. Lys-34, dispensable for integrase catalysis, is required for preintegration complex function and human immunodeficiency virus type 1 replication. J Virol. 2005;79:12584–12591. doi: 10.1128/JVI.79.19.12584-12591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B, Daecke J, Fackler OT, Dittmar MT, Zentgraf H, Krausslich HG. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J Virol. 2004;78:10803–10813. doi: 10.1128/JVI.78.19.10803-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva LO, Keene JD. Biotinylated tags for recovery and characterization of ribonucleoprotein complexes. Biotechniques. 2004;37:604–606. 608–610. doi: 10.2144/04374ST05. [DOI] [PubMed] [Google Scholar]
- Pirounaki M, Heyden NA, Arens M, Ratner L. Rapid phenotypic drug susceptibility assay for HIV-1 with a CCR5 expressing indicator cell line. J Virol Methods. 2000;85:151–61. doi: 10.1016/s0166-0934(99)00163-9. [DOI] [PubMed] [Google Scholar]
- Puglia J, Wang T, Smith-Snyder C, Cote M, Scher M, Pelletier JN, John S, Jonsson CB, Roth MJ. Revealing domain structure through linker-scanning analysis of the murine leukemia virus (MuLV) RNase H and MuLV and human immunodeficiency virus type 1 integrase proteins. J Virol. 2006;80:9497–9510. doi: 10.1128/JVI.00856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: A 13 residue consensus peptide specifies biotinylation in Escherichia coli. Nat Biotech. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]